Mass Spectrometric Analysis of Cucurbitacins and Dihydrocucurbitacins from the Tuber of Citrullus naudinianus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Plant Material, Extraction and Sample Preparation
2.3. Flow Injection and HPLC Conditions
2.4. ESI-QTOF-MS/MS and ESI-Orbitrap-MS conditions
2.5. Data Analysis
3. Results
3.1. Extraction of Natural Products
3.2. QTOF-MS/MS Behavior of Flow-Injected Cucurbitacin and 23,24-Dihydrocucurbitacin Standards
3.3. HPLC-QTOF-MS Analysis of the Extract, HPLC-QTOF-MS/MS Screening of Cucurbitacins and 23,24-Dihydrocucurbitacins, and HPLC-Orbitrap-MS Analysis of the Extract
4. Discussion
4.1. Extraction of Natural Products
4.2. QTOF-MS/MS Behavior of Flow-Injected Cucurbitacin and 23,24-Dihydrocucurbitacin Standards
4.3. HPLC-QTOF-MS Analysis of the Extract, HPLC-QTOF-MS/MS Screening of Cucurbitacins and 23,24-Dihydrocucurbitacins, and HPLC-Orbitrap-MS Analysis of the Extract
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Grubben, G.J.H.; Denton, O.A. Plant Resources of Tropical Africa 2, Vegetables; PROTA Foundation: Wageningen, The Netherlands, 2004; pp. 34–35. [Google Scholar]
- Du Preez, C.I.; Gründemann, C.; Reinhardt, J.K.; Mumbengegwi, D.R.; Huber, R. Immunomodulatory effects of some Namibian plants traditionally used for treating inflammatory diseases. J. Ethnopharmacol. 2020, 254, 112683. [Google Scholar] [CrossRef] [PubMed]
- Rehm, S.; Enslin, P.R.; Meeuse, A.D.J.; Wessels, J.H. Bitter principles of the cucurbitaceae. VII. †—The distribution of bitter principles in this plant family. J. Sci. Food Agric. 1957, 8, 679–686. [Google Scholar] [CrossRef]
- Miró, M. Cucurbitacins and their pharmacological effects. Phytother. Res. 1995, 9, 159–168. [Google Scholar] [CrossRef]
- Chen, J.C.; Chiu, M.H.; Nie, R.L.; Cordell, G.A.; Qiu, S.X. Cucurbitacins and cucurbitane glycosides: Structures and biological activities. Nat. Prod. Rep. 2005, 22, 386–399. [Google Scholar] [CrossRef] [PubMed]
- Sallam, A.A.; Hitotsuyanagi, Y.; Mansour, E.S.S.; Ahmed, A.F.; Gedara, S.; Fukaya, H.; Takeya, K. Cucurbitacins from Bryonia cretica. Phytochem. Lett. 2010, 3, 117–121. [Google Scholar] [CrossRef]
- Jayaprakasam, B.; Seeram, N.P.; Nair, M.G. Anticancer and antiinflammatory activities of cucurbitacins from Cucurbita andreana. Canc. Lett. 2003, 189, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.B.; Chen, G.Y.; Liu, J.H.; Lei, M.; Meng, Y.H.; Guo, D.A.; Liu, X.; Ho, L.H. Cytotoxic cucurbitane triterpenoids isolated from the rhizomes of Hemsleya amabilis. Fitoterapia 2014, 94, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Escandell, J.M.; Recio, M.C.; Mañez, S.; Giner, R.M.; Cerdá-Nicolás, M.; Ríos, J.L. Dihydrocucurbitacin B, isolated from Cayaponia tayuya, reduces damage in adjuvant-induced arthritis. Eur. J. Pharmacol. 2006, 532, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Escandell, J.M.; Recio, M.C.; Mañez, S.; Giner, R.M.; Cerdá-Nicolás, M.; Ríos, J.L. Cucurbitacin R Reduces the Inflammation and Bone Damage Associated with Adjuvant Arthritis in Lewis Rats by Suppression of Tumor Necrosis Factor-α in T Lymphocytes and Macrophages. J. Pharmacol. Exp. Ther. 2007, 320, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Agil, M.A.; Risco, S.; Miró, M.; Navarro, M.C.; Ocete, M.A.; Jiménez, J. Analgesic and antipyretic effects of Ecballium elaterium (L.) A. Richard. Extract in rodents. Phytother. Res. 1995, 9, 135–138. [Google Scholar] [CrossRef]
- Afifi, M.S.; Ross, S.A.; ElSohly, M.A.; Naeem, Z.E.; Halaweish, F.T. Cucurbitacins of Cucumis prophetarum and Cucumis prophetarum. J. Chem. Ecol. 1999, 25, 847–859. [Google Scholar] [CrossRef]
- Bajcsik, N.; Pfab, R.; Pietsch, J. Simultaneous determination of cucurbitacin B, E, I and E-glucoside in plant material and body fluids by HPLC-MS. J. Chromatogr. B 2017, 1052, 128–134. [Google Scholar] [CrossRef] [PubMed]


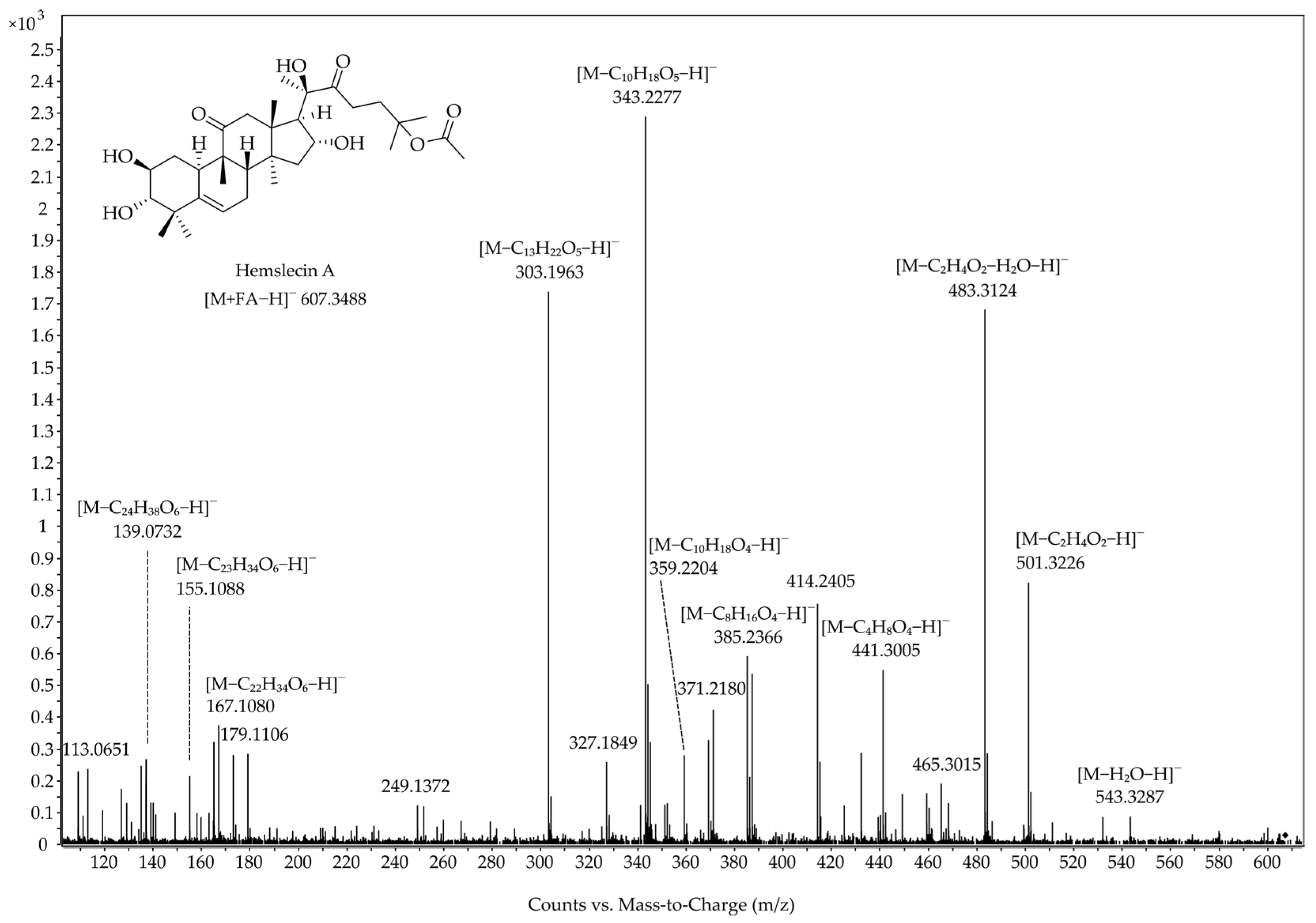
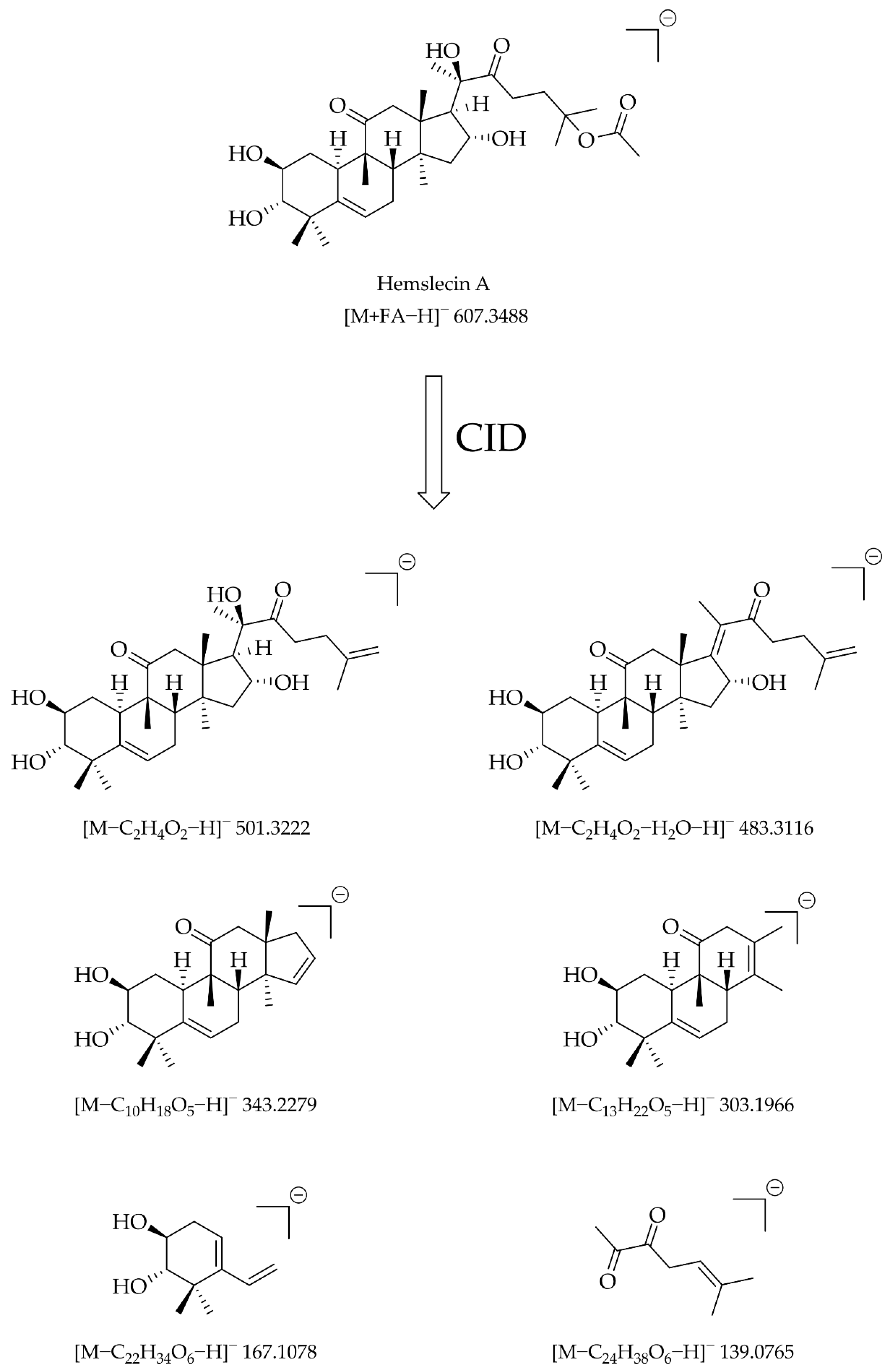



| Cucurbitacin | Position and Type of O-Functionalities in the Cucurbitane Skeleton | Position of C-C Double Bonds in the Cucurbitane Skeleton | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 3 | 11 | 16 | 19 | 20 | 22 | 24 | 25 | 1 | 5 | 23 | |
| A | OHβ | =O | =O | OHα | OH | OH | =O | OAc | Δ5 | Δ23 | ||
| B | OHβ | =O | =O | OHα | OH | =O | OAc | Δ5 | Δ23 | |||
| C | OHα | =O | OHα | OH | OH | =O | OAc | Δ5 | Δ23 | |||
| D | OHβ | =O | =O | OHα | OH | =O | OH | Δ5 | Δ23 | |||
| E | OH | =O | =O | OHα | OH | =O | OAc | Δ1 | Δ5 | Δ23 | ||
| F | OHβ | OHα | =O | OHα | OH | =O | OH | Δ5 | Δ23 | |||
| G | OHβ | =O | =O | OHα | OH | =O | (24R)-OH | OH | Δ5 | |||
| H | OHβ | =O | =O | OHα | OH | =O | (24S)-OH | OH | Δ5 | |||
| I | OH | =O | =O | OHα | OH | =O | OH | Δ1 | Δ5 | Δ23 | ||
| J | OH | =O | =O | OHα | OH | =O | (24R)-OH | OH | Δ1 | Δ5 | ||
| K | OH | =O | =O | OHα | OH | =O | (24S)-OH | OH | Δ1 | Δ5 | ||
| L | OH | =O | =O | OHα | OH | =O | OH | Δ1 | Δ5 | |||
| O | OHα | OHα | =O | OHα | OH | =O | OH | Δ5 | Δ23 | |||
| P | OHα | OHα | =O | OHα | OH | =O | OH | Δ5 | ||||
| Q | OHα | OHα | =O | OHα | OH | =O | OAc | Δ5 | Δ23 | |||
| R | OHβ | =O | =O | OHα | OH | =O | OH | Δ5 | ||||
| S | OH | =O | =O | Oα-C24 | =O | (24S)-O-C16 | OH | Δ1 | Δ5 | |||
| T | OH | =O | =O | Oα-C24 | OH | =O | (24S)-O-C16 | OMe | Δ1 | Δ5 | ||
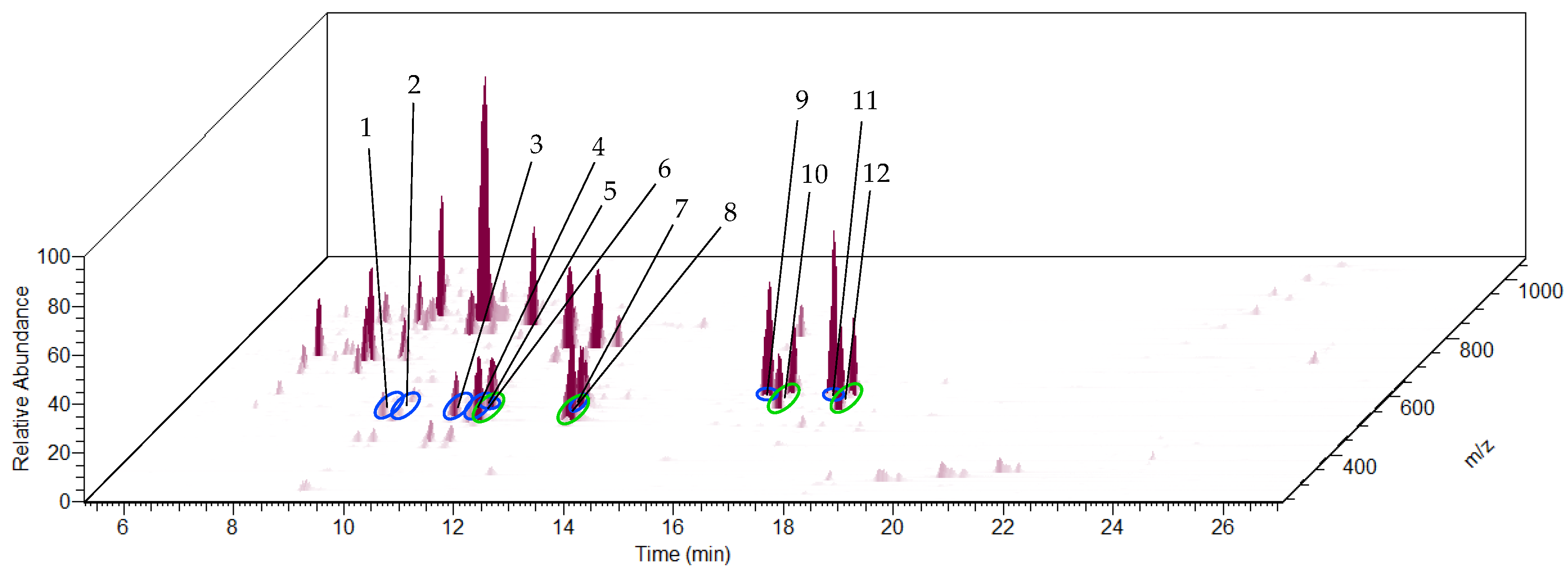
| Peak No. in Figure 5/Figure 6 | Name | Molecular Formula | RT (min) | m/z | Ion Type | Exact m/z | Error (ppm) | MS/MS m/z of [M + FA − H]− |
|---|---|---|---|---|---|---|---|---|
| 1 | Cucurbitacin G/H | C30H46O8 | 9.93 | 579.3178 | [M + FA − H]− | 579.3175 | 0.52 | 497.2708; |
| 569.2898 | [M + Cl]− | 569.2887 | 1.93 | 301.1792; | ||||
| 533.3130 | [M − H]− | 533.3120 | 1.88 | 165.0914. | ||||
| 2 | Cucurbitacin G/H | C30H46O8 | 10.19 | 579.3179 | [M + FA − H]− | 579.3175 | 0.69 | 497.2931; |
| 569.2895 | [M + Cl]− | 569.2887 | 1.41 | 301.1826; | ||||
| 533.3127 | [M − H]− | 533.3120 | 1.31 | 165.0934. | ||||
| 3 | Cucurbitacin J/K | C30H44O8 | 11.21 | 577.3029 | [M + FA − H]− | 577.3018 | 1.91 | 495.2697; |
| 567.2749 | [M + Cl]− | 567.2730 | 3.35 | 299.1601; | ||||
| 531.2980 | [M − H]− | 531.2963 | 3.20 | 163.0751. | ||||
| 4 | Cucurbitacin J/K | C30H44O8 | 11.57 | 577.3030 | [M + FA − H]− | 577.3018 | 2.08 | 495.2789; |
| 567.2746 | [M + Cl]− | 567.2730 | 2.82 | 299.1673; | ||||
| 531.2974 | [M − H]− | 531.2963 | 2.07 | 163.0775. | ||||
| 5 | Cucurbitacin D | C30H44O7 | 11.77 | 561.3090 | [M + FA − H]− | 561.3069 | 3.74 | 497.2872; |
| 551.2796 | [M + Cl]− | 551.2781 | 2.72 | 301.1795; | ||||
| 515.3014 | [M-H]− | 515.3003 | 2.13 | 165.0909. | ||||
| 6 | 23,24-Dihydro-cucurbitacin D | C30H46O7 | 11.80 | 563.3243 | [M + FA − H]− | 563.3226 | 3.02 | 499.3082; |
| 553.2935 | [M + Cl]− | 553.2938 | −0.54 | 4301.1792; | ||||
| 517.3187 | [M − H]− | 517.3171 | 3.09 | 165.0918. | ||||
| 7 | Cucurbitacin I | C30H42O7 | 13.31 | 559.2930 | [M + FA − H]− | 559.2913 | 3.04 | 495.2708; |
| 549.2641 | [M + Cl]− | 549.2625 | 2.91 | 299.1670; | ||||
| 513.2864 | [M − H]− | 513.2858 | 1.17 | 163.0743. | ||||
| 8 | 23,24-Dihydro-cucurbitacin I | C30H44O7 | 13.41 | 561.3085 | [M + FA − H]− | 561.3069 | 2.85 | 497.2943; |
| 551.2783 | [M + Cl]− | 551.2781 | 0.36 | 299.1745; | ||||
| 515.3030 | [M − H]− | 515.3014 | 3.10 | 163.0764. | ||||
| 9 | Cucurbitacin B | C32H46O8 | 16.24 | 603.3190 | [M + FA − H]− | 603.3175 | 2.49 | 497.2901; |
| 593.2902 | [M + Cl]− | 593.2887 | 2.53 | 301.1423; | ||||
| 557.3102 | [M − H]− | 557.3120 | −3.23 | 165.0911. | ||||
| 10 | 23,24-Dihydro-cucurbitacin B | C32H48O8 | 16.63 | 605.3345 | [M + FA − H]− | 605.3331 | 2.31 | 499.3121; |
| 595.3055 | [M + Cl]− | 595.3043 | 2.02 | 301.1763; | ||||
| 559.3282 | [M − H]− | 559.3276 | 1.07 | 165.0914. | ||||
| 11 | Cucurbitacin E | C32H44O8 | 17.29 | 601.3034 | [M + FA − H]− | 601.3018 | 2.66 | 495.2737; |
| 591.2745 | [M + Cl]− | 591.2730 | 2.54 | 299.1276; | ||||
| 555.2960 | [M − H]− | 555.2963 | −0.54 | 163.0751. | ||||
| 12 | 23,24-Dihydro-cucurbitacin E | C32H46O8 | 17.64 | 603.3185 | [M + FA − H]− | 603.3175 | 1.66 | 497.2902; |
| 593.2895 | [M + Cl]− | 593.2887 | 1.35 | 299.1663; | ||||
| 557.3126 | [M − H]− | 557.3120 | 1.08 | 163.0771. |
| Peak No. in Figure 8 | Name | Molecular Formula | RT (min) | m/z | Ion Type | Exact m/z | Error (ppm) |
|---|---|---|---|---|---|---|---|
| 1 | Cucurbitacin G/H | C30H46O8 | 9.18 | 579.3170 | [M + FA − H]− | 579.3175 | −0.86 |
| 2 | Cucurbitacin G/H | C30H46O8 | 9.48 | 579.3172 | [M + FA − H]− | 579.3175 | −0.52 |
| 3 | Cucurbitacin J/K | C30H44O8 | 10.50 | 577.3014 | [M + FA − H]− | 577.3018 | −0.69 |
| 4 | Cucurbitacin J/K | C30H44O8 | 10.88 | 577.3013 | [M + FA − H]− | 577.3018 | −0.87 |
| 5 | Cucurbitacin D | C30H44O7 | 11.01 | 561.3066 | [M + FA − H]− | 561.3069 | −0.53 |
| 6 | 23,24-Dihydrocucurbitacin D | C30H46O7 | 11.01 | 563.3220 | [M + FA − H]− | 563.3226 | −1.07 |
| 7 | Cucurbitacin I | C30H42O7 | 12.63 | 559.2911 | [M + FA − H]− | 559.2913 | −0.36 |
| 8 | 23,24-Dihydrocucurbitacin I | C30H44O7 | 12.71 | 561.3064 | [M + FA − H]− | 561.3069 | −0.89 |
| 9 | Cucurbitacin B | C32H46O8 | 15.83 | 603.3172 | [M + FA − H]− | 603.3175 | −0.50 |
| 10 | 23,24-Dihydrocucurbitacin B | C32H48O8 | 16.26 | 605.3329 | [M + FA − H]− | 605.3331 | −0.33 |
| 11 | Cucurbitacin E | C32H44O8 | 17.03 | 601.3018 | [M + FA − H]− | 601.3018 | 0.00 |
| 12 | 23,24-Dihydrocucurbitacin E | C32H46O8 | 17.36 | 603.3175 | [M + FA − H]− | 603.3175 | 0.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Benka, M.; Görlitz, K.; Schöttgen, M.C.; Lagies, S.; Mohl, D.A.; Kather, M.; Du Preez-Bruwer, I.; Mumbengegwi, D.; Teufel, R.; Kowarschik, S.; et al. Mass Spectrometric Analysis of Cucurbitacins and Dihydrocucurbitacins from the Tuber of Citrullus naudinianus. Biomolecules 2023, 13, 1168. https://doi.org/10.3390/biom13081168
Benka M, Görlitz K, Schöttgen MC, Lagies S, Mohl DA, Kather M, Du Preez-Bruwer I, Mumbengegwi D, Teufel R, Kowarschik S, et al. Mass Spectrometric Analysis of Cucurbitacins and Dihydrocucurbitacins from the Tuber of Citrullus naudinianus. Biomolecules. 2023; 13(8):1168. https://doi.org/10.3390/biom13081168
Chicago/Turabian StyleBenka, Moritz, Kristof Görlitz, Michael C. Schöttgen, Simon Lagies, Daniel A. Mohl, Michel Kather, Iwanette Du Preez-Bruwer, Davis Mumbengegwi, Robin Teufel, Stefanie Kowarschik, and et al. 2023. "Mass Spectrometric Analysis of Cucurbitacins and Dihydrocucurbitacins from the Tuber of Citrullus naudinianus" Biomolecules 13, no. 8: 1168. https://doi.org/10.3390/biom13081168
APA StyleBenka, M., Görlitz, K., Schöttgen, M. C., Lagies, S., Mohl, D. A., Kather, M., Du Preez-Bruwer, I., Mumbengegwi, D., Teufel, R., Kowarschik, S., Huber, R., Plattner, D. A., & Kammerer, B. (2023). Mass Spectrometric Analysis of Cucurbitacins and Dihydrocucurbitacins from the Tuber of Citrullus naudinianus. Biomolecules, 13(8), 1168. https://doi.org/10.3390/biom13081168






