Identification of Potential Inflammation-Related Genes and Key Pathways Associated with Complex Regional Pain Syndrome
Abstract
1. Introduction
2. Materials and Methods
2.1. Microarray Datasets of CRPS
2.2. Differential Expression Analysis
2.3. Gene Ontology and Pathway Enrichment Analysis
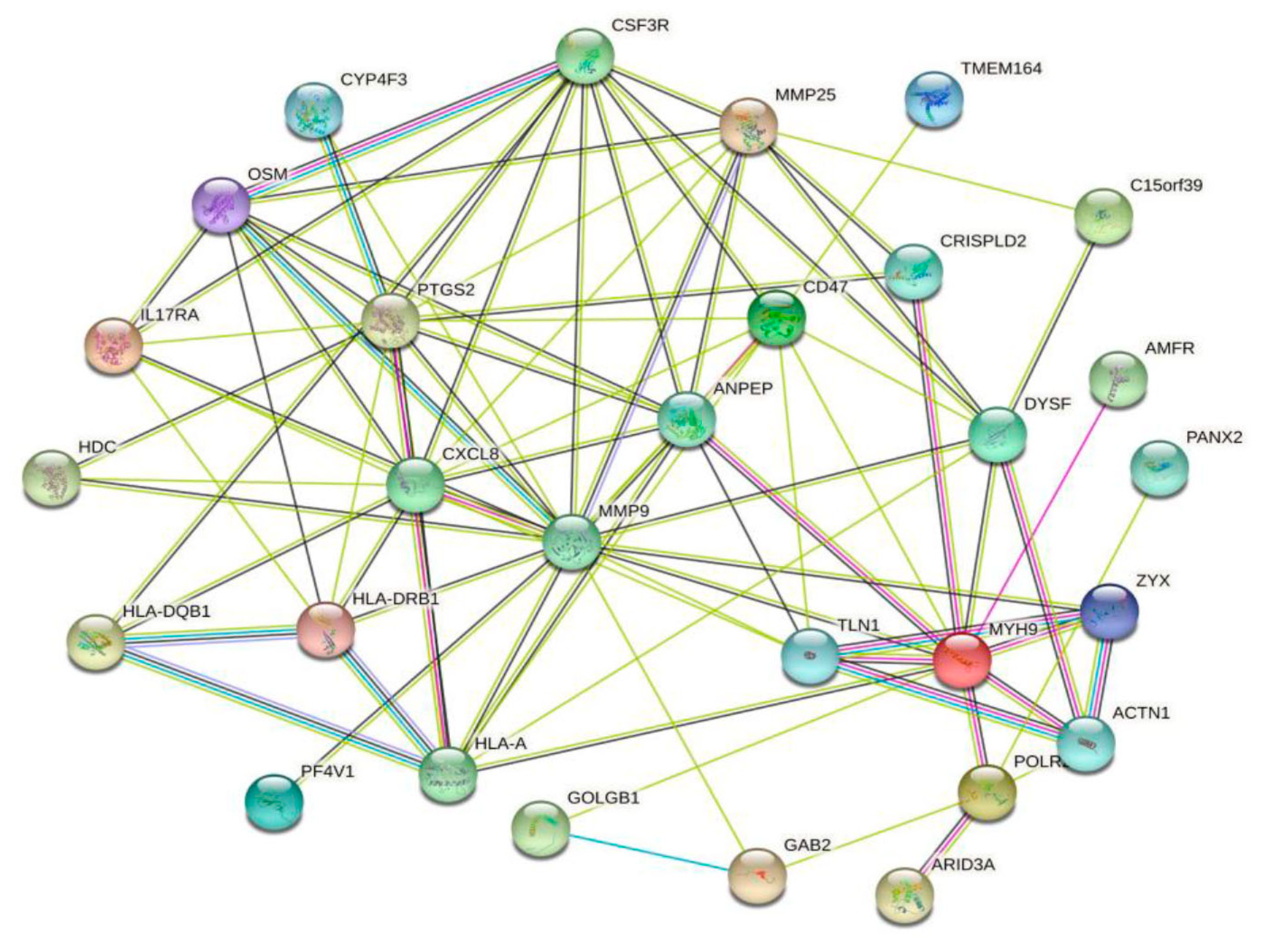
2.4. Protein–Protein Interaction (PPI) Network Analysis
2.5. Gene Set Enrichment Analyses (GSEA)
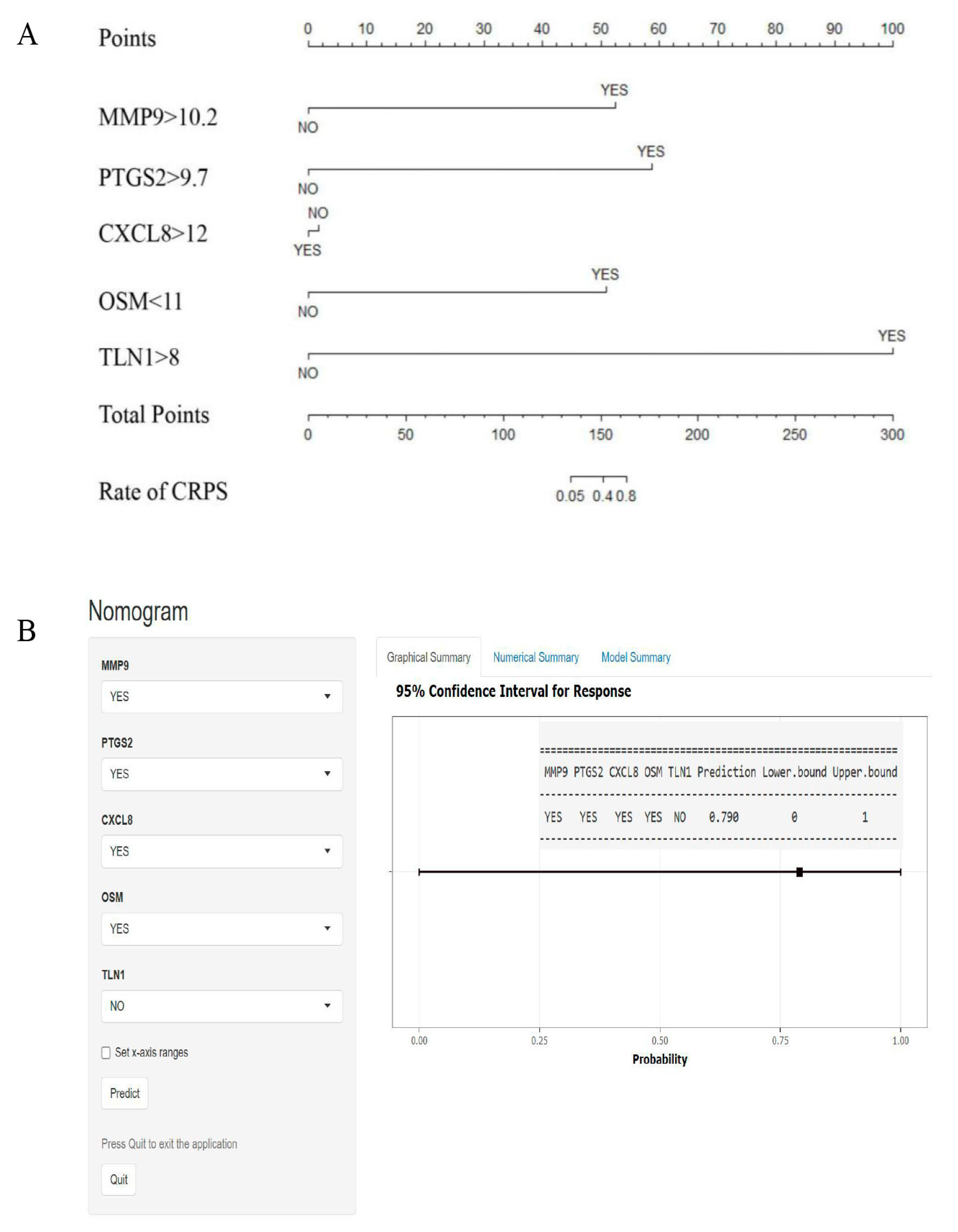
2.6. Establishment of Predictive Models
2.7. Ethical Declaration
3. Results
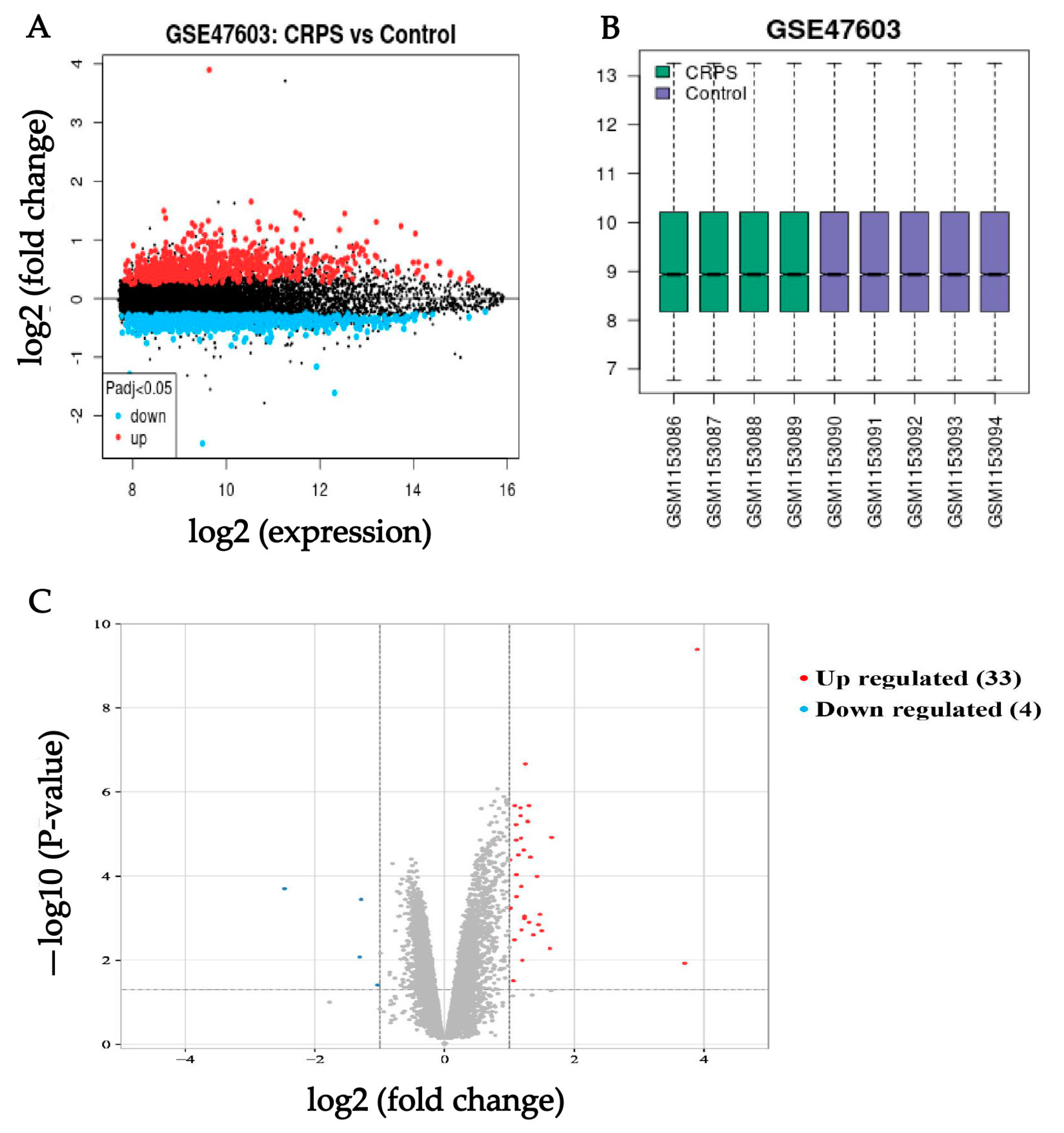
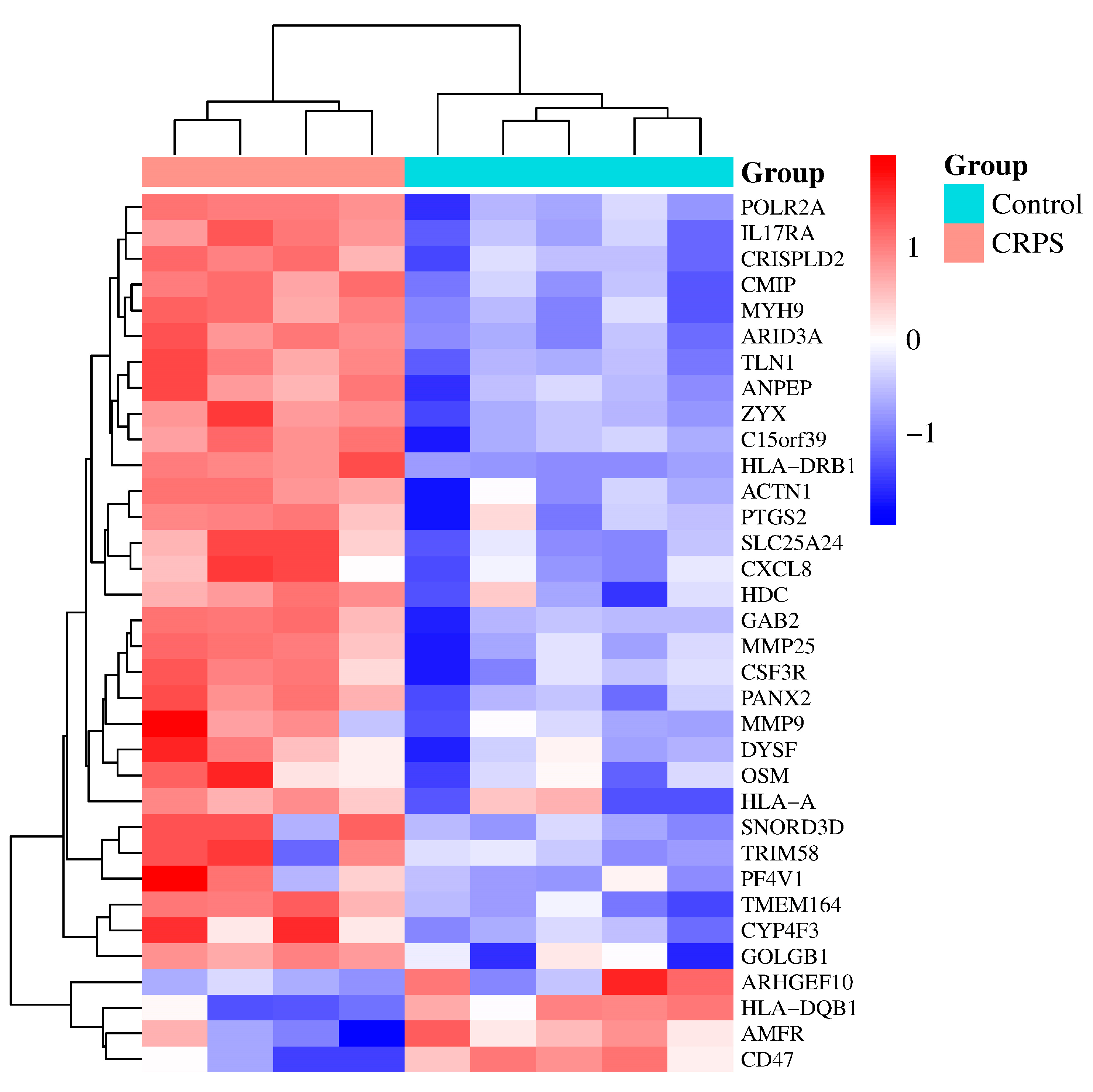
3.1. Identification of CRPS-Associated Genes
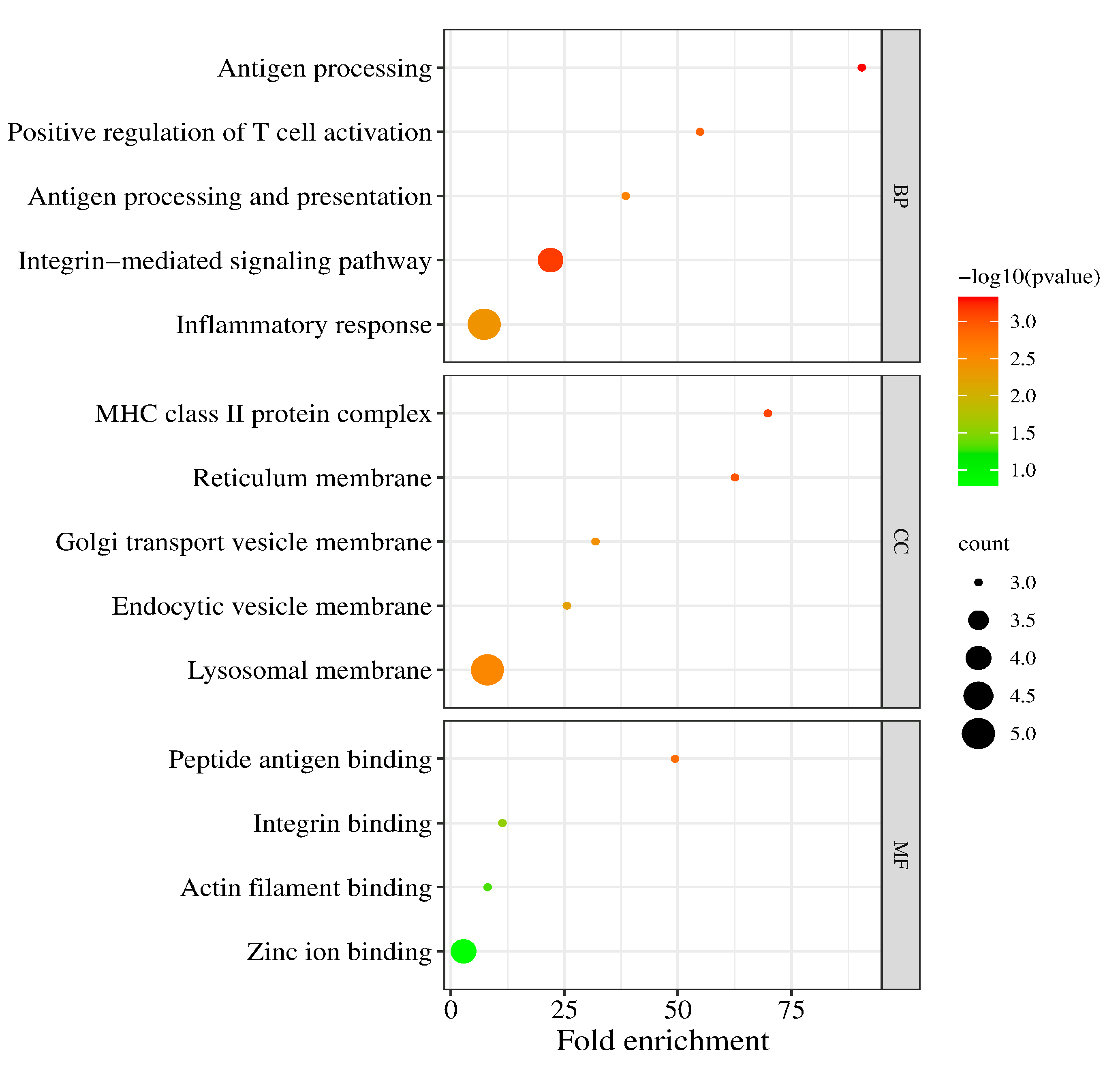
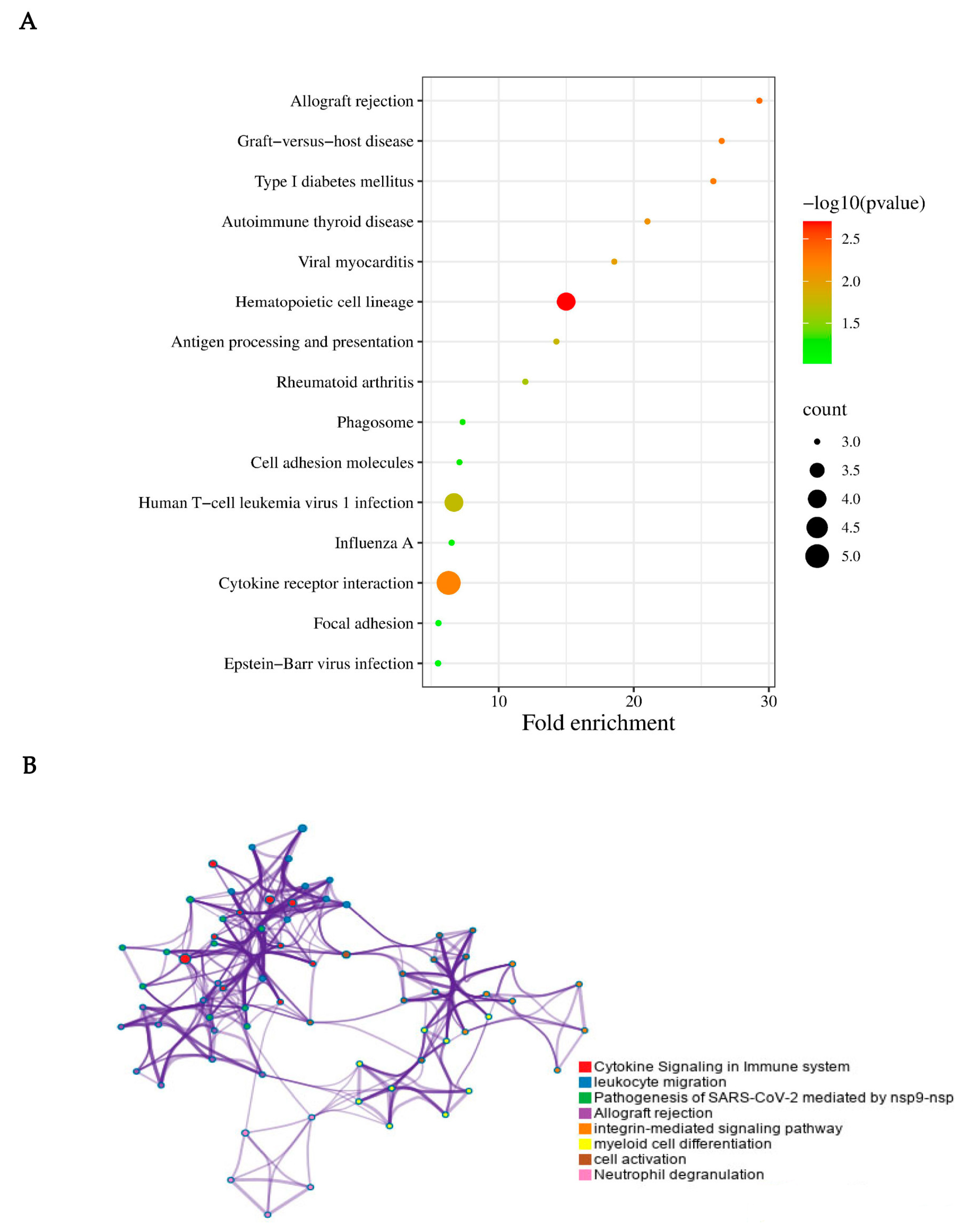
3.2. Functional Annotation
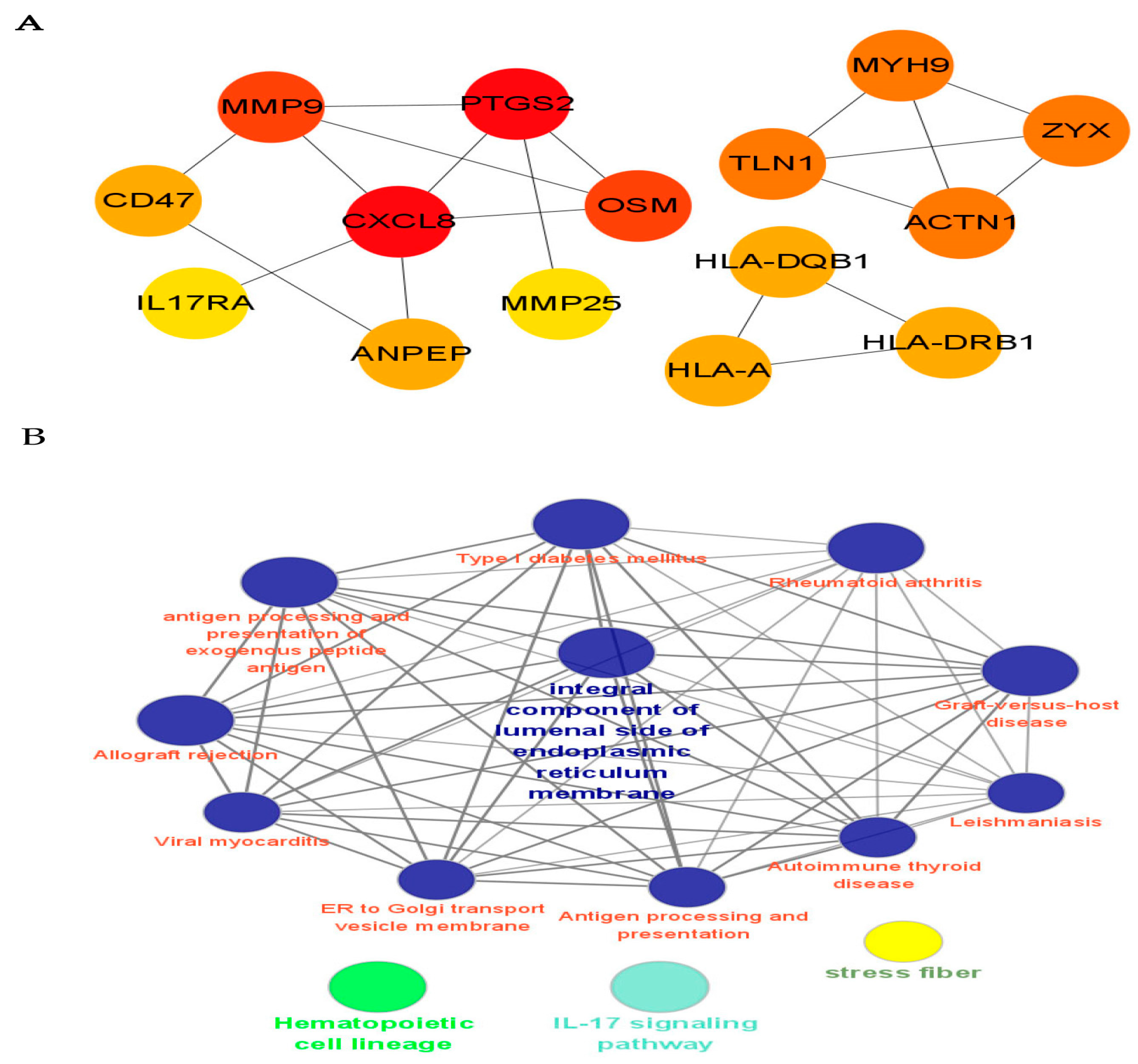
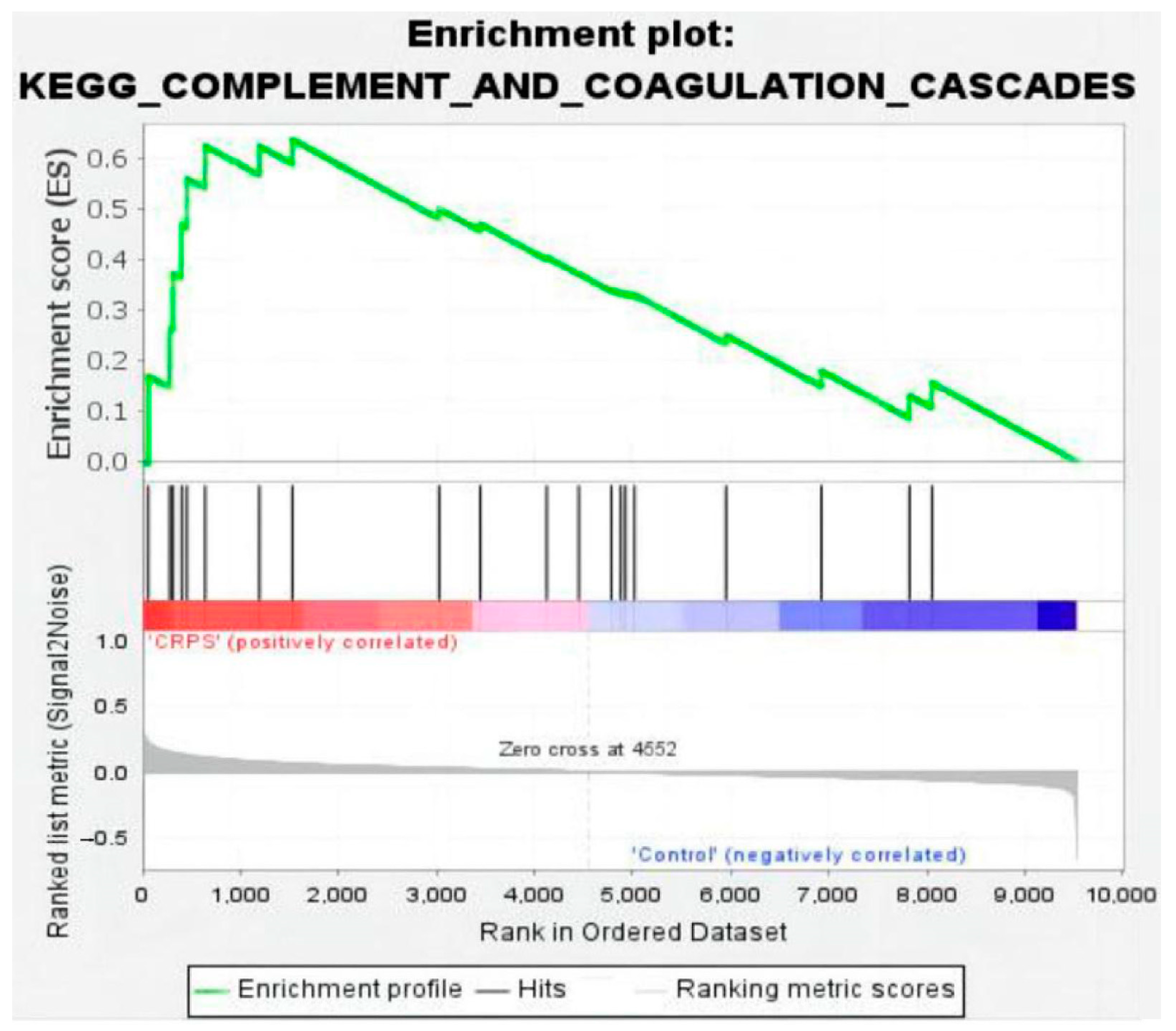
3.3. PPI Network Analysis and GSEA Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Petersen, P.B.; Mikkelsen, K.L.; Lauritzen, J.B.; Krogsgaard, M.R. Risk Factors for Post-treatment Complex Regional Pain Syndrome (CRPS): An Analysis of 647 Cases of CRPS from the Danish Patient Compensation Association. Pain Pract. 2018, 18, 341–349. [Google Scholar] [CrossRef] [PubMed]
- Moseley, G.L.; Herbert, R.D.; Parsons, T.; Lucas, S.; Van Hilten, J.J.; Marinus, J. Intense pain soon after wrist fracture strongly predicts who will develop complex regional pain syndrome: Prospective cohort study. J. Pain 2014, 15, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Beerthuizen, A.; Stronks, D.L.; Van’t Spijker, A.; Yaksh, A.; Hanraets, B.M.; Klein, J.; Huygen, F.J.P.M. Demographic and medical parameters in the development of complex regional pain syndrome type 1 (CRPS1): Prospective study on 596 patients with a fracture. Pain 2012, 153, 1187–1192. [Google Scholar] [CrossRef]
- Ott, S.; Maihöfner, C. Signs and Symptoms in 1043 Patients with Complex Regional Pain Syndrome. J. Pain 2018, 19, 599–611. [Google Scholar] [CrossRef] [PubMed]
- Shim, H.; Rose, J.; Halle, S.; Shekane, P. Complex regional pain syndrome: A narrative review for the practising clinician. Br. J. Anaesth. 2019, 123, e424–e433. [Google Scholar] [CrossRef]
- Harden, N.R.; Bruehl, S.; Perez, R.S.G.M.; Birklein, F.; Marinus, J.; Maihofner, C.; Lubenow, T.; Buvanendran, A.; Mackey, S.; Graciosa, J.; et al. Validation of proposed diagnostic criteria (the “Budapest Criteria”) for Complex Regional Pain Syndrome. Pain 2010, 150, 268–274. [Google Scholar] [CrossRef]
- Birklein, F.; Ajit, S.K.; Goebel, A.; Perez, R.S.G.M.; Sommer, C. Complex regional pain syndrome—Phenotypic characteristics and potential biomarkers. Nat. Rev. Neurol. 2018, 14, 272–284. [Google Scholar] [CrossRef]
- Bharwani, K.D.; Dik, W.A.; Dirckx, M.; Huygen, F.J.P.M. Highlighting the Role of Biomarkers of Inflammation in the Diagnosis and Management of Complex Regional Pain Syndrome. Mol. Diagn. Ther. 2019, 23, 615–626. [Google Scholar] [CrossRef]
- Jin, E.H.; Zhang, E.; Ko, Y.; Sim, W.S.; Moon, D.E.; Yoon, K.J.; Hong, J.H.; Lee, W.H. Genome-wide expression profiling of complex regional pain syndrome. PLoS ONE 2013, 8, e79435. [Google Scholar] [CrossRef]
- Wang, K.; Yi, D.; Yu, Z.; Zhu, B.; Li, S.; Liu, X. Identification of the Hub Genes Related to Nerve Injury-Induced Neuropathic Pain. Front. Neurosci. 2020, 14, 488. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets–update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Chaturvedi, S.; Braunstein, E.M.; Brodsky, R.A. Antiphospholipid syndrome: Complement activation, complement gene mutations, and therapeutic implications. J. Thromb. Haemost. 2021, 19, 607–616. [Google Scholar] [CrossRef]
- Bruehl, S.; Maihöfner, C.; Stanton-Hicks, M.; Perez, R.S.; Vatine, J.J.; Brunner, F.; Birklein, F.; Schlereth, T.; Mackey, S.; Mailis-Gagnon, A.; et al. Complex regional pain syndrome: Evidence for warm and cold subtypes in a large prospective clinical sample. Pain 2016, 157, 1674–1681. [Google Scholar] [CrossRef]
- Cha, M.; Lee, K.H.; Kwon, M.; Lee, B. Possible Therapeutic Options for Complex Regional Pain Syndrome. Biomedicines 2021, 9, 596. [Google Scholar] [CrossRef]
- Torta, D.M.; Legrain, V.; Rossetti, Y.; Mouraux, A. Prisms for pain. Can visuo-motor rehabilitation strategies alleviate chronic pain? Eur. J. Pain 2016, 20, 64–69. [Google Scholar] [CrossRef]
- Catley, M.J.; O’Connell, N.E.; Berryman, C.; Ayhan, F.F.; Moseley, G.L. Is tactile acuity altered in people with chronic pain? a systematic review and meta-analysis. J. Pain 2014, 15, 985–1000. [Google Scholar] [CrossRef]
- Roh, Y.H.; Lee, B.K.; Noh, J.H.; Baek, J.R.; Oh, J.H.; Gong, H.S.; Baek, G.H. Factors associated with complex regional pain syndrome type I in patients with surgically treated distal radius fracture. Arch. Orthop. Trauma Surg. 2014, 134, 1775–1781. [Google Scholar] [CrossRef]
- David Clark, J.; Tawfik, V.L.; Tajerian, M.; Kingery, W.S. Autoinflammatory and autoimmune contributions to complex regional pain syndrome. Mol. Pain 2018, 14, 1744806918799127. [Google Scholar] [CrossRef] [PubMed]
- Lenz, M.; Üçeyler, N.; Frettlöh, J.; Höffken, O.; Krumova, E.K.; Lissek, S.; Reinersmann, A.; Sommer, C.; Stude, P.; Waaga-Gasser, A.M.; et al. Local cytokine changes in complex regional pain syndrome type I (CRPS I) resolve after 6 months. Pain 2013, 154, 2142–2149. [Google Scholar] [CrossRef] [PubMed]
- Morellini, N.; Finch, P.M.; Goebel, A.; Drummond, P.D. Dermal nerve fibre and mast cell density, and proximity of mast cells to nerve fibres in the skin of patients with complex regional pain syndrome. Pain 2018, 159, 2021–2029. [Google Scholar] [CrossRef] [PubMed]
- Heyn, J.; Azad, S.C.; Luchting, B. Altered regulation of the T-cell system in patients with CRPS. Inflamm. Res. 2019, 68, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Ji, R.R.; Xu, Z.Z.; Gao, Y.J. Emerging targets in neuroinflammation-driven chronic pain. Nat. Rev. Drug Discov. 2014, 13, 533–548. [Google Scholar] [CrossRef]
- Escolano-Lozano, F.; Gries, E.; Schlereth, T.; Dimova, V.; Baka, P.; Vlckova, E.; König, S.; Birklein, F. Local and Systemic Expression Pattern of MMP-2 and MMP-9 in Complex Regional Pain Syndrome. J. Pain 2021, 22, 1294–1302. [Google Scholar] [CrossRef]
- Hossaini, M.; Sarac, C.; Jongen, J.L.; Holstege, J.C. Spinal glycinergic and GABAergic neurons expressing C-fos after capsaicin stimulation are increased in rats with contralateral neuropathic pain. Neuroscience 2011, 196, 265–275. [Google Scholar] [CrossRef]
- Linnman, C.; Becerra, L.; Borsook, D. Inflaming the brain: CRPS a model disease to understand neuroimmune interactions in chronic pain. J. Neuroimmune Pharmacol. 2013, 8, 547–563. [Google Scholar] [CrossRef]
- Ji, R.R.; Chamessian, A.; Zhang, Y.Q. Pain regulation by non-neuronal cells and inflammation. Science 2016, 354, 572–577. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, H.; Hua, L.; Hou, C.; Jia, Q.; Chen, J.; Zhang, S.; Wang, Y.; He, S.; Jia, E. Verification of ferroptosis and pyroptosis and identification of PTGS2 as the hub gene in human coronary artery atherosclerosis. Free Radic. Biol. Med. 2021, 171, 55–68. [Google Scholar] [CrossRef]
- de Oliveira, S.; Reyes-Aldasoro, C.C.; Candel, S.; Renshaw, S.A.; Mulero, V.; Calado, A. Cxcl8 (IL-8) mediates neutrophil recruitment and behavior in the zebrafish inflammatory response. J. Immunol. 2013, 190, 4349–4359. [Google Scholar] [CrossRef] [PubMed]
- Liubomirski, Y.; Lerrer, S.; Meshel, T.; Rubinstein-Achiasaf, L.; Morein, D.; Wiemann, S.; Körner, C.; Ben-Baruch, A. Tumor-Stroma-Inflammation Networks Promote Pro-metastatic Chemokines and Aggressiveness Characteristics in Triple-Negative Breast Cancer. Front. Immunol. 2019, 12, 757. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Du, G.; Huang, X.; Han, L.; Han, X.; Xu, B.; Zhang, Y.; Yu, M.; Qin, Y.; Xia, Y. The enhancer RNA lnc-SLC4A1-1 epigenetically regulates unexplained recurrent pregnancy loss (URPL) by activating CXCL8 and NF-kB pathway. EBioMedicine 2018, 38, 162–170. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Ren, R.; Lin, W.; Xiang, L.; Zhao, Z.; Shao, B. Exploring the oncostatin M (OSM) feed-forward signaling of glioblastoma via STAT3 in pan-cancer analysis. Cancer Cell Int. 2021, 21, 565. [Google Scholar] [CrossRef]
- Sanchez-Infantes, D.; Stephens, J.M. Adipocyte Oncostatin Receptor Regulates Adipose Tissue Homeostasis and Inflammation. Front. Immunol. 2021, 29, 612013. [Google Scholar] [CrossRef]
- Elks, C.M.; Zhao, P.; Grant, R.W.; Hang, H.; Bailey, J.L.; Burk, D.H.; McNulty, M.A.; Mynatt, R.L.; Stephens, J.M. Loss of Oncostatin M Signaling in Adipocytes Induces Insulin Resistance and Adipose Tissue Inflammation In Vivo. J. Biol. Chem. 2016, 291, 17066–17076. [Google Scholar] [CrossRef]
- van Rooijen, D.E.; Roelen, D.L.; Verduijn, W.; Haasnoot, G.W.; Huygen, F.J.; Perez, R.S.; Claas, F.H.; Marinus, J.; van Hilten, J.J.; van den Maagdenberg, A.M. Genetic HLA associations in complex regional pain syndrome with and without dystonia. J. Pain 2012, 13, 784–789. [Google Scholar] [CrossRef]
- Sawcer, S. The complex genetics of multiple sclerosis: Pitfalls and prospects. Brain 2008, 131, 3118–3131. [Google Scholar] [CrossRef]
- de Rooij, A.M.; Florencia Gosso, M.; Haasnoot, G.W.; Marinus, J.; Verduijn, W.; Claas, F.H.; van den Maagdenberg, A.M.; van Hilten, J.J. HLA-B62 and HLA-DQ8 are associated with Complex Regional Pain Syndrome with fixed dystonia. Pain 2009, 145, 82–85. [Google Scholar] [CrossRef]
- Tang, S.C.W.; You, W.H. Innate immunity in diabetic kidney disease. Nat. Rev. Nephrol. 2020, 16, 206–222. [Google Scholar] [CrossRef]
- Harden, R.N.; Oaklander, A.L.; Burton, A.W.; Perez, R.S.; Richardson, K.; Swan, M.; Barthel, J.; Costa, B.; Graciosa, J.R.; Bruehl, S. Reflex Sympathetic Dystrophy Syndrome Association. Complex regional pain syndrome: Practical diagnostic and treatment guidelines, 4th edition. Pain Med. 2013, 14, 180–229. [Google Scholar] [CrossRef] [PubMed]
- Urits, I.; Shen, A.H.; Jones, M.R.; Viswanath, O.; Kaye, A.D. Complex Regional Pain Syndrome, Current Concepts and Treatment Options. Curr. Pain Headache Rep. 2018, 22, 10. [Google Scholar] [CrossRef] [PubMed]
- Breuer, A.J.; Mainka, T.; Hansel, N.; Maier, C.; Krumova, E.K. Short-term treatment with parecoxib for complex regional pain syndrome: A randomized, placebo-controlled double-blind trial. Pain Physician 2014, 17, 127–137. [Google Scholar]
- Eckmann, M.S.; Ramamurthy, S.; Griffin, J.G. Intravenous regional ketorolac and lidocaine in the treatment of complex regional pain syndrome of the lower extremity: A randomized, double-blinded, crossover study. Clin. J. Pain 2011, 27, 203–206. [Google Scholar] [CrossRef] [PubMed]
- Visnjevac, O.; Costandi, S.; Patel, B.A.; Azer, G.; Agarwal, P.; Bolash, R.; Mekhail, N.A. A Comprehensive Outcome-Specific Review of the Use of Spinal Cord Stimulation for Complex Regional Pain Syndrome. Pain Pract. 2017, 17, 533–545. [Google Scholar] [CrossRef]
- Deer, T.R.; Levy, R.M.; Kramer, J.; Poree, L.; Amirdelfan, K.; Grigsby, E.; Staats, P.; Burton, A.W.; Burgher, A.H.; Obray, J.; et al. Dorsal root ganglion stimulation yielded higher treatment success rate for complex regional pain syndrome and causalgia at 3 and 12 months: A randomized comparative trial. Pain 2017, 158, 669–681. [Google Scholar] [CrossRef]
- Zhao, B.; Pan, Y.; Xu, H.; Song, X. Hyperbaric oxygen attenuates neuropathic pain and reverses inflammatory signaling likely via the Kindlin-1/Wnt-10a signaling pathway in the chronic pain injury model in rats. J. Headache Pain 2017, 18, 1. [Google Scholar] [CrossRef]
- Zollinger, P.E.; Tuinebreijer, W.E.; Kreis, R.W.; Breederveld, R.S. Effect of vitamin C on frequency of reflex sympathetic dystrophy in wrist fractures: A randomised trial. Lancet 1999, 354, 2025–2028. [Google Scholar] [CrossRef]
- Evaniew, N.; McCarthy, C.; Kleinlugtenbelt, Y.V.; Ghert, M.; Bhandari, M. Vitamin C to Prevent Complex Regional Pain Syndrome in Patients with Distal Radius Fractures: A Meta-Analysis of Randomized Controlled Trials. J. Orthop. Trauma 2015, 29, e235–e241. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, Y.; Li, Q.; Liu, K.; Hou, J.; Shao, C.; Wang, Y. Immunoregulatory mechanisms of mesenchymal stem and stromal cells in inflammatory diseases. Nat. Rev. Nephrol. 2018, 4, 493–507. [Google Scholar] [CrossRef]
| ID | Gene Symbol | p Value | Log Fc | Gene Title |
|---|---|---|---|---|
| ILMN_1715169 | HLA-DRB1 | 4.06 × 10−10 | 3.90 | Major histocompatibility complex, class II, DR beta 1 |
| ILMN_1670130 | ARID3A | 2.17 × 10−7 | 3.71 | AT-rich interaction domain 3A |
| ILMN_1738075 | CMIP | 2.14 × 10−6 | 1.65 | c-Maf inducing protein |
| ILMN_1696643 | TLN1 | 2.14 × 10−6 | 1.63 | Talin 1 |
| ILMN_2371169 | ZYX | 2.42 × 10−6 | 1.50 | Zyxin |
| ILMN_1722872 | MYH9 | 3.71 × 10−6 | 1.47 | Myosin, heavy chain 9, non-muscle |
| ILMN_1782385 | POLR2A | 5.04 × 10−6 | 1.45 | RNA polymerase II subunit A |
| ILMN_1728724 | IL17RA | 5.91 × 10−6 | 1.42 | Interleukin 17 receptor A |
| ILMN_1790689 | CRISPLD2 | 1.19 × 10−5 | 1.37 | Cysteine rich secretory protein LCCL domain containing 2 |
| ILMN_1701875 | ZYX | 1.24 × 10−5 | 1.33 | Zyxin |
| ILMN_1694810 | PANX2 | 1.39 × 10−5 | 1.31 | Pannexin 2 |
| ILMN_3241091 | TM × 10M164 | 2.38 × 10−5 | 1.30 | Transmembrane protein 164 |
| ILMN_1793729 | C15orf39 | 3.11 × 10−5 | 1.28 | Chromosome 15 open reading frame 39 |
| ILMN_1763837 | ANPEP | 3.54 × 10−5 | 1.25 | Alanyl aminopeptidase, membrane |
| ILMN_1665964 | GAB2 | 4.09 × 10−5 | 1.24 | GRB2 associated binding protein 2 |
| ILMN_1711838 | SLC25A24 | 9.17 × 10−5 | 1.23 | Solute carrier family 25 member 24 |
| ILMN_1717207 | MMP25 | 1.01 × 10−4 | 1.22 | Matrix metallopeptidase 25 |
| ILMN_2232177 | ACTN1 | 1.75 × 10−4 | 1.20 | Actinin alpha 1 |
| ILMN_1661266 | HLA-DQB1 | 1.97 × 10−4 | 1.19 | Major histocompatibility complex, class II, DQ beta 1 |
| ILMN_2371280 | CSF3R | 3.06 × 10−4 | 1.18 | Colony stimulating factor 3 receptor |
| ILMN_2356991 | CD47 | 3.56 × 10−4 | 1.18 | CD47 molecule |
| ILMN_1736190 | CYP4F3 | 5.75 × 10−4 | 1.17 | Cytochrome P450 family 4 subfamily F member 3 |
| ILMN_1677511 | PTGS2 | 8.13 × 10−4 | 1.17 | Prostaglandin-endoperoxide synthase 2 |
| ILMN_2184373 | CXCL8 | 8.98 × 10−4 | 1.13 | C-X-C motif chemokine ligand 8 |
| ILMN_1792323 | HDC | 1.01 × 10−3 | 1.11 | Histidine decarboxylase |
| ILMN_2054297 | PTGS2 | 1.25 × 10−3 | 1.10 | Prostaglandin-endoperoxide synthase 2 |
| ILMN_1666733 | CXCL8 | 1.43 × 10−3 | 1.10 | C-X-C motif chemokine ligand 8 |
| ILMN_1810420 | DYSF | 1.90 × 10−3 | 1.10 | Dysferlin |
| ILMN_3242315 | SNORD3D | 1.98 × 10−3 | 1.08 | Small nucleolar RNA, C/D box 3D |
| ILMN_1747935 | GOLGB1 | 2.49 × 10−3 | 1.08 | Olgin B1 |
| ILMN_1780546 | OSM | 3.28 × 10−3 | 1.06 | Oncostatin M |
| ILMN_1796316 | MMP9 | 5.24 × 10−3 | 1.02 | Matrix metallopeptidase 9 |
| ILMN_1723116 | AMFR | 8.38 × 10−3 | 1.01 | Autocrine motility factor receptor |
| ILMN_1745522 | PF4V1 | 1.00 × 10−2 | −1.04 | Platelet factor 4 variant 1 |
| ILMN_2165753 | HLA-A | 1.18 × 10−2 | −1.29 | Major histocompatibility complex, class I, A |
| ILMN_1705458 | TRIM58 | 3.08 × 10−2 | −1.31 | Tripartite motif containing 58 |
| ILMN_2132809 | ARHGEF10 | 3.88 × 10−2 | −2.47 | Rho guanine nucleotide exchange factor 10 |
| GO ID | Term | Count | p Value | Fold Enrichment |
|---|---|---|---|---|
| BP | ||||
| GO:0002504 | Antigen processing | 3 | 0.0005 | 90.5 |
| GO:0050870 | Positive regulation of T cell activation | 3 | 0.0013 | 54.9 |
| GO:0019882 | Antigen processing and presentation | 3 | 0.0026 | 38.5 |
| GO:0007229 | Integrin-mediated signaling pathway | 4 | 0.0007 | 21.9 |
| GO:0006954 | Inflammatory response | 5 | 0.0041 | 7.3 |
| CC | ||||
| GO:0042613 | MHC class II protein complex | 3 | 0.0008 | 69.8 |
| GO:0071556 | Reticulum membrane | 3 | 0.0010 | 62.6 |
| GO:0012507 | Golgi transport vesicle membrane | 3 | 0.0038 | 31.8 |
| GO:0030666 | Endocytic vesicle membrane | 3 | 0.0058 | 25.6 |
| GO:0005765 | Lysosomal membrane | 5 | 0.0030 | 8.0 |
| MF | ||||
| GO:0042605 | Peptide antigen binding | 3 | 0.0016 | 49.4 |
| GO:0005178 | Integrin binding | 3 | 0.0269 | 11.3 |
| GO:0051015 | Actin filament binding | 3 | 0.0499 | 8.1 |
| KEGG Pathway ID | Term | Count | p Value | Fold Enrichment |
|---|---|---|---|---|
| hsa05330 | Allograft rejection | 3 | 0.0042 | 29.3 |
| hsa05332 | Graft-versus-host disease | 3 | 0.0051 | 26.5 |
| hsa04940 | Type I diabetes mellitus | 3 | 0.0053 | 25.9 |
| hsa05320 | Autoimmune thyroid disease | 3 | 0.0080 | 21.0 |
| hsa05416 | Viral myocarditis | 3 | 0.0102 | 18.6 |
| hsa04640 | Hematopoietic cell lineage | 4 | 0.0020 | 15.0 |
| hsa04612 | Antigen processing and presentation | 3 | 0.0168 | 14.3 |
| hsa05323 | Rheumatoid arthritis | 3 | 0.0234 | 12.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, H.; Wen, B.; Xu, L.; Huang, Y. Identification of Potential Inflammation-Related Genes and Key Pathways Associated with Complex Regional Pain Syndrome. Biomolecules 2023, 13, 772. https://doi.org/10.3390/biom13050772
Zhu H, Wen B, Xu L, Huang Y. Identification of Potential Inflammation-Related Genes and Key Pathways Associated with Complex Regional Pain Syndrome. Biomolecules. 2023; 13(5):772. https://doi.org/10.3390/biom13050772
Chicago/Turabian StyleZhu, He, Bei Wen, Li Xu, and Yuguang Huang. 2023. "Identification of Potential Inflammation-Related Genes and Key Pathways Associated with Complex Regional Pain Syndrome" Biomolecules 13, no. 5: 772. https://doi.org/10.3390/biom13050772
APA StyleZhu, H., Wen, B., Xu, L., & Huang, Y. (2023). Identification of Potential Inflammation-Related Genes and Key Pathways Associated with Complex Regional Pain Syndrome. Biomolecules, 13(5), 772. https://doi.org/10.3390/biom13050772






