Flaxseed Supplementation in Chicken Feed Accelerates Salmonella enterica subsp. enterica Serovar Enteritidis Clearance, Modulates Cecum Microbiota, and Influences Ovarian Gene Expression in Laying Hens
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Animal Experiments
2.3. DNA and RNA Extraction and RNA Reverse Transcription
2.4. Detection of S. Enteritidis Abundance via Plate Counting
2.5. Histopathological Examination of the Ileum
2.6. 16S rRNA Amplicon Sequencing and Bioinformatics Analysis
2.7. RNA Sequence Library Construction and Sequencing
2.8. Quantitative RT-PCR Validation
2.9. Statistical Analysis
3. Results
3.1. Flaxseed Reduces S. Enteritidis Colonisation in Chickens
3.2. Flaxseed Attenuates Barrier Damage to the Ileum Caused by S. Enteritidis
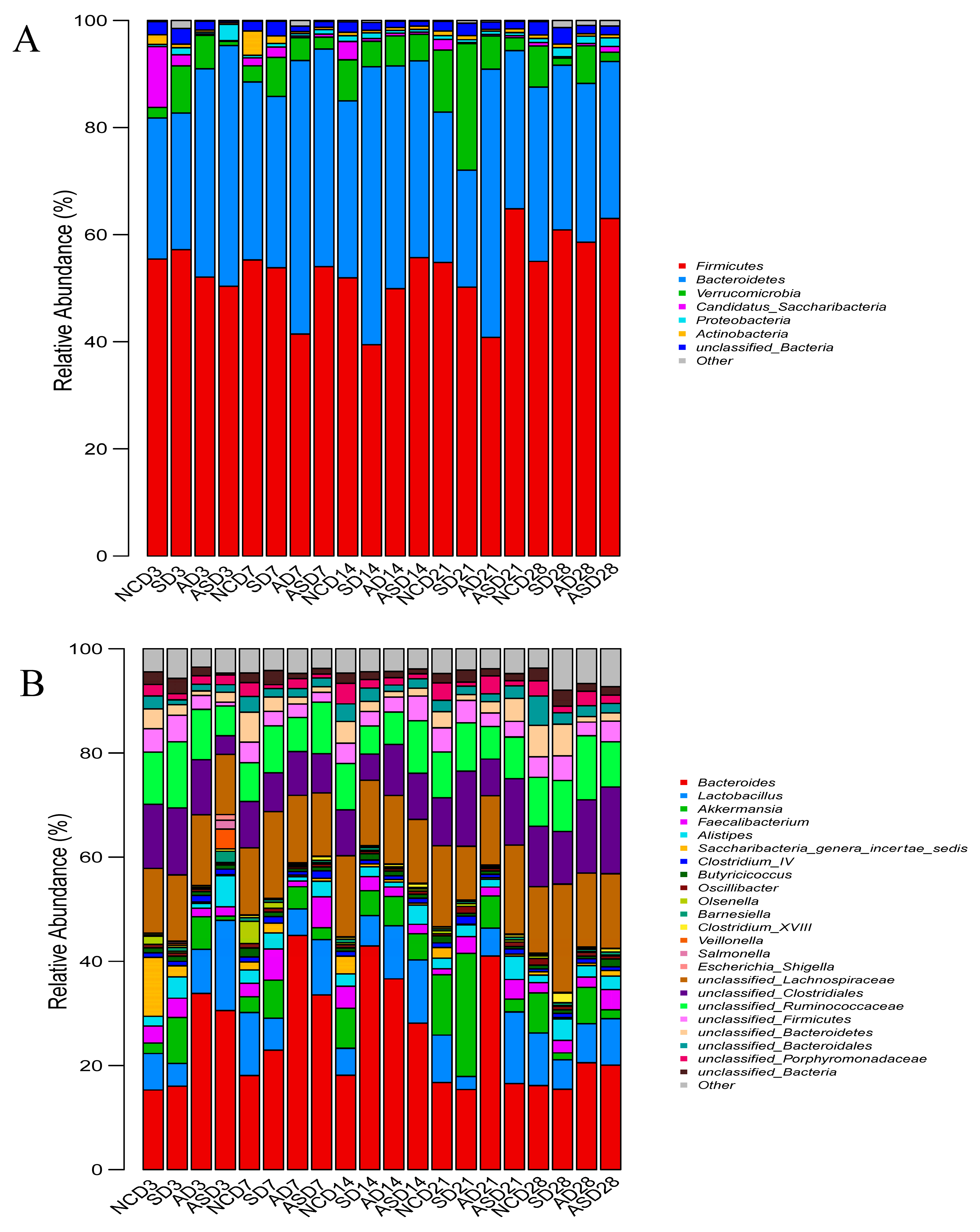
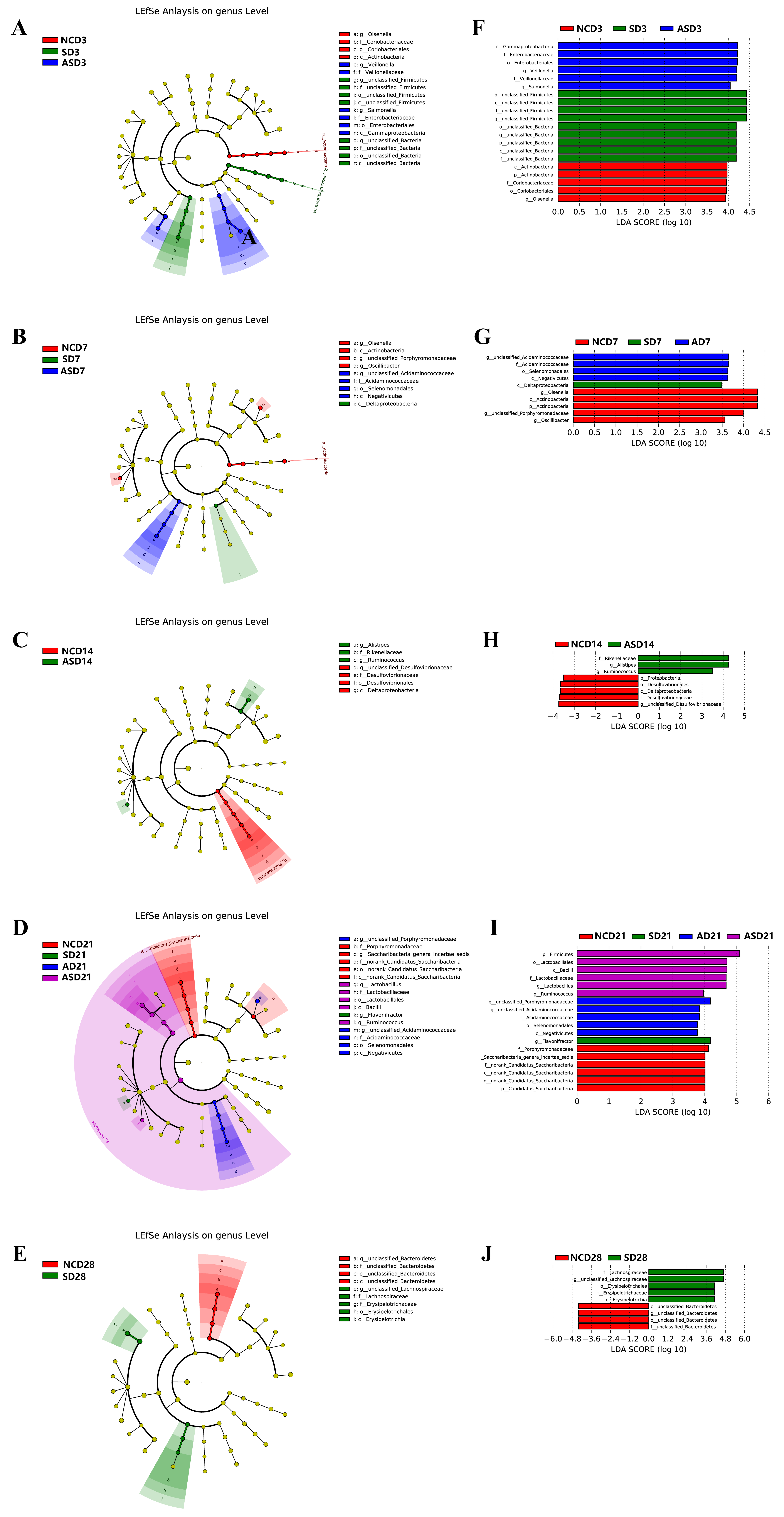
3.3. Effect of Flaxseed on the Microbial Composition of the Cecum Contents of Laying Hens
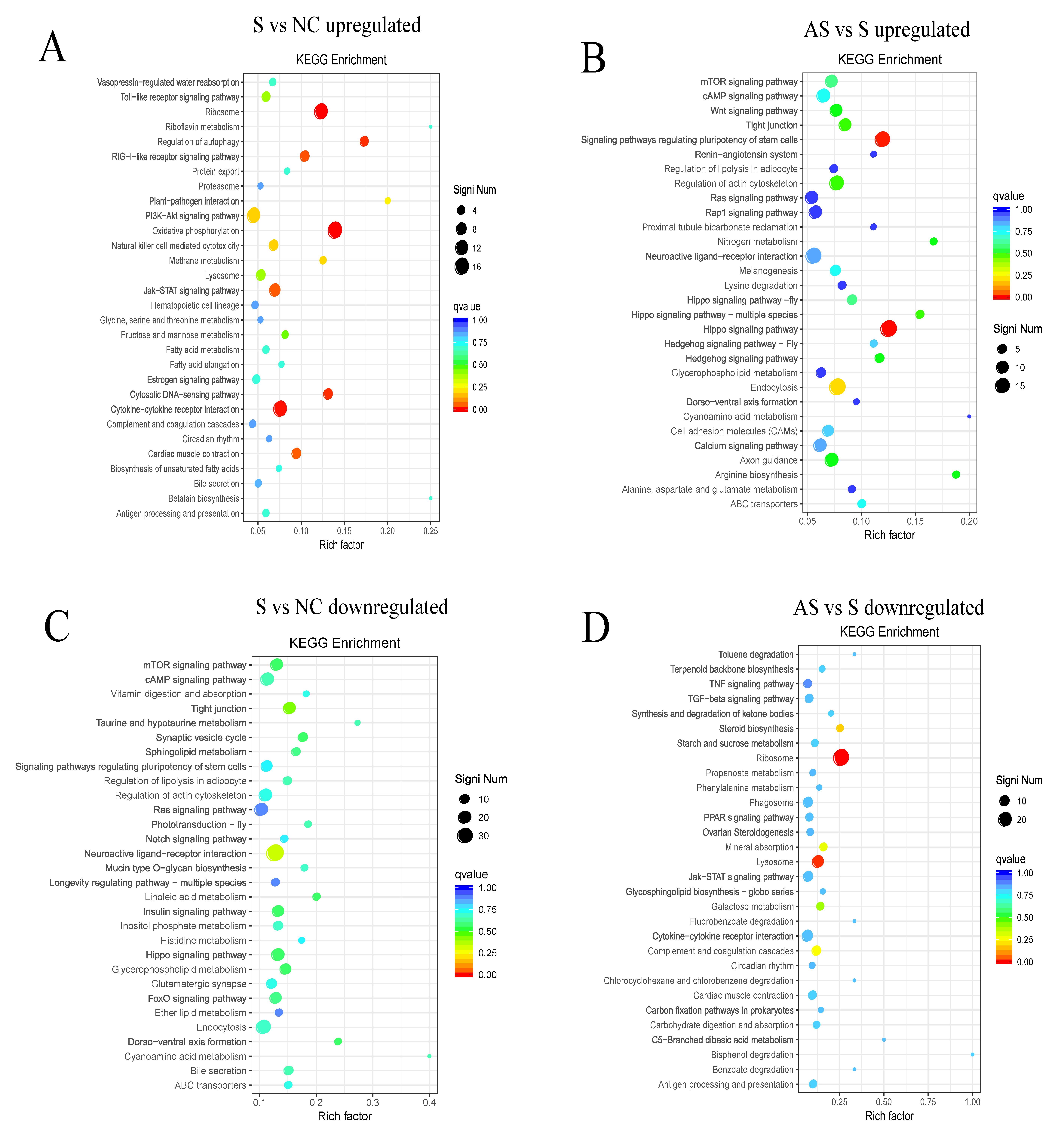
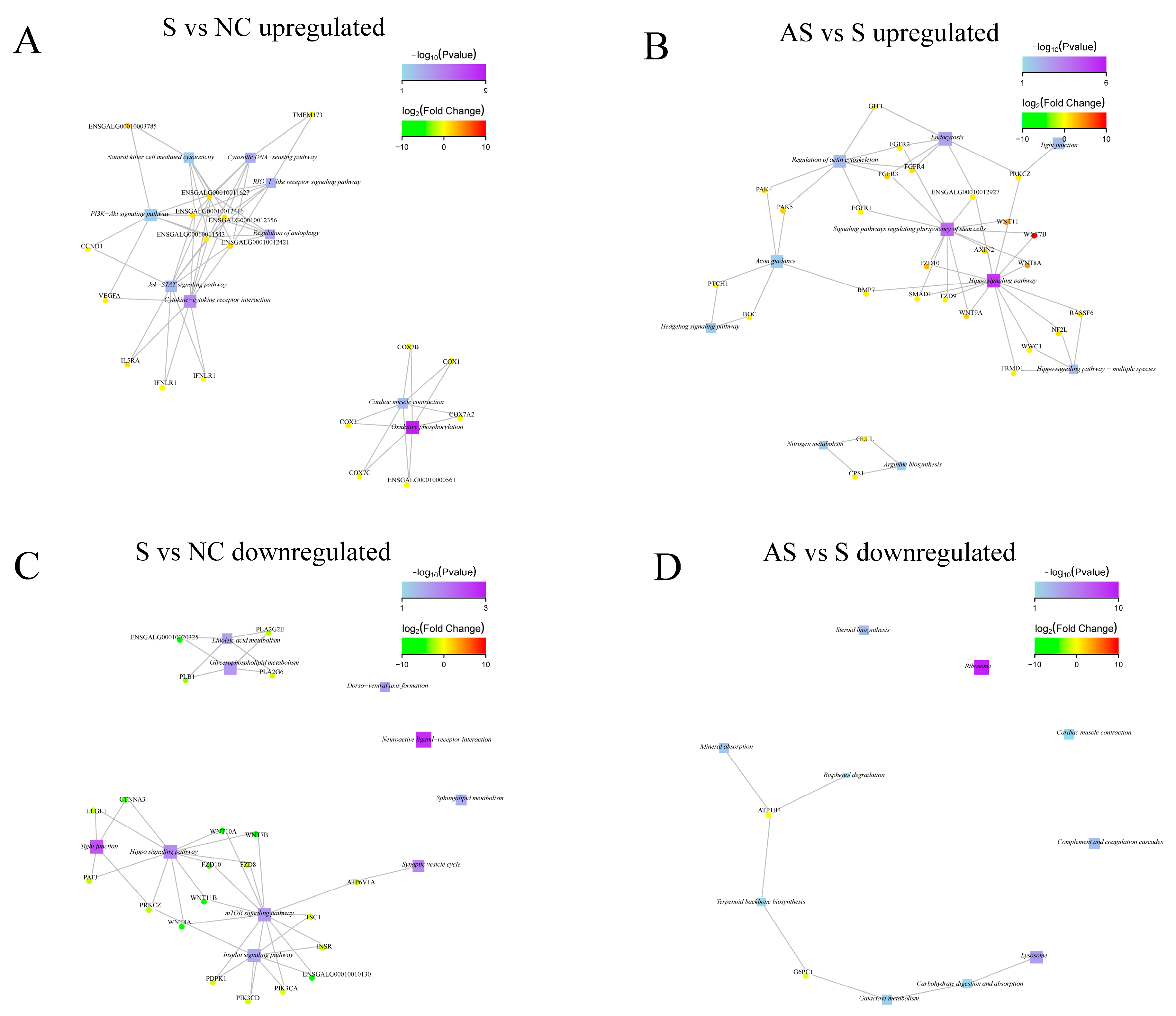
3.4. S. Enteritidis Alters Gene Expression in Ovaries
3.5. Validation of RNA-seq DEGs by Reverse Transcription Quantitative PCR
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mei, X.; Ma, B.; Zhai, X.; Zhang, A.; Lei, C.; Zuo, L.; Yang, X.; Zhou, C.; Wang, H. Florfenicol Enhances Colonization of a Salmonella enterica Serovar Enteritidis floR Mutant with Major Alterations to the Intestinal Microbiota and Metabolome in Neonatal Chickens. Appl. Environ. Microbiol. 2021, 87, e0168121. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, P.; Jiang, X.; Du, H.; Yang, C.; Zhang, Z.; Men, S.; Zhang, Z.; Jiang, W.; Wang, H. Differences in expression of genes in the MyD88 and TRIF signalling pathways and methylation of TLR4 and TRIF in Tibetan chickens and DaHeng S03 chickens infected with Salmonella enterica serovar enteritidis. Vet. Immunol. Immunopathol. 2017, 189, 28–35. [Google Scholar] [CrossRef]
- Romeu, M.J.; Rodrigues, D.; Azeredo, J. Effect of sub-lethal chemical disinfection on the biofilm forming ability, resistance to antibiotics and expression of virulence genes of Salmonella Enteritidis biofilm-surviving cells. Biofouling 2020, 36, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, D.; Abdelfattah-Hassan, A.; Badawi, M.; Ismail, T.A.; Bendary, M.M.; Abdelaziz, A.M.; Mosbah, R.A.; Mohamed, D.I.; Arisha, A.H.; El-Hamid, M.I.A. Thymol nanoemulsion promoted broiler chicken’s growth, gastrointestinal barrier and bacterial community and conferred protection against Salmonella Typhimurium. Sci. Rep. 2021, 11, 7742. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Li, G.; Shahid, M.S.; Gan, L.; Fan, H.; Lv, Z.; Yan, S.; Guo, Y. Dietary l-arginine supplementation ameliorates inflammatory response and alters gut microbiota composition in broiler chickens infected with Salmonella enterica serovar Typhimurium. Poult. Sci. 2020, 99, 1862–1874. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, Y.; Lyu, N.; Li, Z.; Ma, S.; Cao, D.; Pan, Y.; Hu, Y.; Huang, H.; Gao, G.F.; et al. The temporal dynamics of antimicrobial-resistant Salmonella enterica and predominant serovars in China. Natl. Sci. Rev. 2023, 10, nwac269. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, H.; Li, T.; Liu, F.; Cheng, Y.; Guo, X.; Wen, G.; Luo, Q.; Shao, H.; Pan, Z.; et al. Characterization of Salmonella spp. isolated from chickens in Central China. BMC Vet. Res. 2020, 16, 299. [Google Scholar] [CrossRef]
- Wang, J.; Wan, C.; Shuju, Z.; Yang, Z.; Celi, P.; Ding, X.; Bai, S.; Zeng, Q.; Mao, X.; Xu, S.; et al. Differential analysis of gut microbiota and the effect of dietary Enterococcus faecium supplementation in broiler breeders with high or low laying performance. Poult. Sci. 2021, 100, 1109–1119. [Google Scholar] [CrossRef]
- Cao, S.; Guo, D.; Yin, H.; Ding, X.; Bai, S.; Zeng, Q.; Liu, J.; Zhang, K.; Mao, X.; Wang, J. Improvement in ovarian function following fecal microbiota transplantation from high-laying rate breeders. Poult. Sci. 2023, 102, 102467. [Google Scholar] [CrossRef]
- Huang, S.; Rong, X.; Liu, M.; Liang, Z.; Geng, Y.; Wang, X.; Zhang, J.; Ji, C.; Zhao, L.; Ma, Q. Intestinal Mucosal Immunity-Mediated Modulation of the Gut Microbiome by Oral Delivery of Enterococcus faecium against Salmonella Enteritidis Pathogenesis in a Laying Hen Model. Front. Immunol. 2022, 13, 853954. [Google Scholar] [CrossRef]
- Ma, B.; Mei, X.; Lei, C.; Li, C.; Gao, Y.; Kong, L.; Zhai, X.; Wang, H. Enrofloxacin Shifts Intestinal Microbiota and Metabolic Profiling and Hinders Recovery from Salmonella enterica subsp. enterica Serovar Typhimurium Infection in Neonatal Chickens. mSphere 2020, 5, e00725-20. [Google Scholar] [CrossRef]
- Zhang, Y.; Gu, T.T.; Chen, Y.; Huang, Y.; Du, J.; Lu, L.; Zhu, G.Q.; Xu, Q.; Chen, G.H. Comparative transcriptome analysis reveals PERP upregulated during Salmonella Enteritidis challenge in laying ducks. J. Cell. Physiol. 2019, 234, 11330–11347. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, T.; Baba, E.; Tanaka, T.; Sasai, K.; Fukata, T.; Arakawa, A. Salmonella enteritidis contamination of eggs from hens inoculated by vaginal, cloacal, and intravenous routes. Avian Dis. 1997, 41, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Gast, R.K.; Guraya, R.; Guard-Bouldin, J.; Holt, P.S.; Moore, R.W. Colonization of specific regions of the reproductive tract and deposition at different locations inside eggs laid by hens infected with Salmonella enteritidis or Salmonella heidelberg. Avian Dis. 2007, 51, 40–44. [Google Scholar] [CrossRef] [PubMed]
- Sadat Masjedi, M.; Mohammadi Pour, P.; Shokoohinia, Y.; Asgary, S. Effects of Flaxseed on Blood Lipids in Healthy and Dyslipidemic Subjects: A Systematic Review and Meta-Analysis of Randomized Controlled Trials. Curr. Probl. Cardiol. 2022, 47, 100931. [Google Scholar] [CrossRef]
- Parikh, M.; Netticadan, T.; Pierce, G.N. Flaxseed: Its bioactive components and their cardiovascular benefits. Am. J. Physiol. Heart Circ. Physiol. 2018, 314, H146–H159. [Google Scholar] [CrossRef]
- Tadesse, D.; Retta, N.; Girma, M.; Ndiwa, N.; Dessie, T.; Hanotte, O.; Getachew, P.; Dannenberger, D.; Maak, S. In Vitro Antioxidant Activities of Plant Polyphenol Extracts and Their Combined Effect with Flaxseed on Raw and Cooked Breast Muscle Fatty Acid Content, Lipid Health Indices and Oxidative Stability in Slow-Growing Sasso Chickens. Foods 2022, 12, 115. [Google Scholar] [CrossRef]
- Yuan, Q.; Xie, F.; Huang, W.; Hu, M.; Yan, Q.; Chen, Z.; Zheng, Y.; Liu, L. The review of alpha-linolenic acid: Sources, metabolism, and pharmacology. Phytother. Res. 2022, 36, 164–188. [Google Scholar] [CrossRef]
- Sijben, J.W.; Klasing, K.C.; Schrama, J.W.; Parmentier, H.K.; van der Poel, J.J.; Savelkoul, H.F.; Kaiser, P. Early in vivo cytokine genes expression in chickens after challenge with Salmonella typhimurium lipopolysaccharide and modulation by dietary n—3 polyunsaturated fatty acids. Dev. Comp. Immunol. 2003, 27, 611–619. [Google Scholar] [CrossRef]
- Cacciatore, F.A.; Maders, C.; Alexandre, B.; Barreto Pinilla, C.M.; Brandelli, A.; da Silva Malheiros, P. Carvacrol encapsulation into nanoparticles produced from chia and flaxseed mucilage: Characterization, stability and antimicrobial activity against Salmonella and Listeria monocytogenes. Food Microbiol. 2022, 108, 104116. [Google Scholar] [CrossRef]
- Weston, W.C.; Hales, K.H.; Hales, D.B. Flaxseed Increases Animal Lifespan and Reduces Ovarian Cancer Severity by Toxically Augmenting One-Carbon Metabolism. Molecules 2021, 26, 5674. [Google Scholar] [CrossRef] [PubMed]
- Dikshit, A.; Gao, C.; Small, C.; Hales, K.; Hales, D.B. Flaxseed and its components differentially affect estrogen targets in pre-neoplastic hen ovaries. J. Steroid Biochem. Mol. Biol. 2016, 159, 73–85. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dutra, P.A.; Pinto, L.F.B.; Cardoso-Neto, B.M.; Mendes, C.S.; Pinheiro, A.M.; Barbosa, L.P.; de Jesus Pereira, T.C.; de Carvalho, G.G.P. Flaxseed added to the diet of Alpine goats affects the nutrients intake and blood parameters. Trop. Anim. Health Prod. 2022, 54, 104. [Google Scholar] [CrossRef] [PubMed]
- de Souza, M.; Baptista, A.A.S.; Valdiviezo, M.J.J.; Justino, L.; Menck-Costa, M.F.; Ferraz, C.R.; da Gloria, E.M.; Verri, W.A., Jr.; Bracarense, A. Lactobacillus spp. reduces morphological changes and oxidative stress induced by deoxynivalenol on the intestine and liver of broilers. Toxicon 2020, 185, 203–212. [Google Scholar] [CrossRef]
- Kechin, A.; Boyarskikh, U.; Kel, A.; Filipenko, M. cutPrimers: A New Tool for Accurate Cutting of Primers from Reads of Targeted Next Generation Sequencing. J. Comput. Biol. 2017, 24, 1138–1143. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Kobert, K.; Flouri, T.; Stamatakis, A. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 2014, 30, 614–620. [Google Scholar] [CrossRef]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, S.; Li, W. RSeQC: Quality control of RNA-seq experiments. Bioinformatics 2012, 28, 2184–2185. [Google Scholar] [CrossRef] [PubMed]
- Okonechnikov, K.; Conesa, A.; García-Alcalde, F. Qualimap 2: Advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics 2016, 32, 292–294. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang le, L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Chytilová, M.; Nemcová, R.; Gancarčíková, S.; Mudroňová, D.; Tkáčiková, L. Flax-seed oil and Lactobacillus plantarum supplementation modulate TLR and NF-κB gene expression in enterotoxigenic Escherichia coli challenged gnotobiotic pigs. Acta Vet. Hung. 2014, 62, 463–472. [Google Scholar] [CrossRef][Green Version]
- Andrejčáková, Z.; Sopková, D.; Vlčková, R.; Hertelyová, Z.; Gancarčíková, S.; Nemcová, R. The Application of Lactobacillus reuteri CCM 8617 and Flaxseed Positively Improved the Health of Mice Challenged with Enterotoxigenic E. coli O149:F4. Probiotics Antimicrob. Proteins 2020, 12, 937–951. [Google Scholar] [CrossRef]
- Zeng, J.; Lei, C.; Wang, Y.; Chen, Y.; Zhang, X.; Kang, Z.; Zhai, X.; Ye, X.; Wang, H. Distribution of Salmonella Enteritidis in internal organs and variation of cecum microbiota in chicken after oral challenge. Microb. Pathog. 2018, 122, 174–179. [Google Scholar] [CrossRef]
- Husson, M.O.; Ley, D.; Portal, C.; Gottrand, M.; Hueso, T.; Desseyn, J.L.; Gottrand, F. Modulation of host defence against bacterial and viral infections by omega-3 polyunsaturated fatty acids. J. Infect. 2016, 73, 523–535. [Google Scholar] [CrossRef] [PubMed]
- Zając, M.; Kiczorowska, B.; Samolińska, W.; Kowalczyk-Pecka, D.; Andrejko, D.; Kiczorowski, P. Effect of inclusion of micronized camelina, sunflower, and flax seeds in the broiler chicken diet on performance productivity, nutrient utilization, and intestinal microbial populations. Poult. Sci. 2021, 100, 101118. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Yun, C.; Pang, Y.; Qiao, J. The impact of the gut microbiota on the reproductive and metabolic endocrine system. Gut Microbes 2021, 13, 1894070. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, L.; Sun, X.; Wan, X.; Sun, G.; Jiang, R.; Li, W.; Tian, Y.; Liu, X.; Kang, X. Characteristics of the fecal microbiota of high- and low-yield hens and effects of fecal microbiota transplantation on egg production performance. Res. Vet. Sci. 2020, 129, 164–173. [Google Scholar] [CrossRef] [PubMed]
- Elokil, A.A.; Magdy, M.; Melak, S.; Ishfaq, H.; Bhuiyan, A.; Cui, L.; Jamil, M.; Zhao, S.; Li, S. Faecal microbiome sequences in relation to the egg-laying performance of hens using amplicon-based metagenomic association analysis. Animal 2020, 14, 706–715. [Google Scholar] [CrossRef]
- Kleigrewe, K.; Haack, M.; Baudin, M.; Ménabréaz, T.; Crovadore, J.; Masri, M.; Beyrer, M.; Andlauer, W.; Lefort, F.; Dawid, C.; et al. Dietary Modulation of the Human Gut Microbiota and Metabolome with Flaxseed Preparations. Int. J. Mol. Sci. 2022, 23, 473. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, Y.; Yin, D.; Shahid, M.S.; Yuan, J. Flaxseed diet caused inflammation by altering the gut microbiota of Peking ducks. Anim. Biotechnol. 2020, 31, 520–531. [Google Scholar] [CrossRef]
- Xia, H.; Shi, X.; Zhou, B.; Sui, J.; Yang, C.; Liu, H.; Yang, L.; Wang, S.; Sun, G. Milled flaxseed-added diets ameliorated hepatic inflammation by reducing gene expression of TLR4/NF-κB pathway and altered gut microbiota in STZ-induced type 1 diabetic mice. Food Sci. Hum. Wellness 2022, 11, 32–40. [Google Scholar] [CrossRef]
- Jacobson, A.; Lam, L.; Rajendram, M.; Tamburini, F.; Honeycutt, J.; Pham, T.; Van Treuren, W.; Pruss, K.; Stabler, S.R.; Lugo, K.; et al. A Gut Commensal-Produced Metabolite Mediates Colonization Resistance to Salmonella Infection. Cell Host Microbe 2018, 24, 296–307.e297. [Google Scholar] [CrossRef]
- Ganesh, B.P.; Richter, J.F.; Blaut, M.; Loh, G. Enterococcus faecium NCIMB 10415 does not protect interleukin-10 knock-out mice from chronic gut inflammation. Benef. Microbes 2012, 3, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Ducarmon, Q.R.; Zwittink, R.D.; Hornung, B.V.H.; van Schaik, W.; Young, V.B.; Kuijper, E.J. Gut Microbiota and Colonization Resistance against Bacterial Enteric Infection. Microbiol. Mol. Biol. Rev. 2019, 83, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Delday, M.; Mulder, I.; Logan, E.T.; Grant, G. Bacteroides thetaiotaomicron Ameliorates Colon Inflammation in Preclinical Models of Crohn’s Disease. Inflamm. Bowel Dis. 2019, 25, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Wu, F.; Zhou, Q.; Wei, W.; Yue, J.; Xiao, B.; Luo, Z. Lactobacillus and intestinal diseases: Mechanisms of action and clinical applications. Microbiol. Res. 2022, 260, 127019. [Google Scholar] [CrossRef] [PubMed]
- Kiarie, E.; Nyachoti, C.M.; Slominski, B.A.; Blank, G. Growth performance, gastrointestinal microbial activity, and nutrient digestibility in early-weaned pigs fed diets containing flaxseed and carbohydrase enzyme. J. Anim. Sci. 2007, 85, 2982–2993. [Google Scholar] [CrossRef]
- Luijten, M.; Singh, A.V.; Bastian, C.A.; Westerman, A.; Pisano, M.M.; Pennings, J.L.; Verhoef, A.; Green, M.L.; Piersma, A.H.; de Vries, A.; et al. Lasting effects on body weight and mammary gland gene expression in female mice upon early life exposure to n-3 but not n-6 high-fat diets. PLoS ONE 2013, 8, e55603. [Google Scholar] [CrossRef] [PubMed]
- Risha, M.A.; Siengdee, P.; Dannenberger, D.; Wimmers, K.; Ponsuksili, S. PUFA Treatment Affects C2C12 Myocyte Differentiation, Myogenesis Related Genes and Energy Metabolism. Genes 2021, 12, 192. [Google Scholar] [CrossRef]
- Zhu, H.; Wang, H.; Wang, S.; Tu, Z.; Zhang, L.; Wang, X.; Hou, Y.; Wang, C.; Chen, J.; Liu, Y. Flaxseed Oil Attenuates Intestinal Damage and Inflammation by Regulating Necroptosis and TLR4/NOD Signaling Pathways Following Lipopolysaccharide Challenge in a Piglet Model. Mol. Nutr. Food Res. 2018, 62, e1700814. [Google Scholar] [CrossRef]
- Arora, T.; Rudenko, O.; Egerod, K.L.; Husted, A.S.; Kovatcheva-Datchary, P.; Akrami, R.; Kristensen, M.; Schwartz, T.W.; Bäckhed, F. Microbial fermentation of flaxseed fibers modulates the transcriptome of GPR41-expressing enteroendocrine cells and protects mice against diet-induced obesity. Am. J. Physiol. Endocrinol. Metab. 2019, 316, E453–E463. [Google Scholar] [CrossRef]
- Giansanti, F.; Giardi, M.F.; Botti, D. Avian cytokines—An overview. Curr. Pharm. Des. 2006, 12, 3083–3099. [Google Scholar] [CrossRef]
- Rawlings, J.S.; Rosler, K.M.; Harrison, D.A. The JAK/STAT signaling pathway. J. Cell Sci. 2004, 117, 1281–1283. [Google Scholar] [CrossRef]
- Kwon, J.; Bakhoum, S.F. The Cytosolic DNA-Sensing cGAS-STING Pathway in Cancer. Cancer Discov. 2020, 10, 26–39. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.E.; Li, Y.; Nishimura, A.; Jheng, H.F.; Yuliana, A.; Kitano-Ohue, R.; Nomura, W.; Takahashi, N.; Kim, C.S.; Yu, R.; et al. Synthesized enone fatty acids resembling metabolites from gut microbiota suppress macrophage-mediated inflammation in adipocytes. Mol. Nutr. Food Res. 2017, 61, 1700064. [Google Scholar] [CrossRef]
- Guido, M.E.; Monjes, N.M.; Wagner, P.M.; Salvador, G.A. Circadian Regulation and Clock-Controlled Mechanisms of Glycerophospholipid Metabolism from Neuronal Cells and Tissues to Fibroblasts. Mol. Neurobiol. 2022, 59, 326–353. [Google Scholar] [CrossRef] [PubMed]
- Mohseni, A.H.; Casolaro, V.; Bermúdez-Humarán, L.G.; Keyvani, H.; Taghinezhad, S.S. Modulation of the PI3K/Akt/mTOR signaling pathway by probiotics as a fruitful target for orchestrating the immune response. Gut Microbes 2021, 13, 1886844. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Meng, Z.; Chen, R.; Guan, K.L. The Hippo Pathway: Biology and Pathophysiology. Annu. Rev. Biochem. 2019, 88, 577–604. [Google Scholar] [CrossRef]
- Khan, S.; Chousalkar, K.K. Transcriptome profiling analysis of caeca in chicks challenged with Salmonella Typhimurium reveals differential expression of genes involved in host mucosal immune response. Appl. Microbiol. Biotechnol. 2020, 104, 9327–9342. [Google Scholar] [CrossRef]
- Clark, K.L.; George, J.W.; Przygrodzka, E.; Plewes, M.R.; Hua, G.; Wang, C.; Davis, J.S. Hippo Signaling in the Ovary: Emerging Roles in Development, Fertility, and Disease. Endocr. Rev. 2022, 43, 1074–1096. [Google Scholar] [CrossRef]





| Genes | Primer Sequence (5′–3′) | Tm (°C) | Product Length (bp) | |
|---|---|---|---|---|
| GAPDH | F | GCCCAGAACATCATCCCA | 56.4 | 137 |
| R | CGGCAGGTCAGGTCAACA | 57.7 | ||
| CALR | F | GGAAGTTCTACGGCGATGCT | 59.5 | 150 |
| R | CGATGTTCTGCTCGTGTTTGA | 59.5 | ||
| CD24 | F | GATCCCAATGGAACAAGTC | 51.7 | 128 |
| R | CTCGTGGTGGAGTGAAGG | 53.3 | ||
| RPS27A | F | GGAAGACCATCACCCTCG | 55.1 | 195 |
| R | CACGCAGTCTCAGCACAA | 53.3 | ||
| TAGLN | F | CCAGACCGTTGACCTCTTTGA | 60.0 | 155 |
| R | AGAACTCCCGCTTGTGCTCC | 60.0 | ||
| RPL13 | F | GCCCGACTGTCAGATACCA | 56.0 | 102 |
| R | CAATCGTCCGAGCAAACC | 56.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, D.; Ma, B.; Liao, Z.; Li, W.; Zhang, T.; Lei, C.; Wang, H. Flaxseed Supplementation in Chicken Feed Accelerates Salmonella enterica subsp. enterica Serovar Enteritidis Clearance, Modulates Cecum Microbiota, and Influences Ovarian Gene Expression in Laying Hens. Biomolecules 2023, 13, 1353. https://doi.org/10.3390/biom13091353
Wang D, Ma B, Liao Z, Li W, Zhang T, Lei C, Wang H. Flaxseed Supplementation in Chicken Feed Accelerates Salmonella enterica subsp. enterica Serovar Enteritidis Clearance, Modulates Cecum Microbiota, and Influences Ovarian Gene Expression in Laying Hens. Biomolecules. 2023; 13(9):1353. https://doi.org/10.3390/biom13091353
Chicago/Turabian StyleWang, De, Boheng Ma, Ziwei Liao, Wenjing Li, Tiejun Zhang, Changwei Lei, and Hongning Wang. 2023. "Flaxseed Supplementation in Chicken Feed Accelerates Salmonella enterica subsp. enterica Serovar Enteritidis Clearance, Modulates Cecum Microbiota, and Influences Ovarian Gene Expression in Laying Hens" Biomolecules 13, no. 9: 1353. https://doi.org/10.3390/biom13091353
APA StyleWang, D., Ma, B., Liao, Z., Li, W., Zhang, T., Lei, C., & Wang, H. (2023). Flaxseed Supplementation in Chicken Feed Accelerates Salmonella enterica subsp. enterica Serovar Enteritidis Clearance, Modulates Cecum Microbiota, and Influences Ovarian Gene Expression in Laying Hens. Biomolecules, 13(9), 1353. https://doi.org/10.3390/biom13091353








