Role of Lipids and Divalent Cations in Membrane Fusion Mediated by the Heptad Repeat Domain 1 of Mitofusin
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Peptides
2.3. Liposomes
2.4. Multi-Angle Dynamic Light Scattering (MADLS)
2.5. Cryogenic Transmission Electron Microscopy (cryo-TEM)
2.6. Liposome Co-Flotation Assay
2.7. FRET-Based Lipid Mixing Assay
2.8. Sodium Dithionite Assay
3. Results
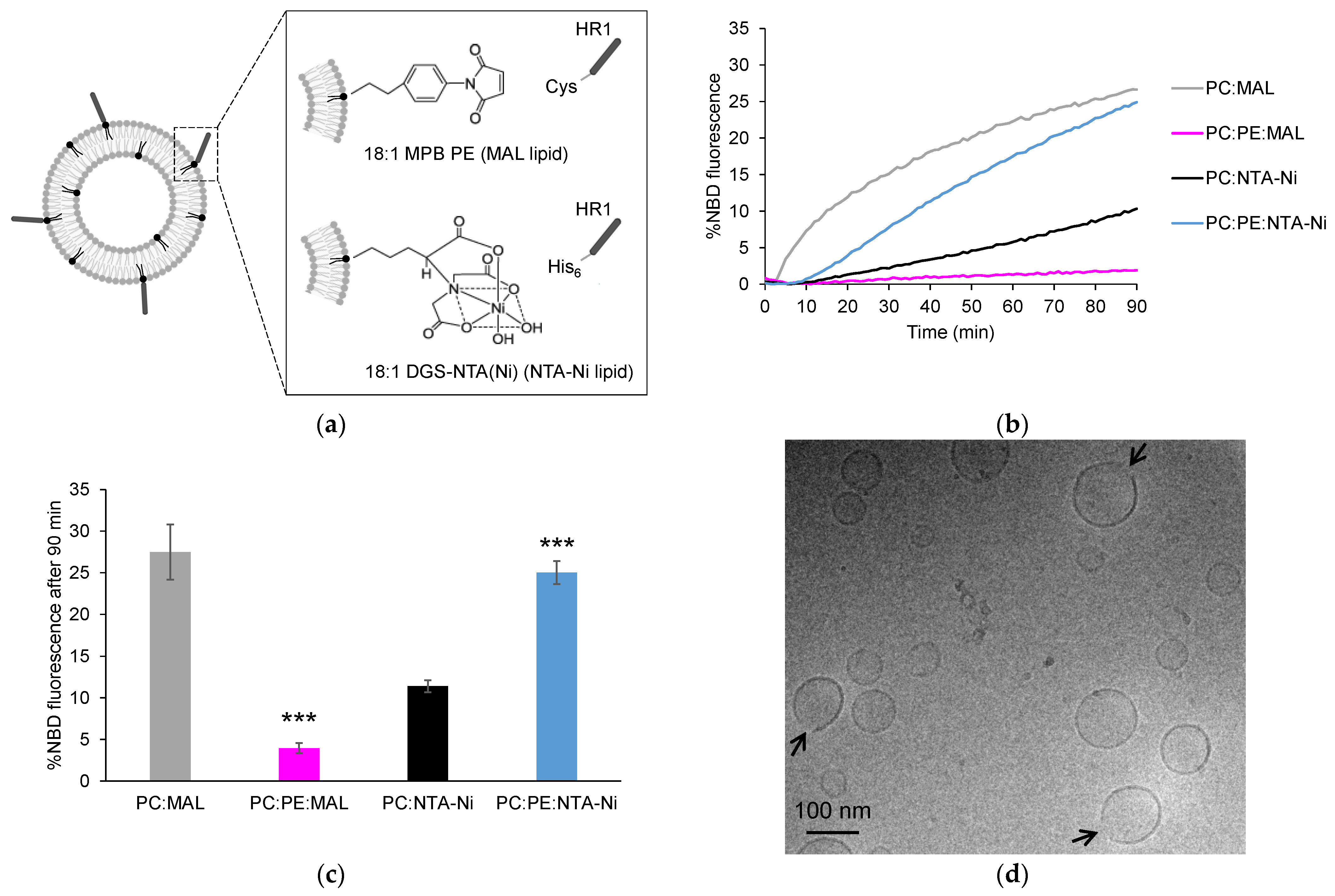
3.1. Exploring HR1-Mediated Fusion with Distinct Lipid Anchors: The Influence of Phosphatidylethanolamine
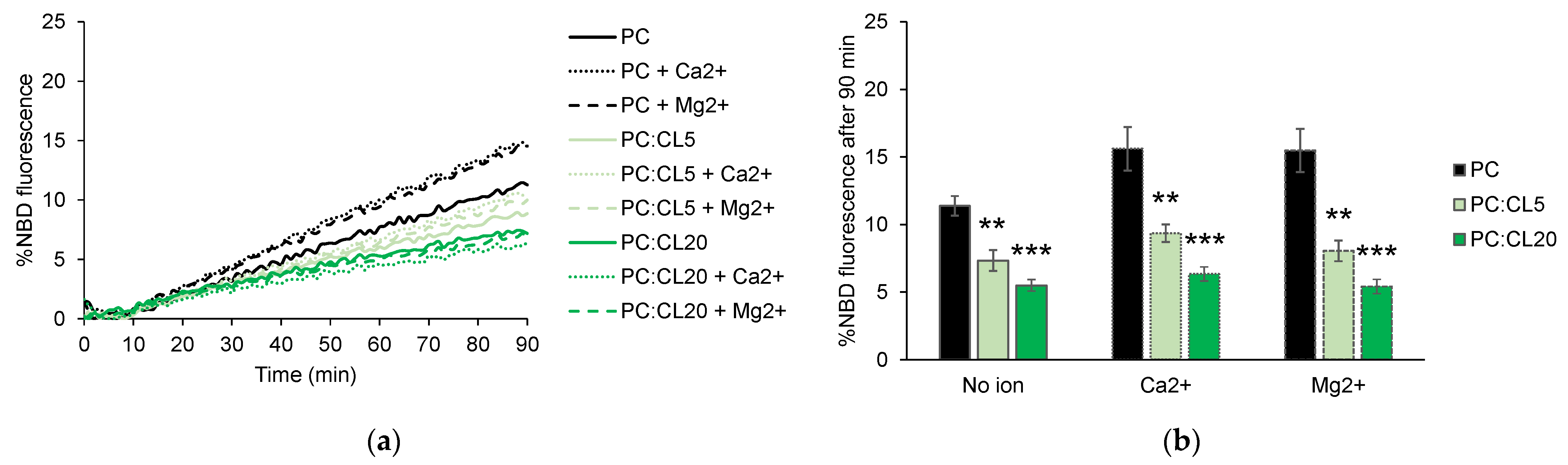
3.2. Cardiolipin Inhibits HR1-Mediated Fusion
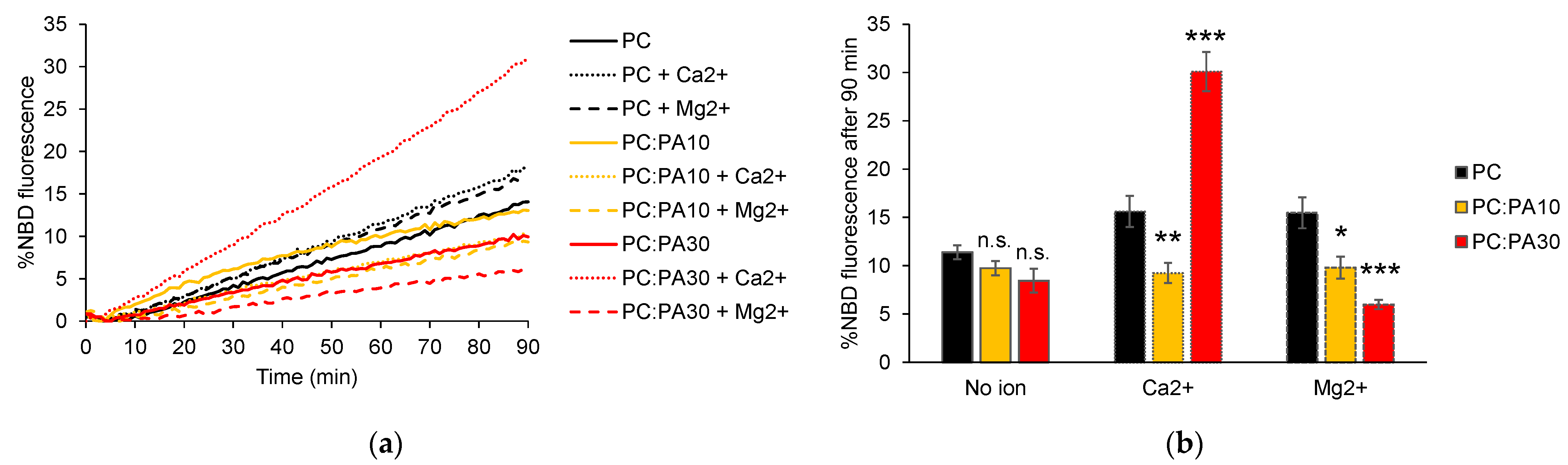
3.3. Phosphatidic Acid Enhances HR1-Mediated Fusion in the Presence of Calcium
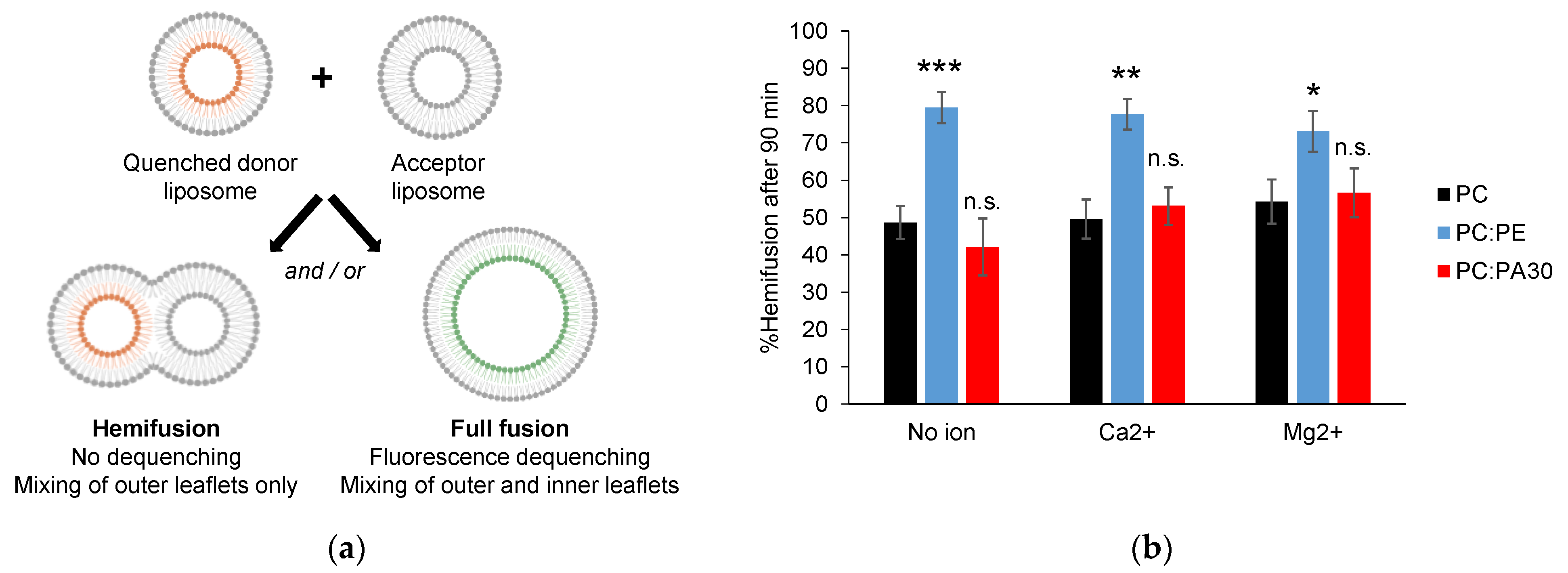
3.4. Phosphatidylethanolamine, but Not Phosphatidic Acid, Facilitates Hemifusion by HR1
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liesa, M.; Palacín, M.; Zorzano, A. Mitochondrial dynamics in mammalian health and disease. Physiol. Rev. 2009, 89, 799–845. [Google Scholar] [CrossRef]
- Chan, D.C. Mitochondrial fusion and fission in mammals. Annu. Rev. Cell Dev. Biol. 2006, 22, 79–99. [Google Scholar] [CrossRef] [PubMed]
- Rojo, M.; Legros, F.; Chateau, D.; Lombès, A. Membrane topology and mitochondrial targeting of mitofusins, ubiquitous mammalian homologs of the transmembrane GTPase Fzo. J. Cell Sci. 2002, 115, 1663–1674. [Google Scholar] [CrossRef]
- Eura, Y.; Ishihara, N.; Yokota, S.; Mihara, K. Two mitofusin proteins, mammalian homologues of FZO, with distinct functions are both required for mitochondrial fusion. J. Biochem. 2003, 134, 333–344. [Google Scholar] [CrossRef] [PubMed]
- Koshiba, T.; Detmer, S.A.; Kaiser, J.T.; Chen, H.; McCaffery, J.M.; Chan, D.C. Structural basis of mitochondrial tethering by mitofusin complexes. Science 2004, 305, 858–862. [Google Scholar] [CrossRef] [PubMed]
- Honda, S.; Aihara, T.; Hontani, M.; Okubo, K.; Hirose, S. Mutational analysis of action of mitochondrial fusion factor mitofusin-2. J. Cell Sci. 2005, 118, 3153–3161. [Google Scholar] [CrossRef]
- Qi, Y.; Yan, L.; Yu, C.; Guo, X.; Zhou, X.; Hu, X.; Huang, X.; Rao, Z.; Lou, Z.; Hu, J. Structures of human mitofusin 1 provide insight into mitochondrial tethering. J. Cell Biol. 2016, 215, 621–629. [Google Scholar] [CrossRef]
- Cao, Y.-L.; Meng, S.; Chen, Y.; Feng, J.-X.; Gu, D.-D.; Yu, B.; Li, Y.-J.; Yang, J.-Y.; Liao, S.; Chan, D.C.; et al. MFN1 structures reveal nucleotide-triggered dimerization critical for mitochondrial fusion. Nature 2017, 542, 372–376. [Google Scholar] [CrossRef]
- Yan, L.; Qi, Y.; Huang, X.; Yu, C.; Lan, L.; Guo, X.; Rao, Z.; Hu, J.; Lou, Z. Structural basis for GTP hydrolysis and conformational change of MFN1 in mediating membrane fusion. Nat. Struct. Mol. Biol. 2018, 25, 233–243. [Google Scholar] [CrossRef]
- Brandt, T.; Cavellini, L.; Kühlbrandt, W.; Cohen, M.M. A mitofusin-dependent docking ring complex triggers mitochondrial fusion in vitro. eLife 2016, 5, e14618. [Google Scholar] [CrossRef]
- Cohen, M.M.; Tareste, D. Recent insights into the structure and function of Mitofusins in mitochondrial fusion. [version 1; peer review: 2 approved]. F1000Res 2018, 7, 1983. [Google Scholar] [CrossRef]
- Daste, F.; Sauvanet, C.; Bavdek, A.; Baye, J.; Pierre, F.; Le Borgne, R.; David, C.; Rojo, M.; Fuchs, P.; Tareste, D. The heptad repeat domain 1 of Mitofusin has membrane destabilization function in mitochondrial fusion. EMBO Rep. 2018, 19, e43637. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Zhou, X.; Hu, X.; Joshi, A.S.; Guo, X.; Zhu, Y.; Chen, Q.; Prinz, W.A.; Hu, J. Sequences flanking the transmembrane segments facilitate mitochondrial localization and membrane fusion by mitofusin. Proc. Natl. Acad. Sci. USA 2017, 114, E9863–E9872. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.Y.; Bian, X.; Sun, S.; Hu, X.; Klemm, R.W.; Prinz, W.A.; Rapoport, T.A.; Hu, J. Lipid interaction of the C terminus and association of the transmembrane segments facilitate atlastin-mediated homotypic endoplasmic reticulum fusion. Proc. Natl. Acad. Sci. USA 2012, 109, E2146–E2154. [Google Scholar] [CrossRef] [PubMed]
- McNew, J.A.; Sondermann, H.; Lee, T.; Stern, M.; Brandizzi, F. GTP-dependent membrane fusion. Annu. Rev. Cell Dev. Biol. 2013, 29, 529–550. [Google Scholar] [CrossRef]
- Faust, J.E.; Desai, T.; Verma, A.; Ulengin, I.; Sun, T.-L.; Moss, T.J.; Betancourt-Solis, M.A.; Huang, H.W.; Lee, T.; McNew, J.A. The Atlastin C-terminal tail is an amphipathic helix that perturbs the bilayer structure during endoplasmic reticulum homotypic fusion. J. Biol. Chem. 2015, 290, 4772–4783. [Google Scholar] [CrossRef]
- Martens, S.; McMahon, H.T. Mechanisms of membrane fusion: Disparate players and common principles. Nat. Rev. Mol. Cell Biol. 2008, 9, 543–556. [Google Scholar] [CrossRef]
- Ban, T.; Ishihara, T.; Kohno, H.; Saita, S.; Ichimura, A.; Maenaka, K.; Oka, T.; Mihara, K.; Ishihara, N. Molecular basis of selective mitochondrial fusion by heterotypic action between OPA1 and cardiolipin. Nat. Cell Biol. 2017, 19, 856–863. [Google Scholar] [CrossRef]
- Tasseva, G.; Bai, H.D.; Davidescu, M.; Haromy, A.; Michelakis, E.; Vance, J.E. Phosphatidylethanolamine deficiency in Mammalian mitochondria impairs oxidative phosphorylation and alters mitochondrial morphology. J. Biol. Chem. 2013, 288, 4158–4173. [Google Scholar] [CrossRef]
- Chan, E.Y.L.; McQuibban, G.A. Phosphatidylserine decarboxylase 1 (Psd1) promotes mitochondrial fusion by regulating the biophysical properties of the mitochondrial membrane and alternative topogenesis of mitochondrial genome maintenance protein 1 (Mgm1). J. Biol. Chem. 2012, 287, 40131–40139. [Google Scholar] [CrossRef]
- Joshi, A.S.; Thompson, M.N.; Fei, N.; Hüttemann, M.; Greenberg, M.L. Cardiolipin and mitochondrial phosphatidylethanolamine have overlapping functions in mitochondrial fusion in Saccharomyces cerevisiae. J. Biol. Chem. 2012, 287, 17589–17597. [Google Scholar] [CrossRef] [PubMed]
- Oemer, G.; Koch, J.; Wohlfarter, Y.; Alam, M.T.; Lackner, K.; Sailer, S.; Neumann, L.; Lindner, H.H.; Watschinger, K.; Haltmeier, M.; et al. Phospholipid Acyl Chain Diversity Controls the Tissue-Specific Assembly of Mitochondrial Cardiolipins. Cell Rep. 2020, 30, 4281–4291.e4. [Google Scholar] [CrossRef] [PubMed]
- Scott, B.L.; Van Komen, J.S.; Liu, S.; Weber, T.; Melia, T.J.; McNew, J.A. Liposome fusion assay to monitor intracellular membrane fusion machines. Meth. Enzymol. 2003, 372, 274–300. [Google Scholar] [CrossRef]
- McNew, J.A.; Weber, T.; Parlati, F.; Johnston, R.J.; Melia, T.J.; Söllner, T.H.; Rothman, J.E. Close is not enough: SNARE-dependent membrane fusion requires an active mechanism that transduces force to membrane anchors. J. Cell Biol. 2000, 150, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Almgren, M.; Edwards, K.; Karlsson, G. Cryo transmission electron microscopy of liposomes and related structures. Colloids Surf. A Physicochem. Eng. Asp. 2000, 174, 3–21. [Google Scholar] [CrossRef]
- Jotwani, A.; Richerson, D.N.; Motta, I.; Julca-Zevallos, O.; Melia, T.J. Approaches to the study of Atg8-mediated membrane dynamics in vitro. Methods Cell Biol. 2012, 108, 93–116. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Coleman, J.; Yang, R.; Melia, T.J.; Rothman, J.E.; Tareste, D. Protein determinants of SNARE-mediated lipid mixing. Biophys. J. 2010, 99, 553–560. [Google Scholar] [CrossRef]
- Nir, S.; Wilschut, J.; Bentz, J. The rate of fusion of phospholipid vesicles and the role of bilayer curvature. Biochim. Biophys. Acta 1982, 688, 275–278. [Google Scholar] [CrossRef]
- Malinin, V.S.; Frederik, P.; Lentz, B.R. Osmotic and curvature stress affect PEG-induced fusion of lipid vesicles but not mixing of their lipids. Biophys. J. 2002, 82, 2090–2100. [Google Scholar] [CrossRef]
- Lee, J.Y.; Schick, M. Calculation of free energy barriers to the fusion of small vesicles. Biophys. J. 2008, 94, 1699–1706. [Google Scholar] [CrossRef]
- Ardail, D.; Privat, J.P.; Egret-Charlier, M.; Levrat, C.; Lerme, F.; Louisot, P. Mitochondrial contact sites. Lipid composition and dynamics. J. Biol. Chem. 1990, 265, 18797–18802. [Google Scholar] [CrossRef] [PubMed]
- Fritz, S.; Rapaport, D.; Klanner, E.; Neupert, W.; Westermann, B. Connection of the mitochondrial outer and inner membranes by Fzo1 is critical for organellar fusion. J. Cell Biol. 2001, 152, 683–692. [Google Scholar] [CrossRef] [PubMed]
- Frohman, M.A. Role of mitochondrial lipids in guiding fission and fusion. J. Mol. Med. 2015, 93, 263–269. [Google Scholar] [CrossRef]
- Wilschut, J.; Holsappel, M.; Jansen, R. Ca2+-induced fusion of cardiolipin/phosphatidylcholine vesicles monitored by mixing of aqueous contents. Biochim. Biophys. Acta 1982, 690, 297–301. [Google Scholar] [CrossRef]
- Ortiz, A.; Killian, J.A.; Verkleij, A.J.; Wilschut, J. Membrane fusion and the lamellar-to-inverted-hexagonal phase transition in cardiolipin vesicle systems induced by divalent cations. Biophys. J. 1999, 77, 2003–2014. [Google Scholar] [CrossRef]
- Horvath, S.E.; Daum, G. Lipids of mitochondria. Prog. Lipid Res. 2013, 52, 590–614. [Google Scholar] [CrossRef] [PubMed]
- Kameoka, S.; Adachi, Y.; Okamoto, K.; Iijima, M.; Sesaki, H. Phosphatidic acid and cardiolipin coordinate mitochondrial dynamics. Trends Cell Biol. 2018, 28, 67–76. [Google Scholar] [CrossRef]
- Choi, S.-Y.; Huang, P.; Jenkins, G.M.; Chan, D.C.; Schiller, J.; Frohman, M.A. A common lipid links Mfn-mediated mitochondrial fusion and SNARE-regulated exocytosis. Nat. Cell Biol. 2006, 8, 1255–1262. [Google Scholar] [CrossRef]
- Adachi, Y.; Itoh, K.; Yamada, T.; Cerveny, K.L.; Suzuki, T.L.; Macdonald, P.; Frohman, M.A.; Ramachandran, R.; Iijima, M.; Sesaki, H. Coincident phosphatidic acid interaction restrains drp1 in mitochondrial division. Mol. Cell 2016, 63, 1034–1043. [Google Scholar] [CrossRef]
- Brock, T.G.; Nagaprakash, K.; Margolis, D.I.; Smolen, J.E. Modeling degranulation with liposomes: Effect of lipid composition on membrane fusion. J. Membr. Biol. 1994, 141, 139–148. [Google Scholar] [CrossRef]
- Blackwood, R.A.; Smolen, J.E.; Transue, A.; Hessler, R.J.; Harsh, D.M.; Brower, R.C.; French, S. Phospholipase D activity facilitates Ca2+-induced aggregation and fusion of complex liposomes. Am. J. Physiol. 1997, 272, C1279-85. [Google Scholar] [CrossRef] [PubMed]
- Chernomordik, L.V.; Kozlov, M.M. Mechanics of membrane fusion. Nat. Struct. Mol. Biol. 2008, 15, 675–683. [Google Scholar] [CrossRef]
- Meers, P.; Ali, S.; Erukulla, R.; Janoff, A.S. Novel inner monolayer fusion assays reveal differential monolayer mixing associated with cation-dependent membrane fusion. Biochim. Biophys. Acta 2000, 1467, 227–243. [Google Scholar] [CrossRef]
- Seddon, J.M. Structure of the inverted hexagonal (HII) phase, and non-lamellar phase transitions of lipids. Biochim. Biophys. Acta 1990, 1031, 1–69. [Google Scholar] [CrossRef] [PubMed]
- Aeffner, S.; Reusch, T.; Weinhausen, B.; Salditt, T. Energetics of stalk intermediates in membrane fusion are controlled by lipid composition. Proc. Natl. Acad. Sci. USA 2012, 109, E1609–E1618. [Google Scholar] [CrossRef] [PubMed]
- Faraudo, J.; Travesset, A. Phosphatidic acid domains in membranes: Effect of divalent counterions. Biophys. J. 2007, 92, 2806–2818. [Google Scholar] [CrossRef]
- Zhukovsky, M.A.; Filograna, A.; Luini, A.; Corda, D.; Valente, C. Phosphatidic acid in membrane rearrangements. FEBS Lett. 2019, 593, 2428–2451. [Google Scholar] [CrossRef]
- Hofbauer, H.F.; Gecht, M.; Fischer, S.C.; Seybert, A.; Frangakis, A.S.; Stelzer, E.H.K.; Covino, R.; Hummer, G.; Ernst, R. The molecular recognition of phosphatidic acid by an amphipathic helix in Opi1. J. Cell Biol. 2018, 217, 3109–3126. [Google Scholar] [CrossRef]
- Tanguy, E.; Kassas, N.; Vitale, N. Protein-phospholipid interaction motifs: A focus on phosphatidic acid. Biomolecules 2018, 8, 20. [Google Scholar] [CrossRef]
- Papahadjopoulos, D.; Vail, W.J.; Pangborn, W.A.; Poste, G. Studies on membrane fusion. II. Induction of fusion in pure phospholipid membranes by calcium ions and other divalent metals. Biochim. Biophys. Acta 1976, 448, 265–283. [Google Scholar] [CrossRef]
- Ohki, S.; Ohshima, H. Divalent cation-induced phosphatidic acid membrane fusion. Effect of ion binding and membrane surface tension. Biochim. Biophys. Acta 1985, 812, 147–154. [Google Scholar] [CrossRef]
- Dudev, T.; Lim, C. Importance of metal hydration on the selectivity of Mg2+ versus Ca2+ in magnesium ion channels. J. Am. Chem. Soc. 2013, 135, 17200–17208. [Google Scholar] [CrossRef] [PubMed]
- Putta, P.; Rankenberg, J.; Korver, R.A.; van Wijk, R.; Munnik, T.; Testerink, C.; Kooijman, E.E. Phosphatidic acid binding proteins display differential binding as a function of membrane curvature stress and chemical properties. Biochim. Biophys. Acta 2016, 1858, 2709–2716. [Google Scholar] [CrossRef]
- Pincet, F.; Cribier, S.; Perez, E. Bilayers of neutral lipids bear a small but significant charge. Eur. Phys. J. B 1999, 11, 127–130. [Google Scholar] [CrossRef]
- Abrisch, R.G.; Gumbin, S.C.; Wisniewski, B.T.; Lackner, L.L.; Voeltz, G.K. Fission and fusion machineries converge at ER contact sites to regulate mitochondrial morphology. J. Cell Biol. 2020, 219, e201911122. [Google Scholar] [CrossRef] [PubMed]
- Monteiro-Cardoso, V.F.; Rochin, L.; Arora, A.; Houcine, A.; Jääskeläinen, E.; Kivelä, A.M.; Sauvanet, C.; Le Bars, R.; Marien, E.; Dehairs, J.; et al. ORP5/8 and MIB/MICOS link ER-mitochondria and intra-mitochondrial contacts for non-vesicular transport of phosphatidylserine. Cell Rep. 2022, 40, 111364. [Google Scholar] [CrossRef] [PubMed]
- Vance, J.E. MAM (mitochondria-associated membranes) in mammalian cells: Lipids and beyond. Biochim. Biophys. Acta 2014, 1841, 595–609. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vlieghe, A.; Niort, K.; Fumat, H.; Guigner, J.-M.; Cohen, M.M.; Tareste, D. Role of Lipids and Divalent Cations in Membrane Fusion Mediated by the Heptad Repeat Domain 1 of Mitofusin. Biomolecules 2023, 13, 1341. https://doi.org/10.3390/biom13091341
Vlieghe A, Niort K, Fumat H, Guigner J-M, Cohen MM, Tareste D. Role of Lipids and Divalent Cations in Membrane Fusion Mediated by the Heptad Repeat Domain 1 of Mitofusin. Biomolecules. 2023; 13(9):1341. https://doi.org/10.3390/biom13091341
Chicago/Turabian StyleVlieghe, Anaïs, Kristina Niort, Hugo Fumat, Jean-Michel Guigner, Mickaël M. Cohen, and David Tareste. 2023. "Role of Lipids and Divalent Cations in Membrane Fusion Mediated by the Heptad Repeat Domain 1 of Mitofusin" Biomolecules 13, no. 9: 1341. https://doi.org/10.3390/biom13091341
APA StyleVlieghe, A., Niort, K., Fumat, H., Guigner, J.-M., Cohen, M. M., & Tareste, D. (2023). Role of Lipids and Divalent Cations in Membrane Fusion Mediated by the Heptad Repeat Domain 1 of Mitofusin. Biomolecules, 13(9), 1341. https://doi.org/10.3390/biom13091341






