Histological Analysis of a Mouse Model of the 22q11.2 Microdeletion Syndrome
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Plasmids
2.3. In Utero Electroporation
2.4. Production of Adeno-Associated Virus
2.5. Intravenous Administration of Adeno-Associated Virus
2.6. Immunofluorescence
2.7. Golgi–Cox Staining and Spine Analysis
2.8. Dendritic Arbor and Spine Density Analyses of GFP-Labeled Neurons
2.9. Statistical Analysis
3. Results
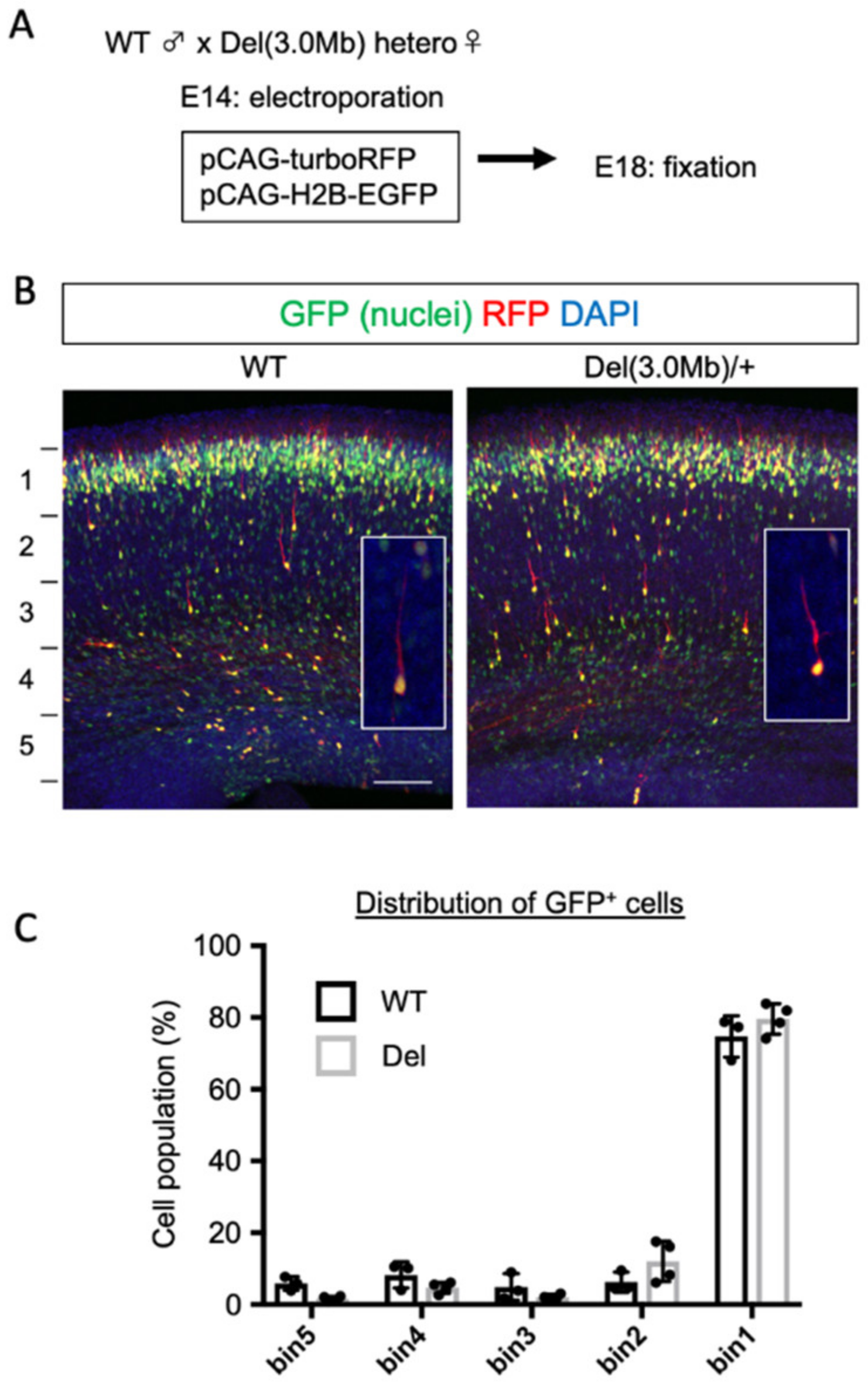
3.1. Cortical Excitatory Neurons Migrate Normally in Embryonic Del(3.0Mb) Heterozygous Mice
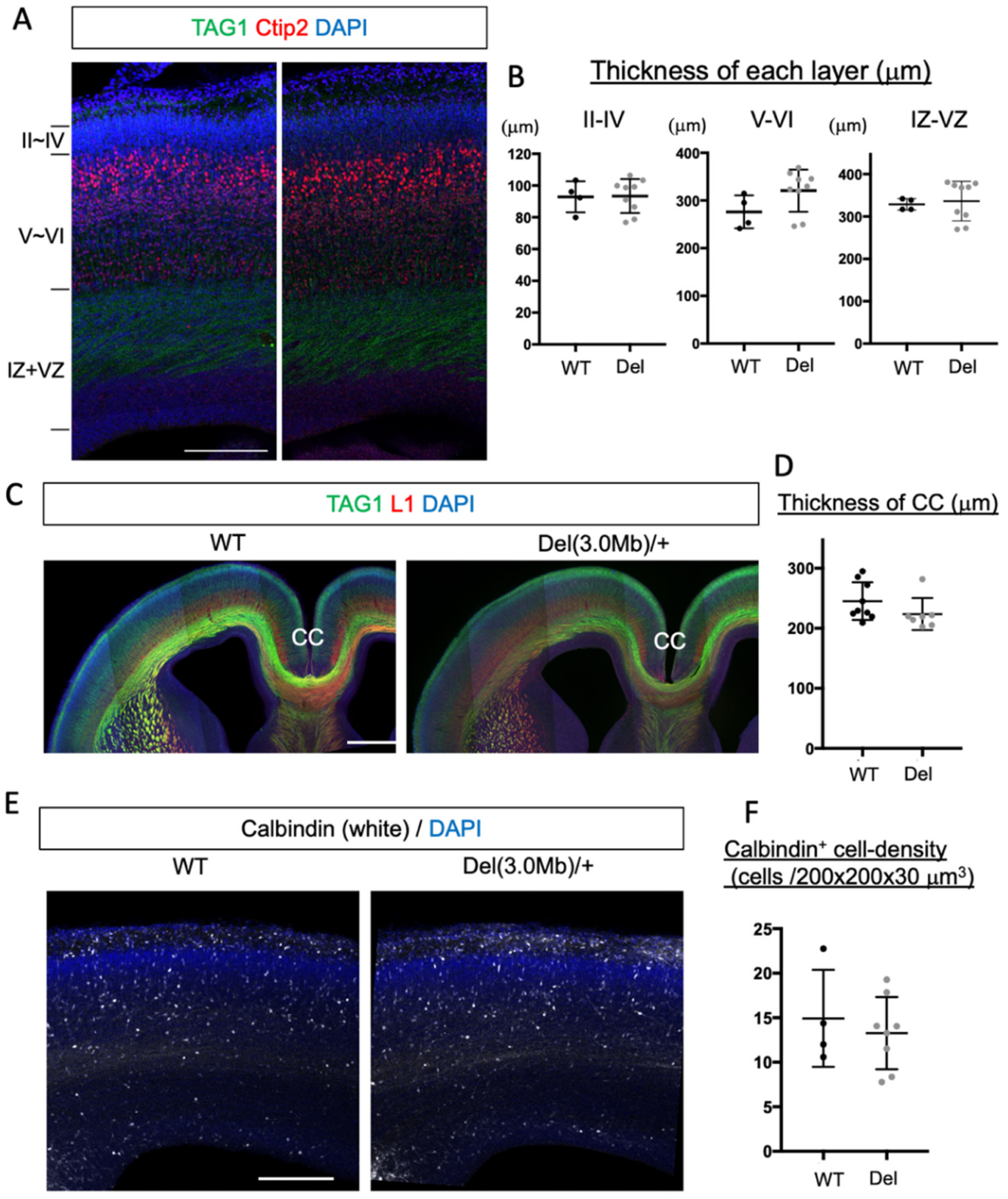
3.2. Overall Histological Organization of Embryonic Del(3.0Mb) Heterozygous Mice Is Normal
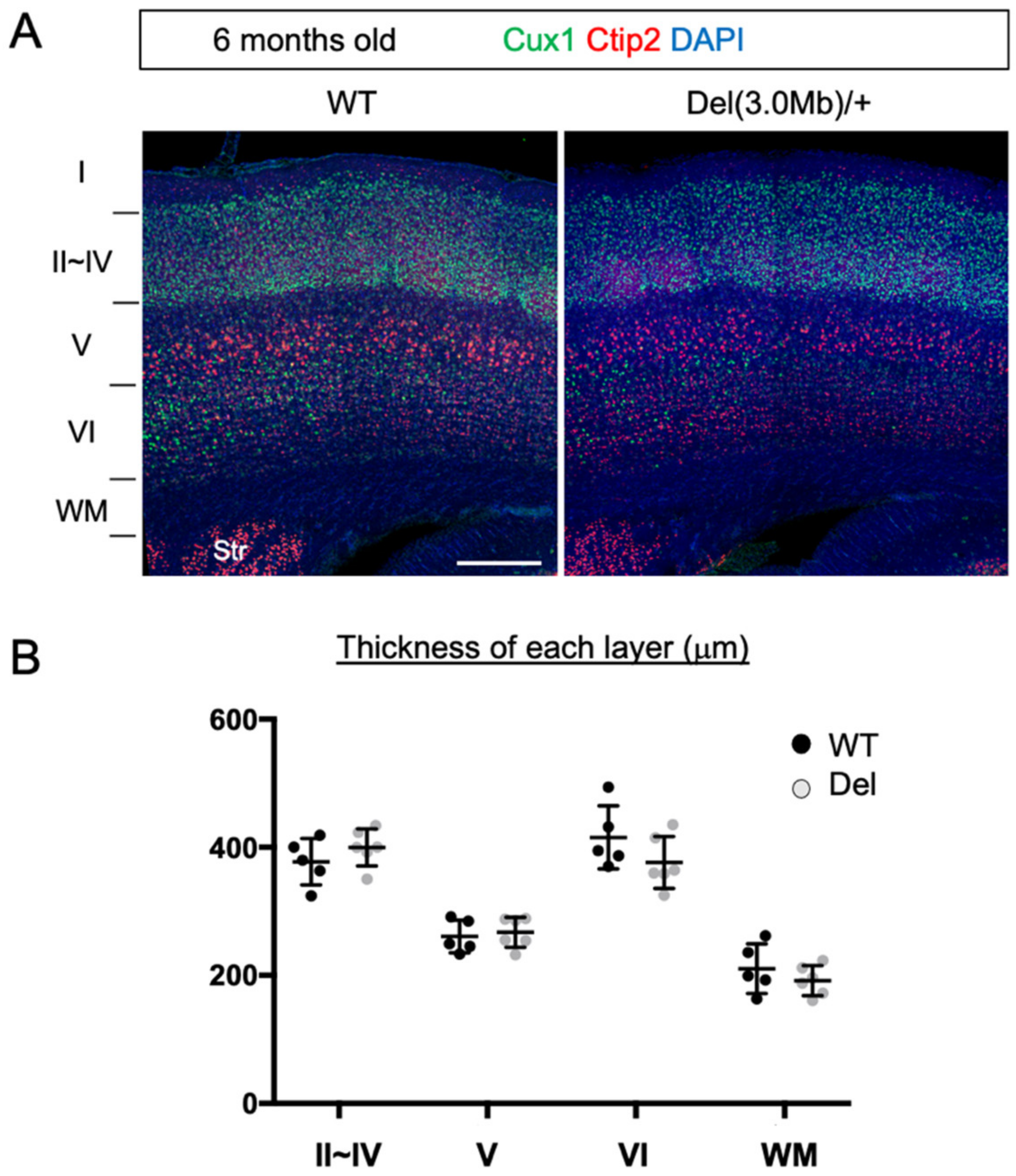
3.3. Gross Layer Structure of Adult Del(3.0Mb)/+ Is Preserved
3.4. Dendritic Arbors of Subsets of Neurons in Adult Del(3.0Mb)/+ Are Slightly but Significantly Reduced
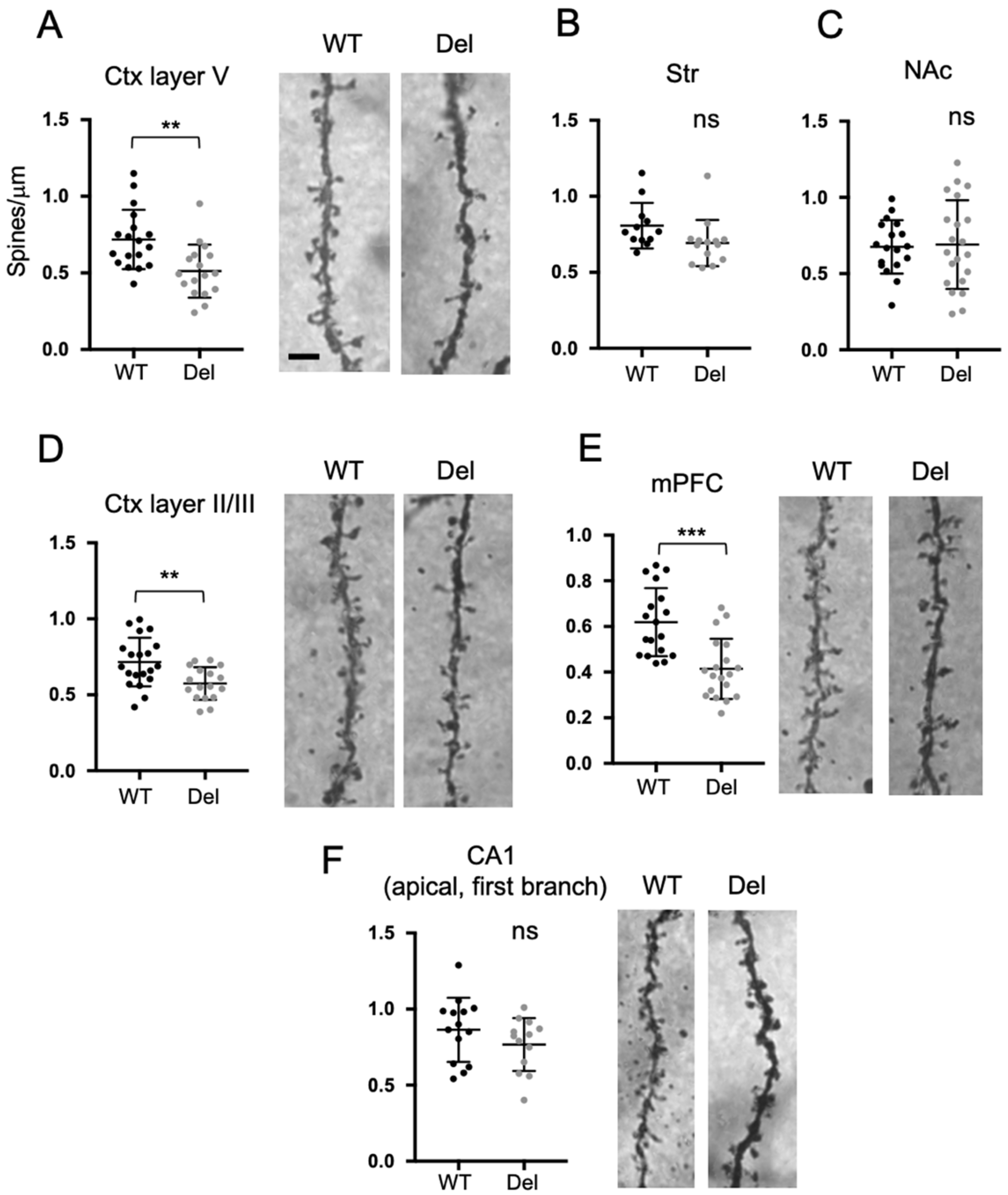
3.5. Dendritic Spine Densities of Subsets of Neurons in Del(3.0Mb)/+ Are Reduced
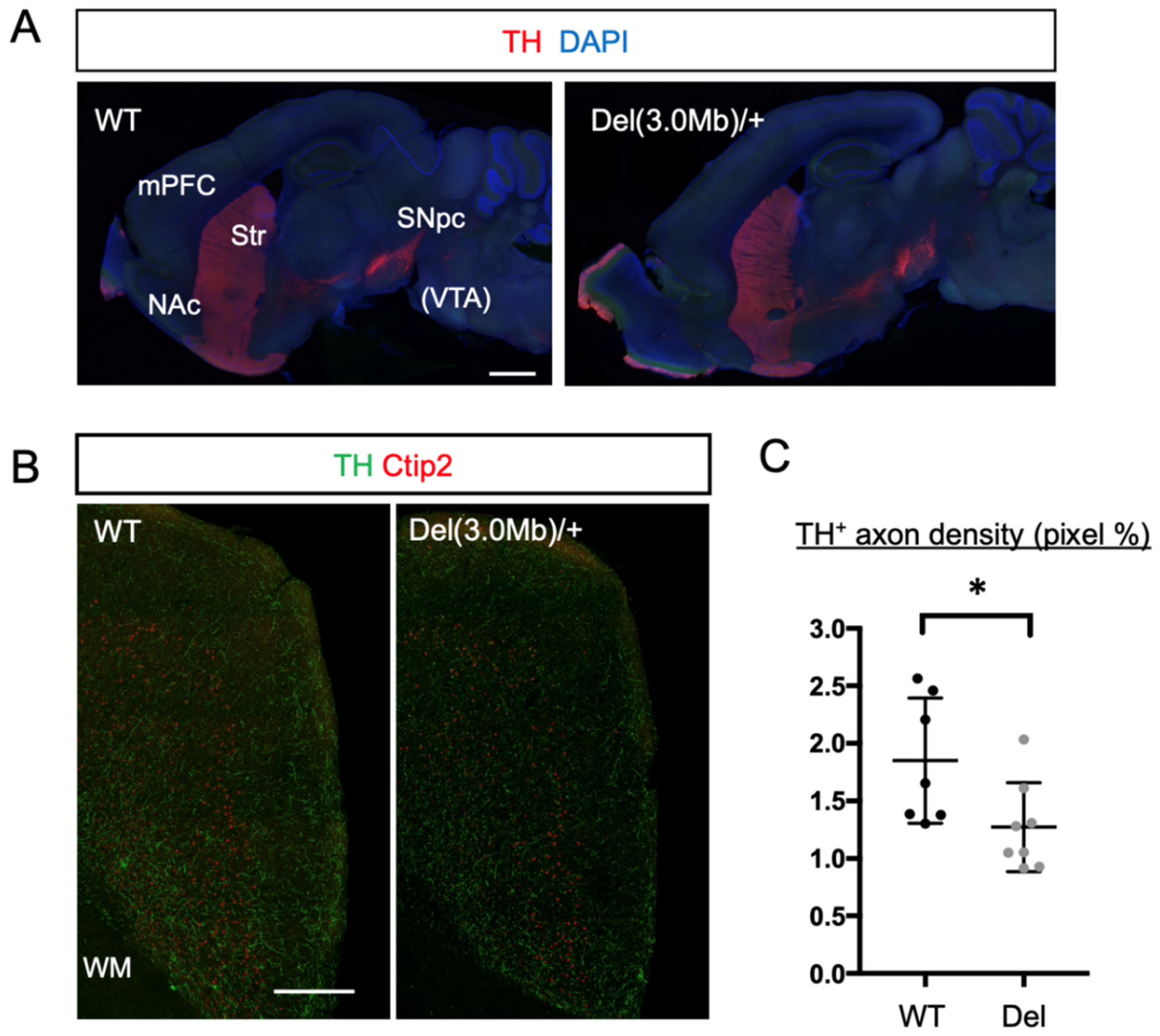
3.6. Axon Innervation of Dopaminergic Neurons into the mPFC in Del(3.0Mb)/+ Are Reduced
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Óskarsdóttir, S.; Holmberg, E.; Fasth, A.; Strömland, K. Facial features in children with the 22q11 deletion syndrome. Acta Paediatr. 2008, 97, 1113–1117. [Google Scholar] [CrossRef] [PubMed]
- Devriendt, K.; Fryns, J.P.; Mortier, G.; van Thienen, M.N.; Keymolen, K. The annual incidence of DiGeorge/velocardiofacial syndrome. J. Med. Genet. 1998, 35, 789–790. [Google Scholar] [CrossRef]
- Guo, T.; Diacou, A.; Nomaru, H.; McDonald-McGinn, D.M.; Hestand, M.; Demaerel, W.; Zhang, L.; Zhao, Y.; Ujueta, F.; Shan, J.; et al. Deletion size analysis of 1680 22q11.2DS subjects identifies a new recombination hotspot on chromosome 22q11.2. Hum. Mol. Genet. 2018, 27, 1150–1163. [Google Scholar] [CrossRef]
- Michaelovsky, E.; Frisch, A.; Carmel, M.; Patya, M.; Zarchi, O.; Green, T.; Basel-Vanagaite, L.; Weizman, A.; Gothelf, D. Genotype-phenotype correlation in 22q11.2 deletion syndrome. BMC Med. Genet. 2012, 13, 122. [Google Scholar] [CrossRef] [PubMed]
- McDonald-McGinn, D.M.; Sullivan, K.E.; Marino, B.; Philip, N.; Swillen, A.; Vorstman, J.A.S.; Zackai, E.H.; Emanuel, B.S.; Vermeesch, J.R.; Morrow, B.E.; et al. 22q11.2 deletion syndrome. Nat. Rev. Dis. Primers 2015, 1, 15071. [Google Scholar] [CrossRef] [PubMed]
- Mok, K.Y.; Sheerin, U.; Simón-Sánchez, J.; Salaka, A.; Chester, L.; Escott-Price, V.; Mantripragada, K.; Doherty, K.M.; Noyce, A.J.; Mencacci, N.E.; et al. Deletions at 22q11.2 in idiopathic Parkinson’s disease: A combined analysis of genome-wide association data. Lancet Neurol. 2016, 15, 585–596. [Google Scholar] [CrossRef] [PubMed]
- Schneider, M.; Debbané, M.; Bassett, A.S.; Chow, E.W.; Fung, W.L.A.; van den Bree, M.B.; Owen, M.; Murphy, K.C.; Niarchou, M.; Kates, W.R.; et al. Psychiatric Disorders from Childhood to Adulthood in 22q11.2 Deletion Syndrome: Results From the International Consortium on Brain and Behavior in 22q11.2 Deletion Syndrome. Am. J. Psychiatry 2014, 171, 627–639. [Google Scholar] [CrossRef]
- Nilsson, S.O.; Fejgin, K.; Gastambide, F.; Vogt, M.A.; Kent, B.A.; Nielsen, V.; Nielsen, J.; Gass, P.; Robbins, T.W.; Saksida, L.M.; et al. Assessing the Cognitive Translational Potential of a Mouse Model of the 22q11.2 Microdeletion Syndrome. Cereb. Cortex 2016, 26, 3991–4003. [Google Scholar] [CrossRef] [PubMed]
- Saito, R.; Koebis, M.; Nagai, T.; Shimizu, K.; Liao, J.; Wulaer, B.; Sugaya, Y.; Nagahama, K.; Uesaka, N.; Kushima, I.; et al. Comprehensive analysis of a novel mouse model of the 22q11.2 deletion syndrome: A model with the most common 3.0-Mb deletion at the human 22q11.2 locus. Transl. Psychiatry 2020, 10, 35. [Google Scholar] [CrossRef]
- Saito, R.; Miyoshi, C.; Koebis, M.; Kushima, I.; Nakao, K.; Mori, D.; Ozaki, N.; Funato, H.; Yanagisawa, M.; Aiba, A. Two novel mouse models mimicking minor deletions in 22q11.2 deletion syndrome revealed the contribution of each deleted region to psychiatric disorders. Mol. Brain 2021, 14, 68. [Google Scholar] [CrossRef] [PubMed]
- Crockett, D.J.; Goudy, S.L.; Chinnadurai, S.; Wootten, C.T. Obstructive Sleep Apnea Syndrome in Children with 22q11.2 Deletion Syndrome after Operative Intervention for Velopharyngeal Insufficiency. Front. Pediatr. 2014, 2. [Google Scholar] [CrossRef] [PubMed]
- Heike, C.L.; Avellino, A.M.; Mirza, S.K.; Kifle, Y.; Perkins, J.; Sze, R.; Egbert, M.; Hing, A.V. Sleep Disturbances in 22q11.2 Deletion Syndrome: A Case with Obstructive and Central Sleep Apnea. Cleft Palate-Craniofacial J. 2007, 44, 340–346. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Ching, C.R.K.; Lin, A.; Forsyth, J.K.; Kushan, L.; Vajdi, A.; Jalbrzikowski, M.; Hansen, L.; Villalon-Reina, J.E.; Qu, X.; et al. Large-scale mapping of cortical alterations in 22q11.2 deletion syndrome: Convergence with idiopathic psychosis and effects of deletion size. Mol. Psychiatry 2020, 25, 1822–1834. [Google Scholar] [CrossRef] [PubMed]
- Campbell, L.E.; Daly, E.; Toal, F.; Stevens, A.; Azuma, R.; Catani, M.; Ng, V.; van Amelsvoort, T.; Chitnis, X.; Cutter, W.; et al. Brain and behaviour in children with 22q11.2 deletion syndrome: A volumetric and voxel-based morphometry MRI study. Brain 2006, 129, 1218–1228. [Google Scholar] [CrossRef]
- Bearden, C.E.; Van Erp, T.G.; Dutton, R.A.; Tran, H.; Zimmermann, L.; Sun, D.; Geaga, J.A.; Simon, T.J.; Glahn, D.C.; Cannon, T.D.; et al. Mapping Cortical Thickness in Children with 22q11.2 Deletions. Cereb. Cortex 2007, 17, 1889–1898. [Google Scholar] [CrossRef] [PubMed]
- Rogdaki, M.; Gudbrandsen, M.; McCutcheon, R.A.; Blackmore, C.E.; Brugger, S.; Ecker, C.; Craig, M.C.; Daly, E.; Murphy, D.G.M.; Howes, O. Magnitude and heterogeneity of brain structural abnormalities in 22q11.2 deletion syndrome: A meta-analysis. Mol. Psychiatry 2020, 25, 1704–1717. [Google Scholar] [CrossRef]
- Gudbrandsen, M.; Bletsch, A.; Mann, C.; Daly, E.; Murphy, C.M.; Stoencheva, V.; Blackmore, C.E.; Rogdaki, M.; Kushan, L.; Bearden, C.E.; et al. Neuroanatomical underpinnings of autism symptomatology in carriers and non-carriers of the 22q11.2 microdeletion. Mol. Autism 2020, 11, 46. [Google Scholar] [CrossRef]
- Ellegood, J.; Markx, S.; Lerch, J.P.; Steadman, P.E.; Genç, C.; Provenzano, F.; Kushner, S.A.; Henkelman, R.M.; Karayiorgou, M.; Gogos, J.A. Neuroanatomical phenotypes in a mouse model of the 22q11.2 microdeletion. Mol. Psychiatry 2014, 19, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Moutin, E.; Nikonenko, I.; Stefanelli, T.; Wirth, A.; Ponimaskin, E.; De Roo, M.; Muller, D. Palmitoylation of cdc42 Promotes Spine Stabilization and Rescues Spine Density Deficit in a Mouse Model of 22q11.2 Deletion Syndrome. Cereb. Cortex 2016, 27, 3618–3629. [Google Scholar] [CrossRef]
- Tabata, H.; Sasaki, M.; Agetsuma, M.; Sano, H.; Hirota, Y.; Miyajima, M.; Hayashi, K.; Honda, T.; Nishikawa, M.; Inaguma, Y.; et al. Erratic and blood vessel-guided migration of astrocyte progenitors in the cerebral cortex. Nat. Commun. 2022, 13, 657. [Google Scholar] [CrossRef]
- Niwa, H.; Yamamura, K.; Miyazaki, J. Efficient Selection for High-Expression Transfectants with a Novel Eukaryotic Vector. Gene 1991, 108, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Tabata, H.; Nakajima, K. Efficient in utero gene transfer system to the developing mouse brain using electroporation: Visualization of neuronal migration in the developing cortex. Neuroscience 2001, 103, 865–872. [Google Scholar] [CrossRef]
- Kasama, E.; Moriya, M.; Kamimura, R.; Matsuki, T.; Seki, K. Formation of False Context Fear Memory Is Regulated by Hypothalamic Corticotropin-Releasing Factor in Mice. Int. J. Mol. Sci. 2022, 23, 6286. [Google Scholar] [CrossRef]
- Challis, R.C.; Ravindra Kumar, S.; Chan, K.Y.; Challis, C.; Beadle, K.; Jang, M.J.; Kim, H.M.; Rajendran, P.S.; Tompkins, J.D.; Shivkumar, K.; et al. Systemic AAV vectors for widespread and targeted gene delivery in rodents. Nat. Protoc. 2019, 14, 379–414. [Google Scholar] [CrossRef] [PubMed]
- Yardeni, T.; Eckhaus, M.; Morris, H.D.; Huizing, M.; Hoogstraten-Miller, S. Retro-orbital injections in mice. Lab Anim. 2011, 40, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Kurobe, N.; Inaguma, Y.; Shinohara, H.; Semba, R.; Inagaki, T.; Kato, K. Developmental and Age-Dependent Changes of 28-kDa Calbindin-D in the Central Nervous Tissue Determined with a Sensitive Immunoassay Method. J. Neurochem. 1992, 58, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Zhong, F.; Liu, L.; Wei, J.-L.; Dai, R.-P. Step by Step Golgi-Cox Staining for Cryosection. Front. Neuroanat. 2019, 13, 244–246. [Google Scholar] [CrossRef] [PubMed]
- Maynard, T.M.; Haskell, G.T.; Peters, A.Z.; Sikich, L.; Lieberman, J.A.; LaMantia, A.-S. A comprehensive analysis of 22q11 gene expression in the developing and adult brain. Proc. Natl. Acad. Sci. USA 2003, 100, 14433–14438. [Google Scholar] [CrossRef]
- Tabata, H.; Nagata, K.-I. Decoding the molecular mechanisms of neuronal migration using in utero electroporation. Med. Mol. Morphol. 2016, 49, 63–75. [Google Scholar] [CrossRef]
- Leid, M.; Ishmael, J.E.; Avram, D.; Shepherd, D.; Fraulob, V.; Dollé, P. CTIP1 and CTIP2 are differentially expressed during mouse embryogenesis. Gene Expr. Patterns 2004, 4, 733–739. [Google Scholar] [CrossRef]
- Namba, T.; Kibe, Y.; Funahashi, Y.; Nakamuta, S.; Takano, T.; Ueno, T.; Shimada, A.; Kozawa, S.; Okamoto, M.; Shimoda, Y.; et al. Pioneering Axons Regulate Neuronal Polarization in the Developing Cerebral Cortex. Neuron 2014, 81, 814–829. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, T.; Kawano, H.; Ohyama, K.; Li, H.P.; Takeda, Y.; Oohira, A.; Kawamura, K. Immunohistochemical Localization of Neurocan and L1 in the Formation of Thalamocortical Pathway of Developing Rats. J. Comp. Neurol. 1997, 382, 141–152. [Google Scholar] [CrossRef]
- Toritsuka, M.; Kimoto, S.; Muraki, K.; Landek-Salgado, M.A.; Yoshida, A.; Yamamoto, N.; Horiuchi, Y.; Hiyama, H.; Tajinda, K.; Keni, N.; et al. Deficits in microRNA-mediated Cxcr4/Cxcl12 signaling in neurodevelopmental deficits in a 22q11 deletion syndrome mouse model. Proc. Natl. Acad. Sci. USA 2013, 110, 17552–17557. [Google Scholar] [CrossRef]
- Andrews, W.; Barber, M.; Hernandez-Miranda, L.R.; Xian, J.; Rakic, S.; Sundaresan, V.; Rabbitts, T.H.; Pannell, R.; Rabbitts, P.; Thompson, H.; et al. The role of Slit-Robo signaling in the generation, migration and morphological differentiation of cortical interneurons. Dev. Biol. 2008, 313, 648–658. [Google Scholar] [CrossRef] [PubMed]
- Nieto, M.; Monuki, E.S.; Tang, H.; Imitola, J.; Haubst, N.; Khoury, S.J.; Cunningham, J.; Gotz, M.; Walsh, C.A. Expression of Cux-1 and Cux-2 in the subventricular zone and upper layers II-IV of the cerebral cortex. J. Comp. Neurol. 2004, 479, 168–180. [Google Scholar] [CrossRef] [PubMed]
- Zinkstok, J.R.; Boot, E.; Bassett, A.S.; Hiroi, N.; Butcher, N.J.; Vingerhoets, C.; Vorstman, J.A.S.; van Amelsvoort, T.A.M.J. Neurobiological perspective of 22q11.2 deletion syndrome. Lancet Psychiatry 2019, 6, 951–960. [Google Scholar] [CrossRef] [PubMed]
- Fénelon, K.; Xu, B.; Lai, C.S.; Mukai, J.; Markx, S.; Stark, K.L.; Hsu, P.-K.; Gan, W.-B.; Fischbach, G.D.; MacDermott, A.B.; et al. The Pattern of Cortical Dysfunction in a Mouse Model of a Schizophrenia-Related Microdeletion. J. Neurosci. 2013, 33, 14825–14839. [Google Scholar] [CrossRef]
- Weinstein, J.J.; Chohan, M.O.; Slifstein, M.; Kegeles, L.S.; Moore, H.; Abi-Dargham, A. Pathway-Specific Dopamine Abnormalities in Schizophrenia. Biol. Psychiatry 2017, 81, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Ching, C.R.K.; Gutman, B.A.; Sun, D.; Reina, J.V.; Ragothaman, A.; Isaev, D.; Zavaliangos-Petropulu, A.; Lin, A.; Jonas, R.K.; Kushan, L.; et al. Mapping Subcortical Brain Alterations in 22q11.2 Deletion Syndrome: Effects of Deletion Size and Convergence with Idiopathic Neuropsychiatric Illness. Am. J. Psychiatry 2020, 177, 589–600. [Google Scholar] [CrossRef]
- Abduljawad, K.A.J.; Langley, R.W.; Bradshaw, C.M.; Szabadi, E. Effects of bromocriptine and haloperidol on prepulse inhibition of the acoustic startle response in man. J. Psychopharmacol. 1998, 12, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Braff, D.L.; Geyer, M.A.; Swerdlow, N.R. Human studies of prepulse inhibition of startle: Normal subjects, patient groups, and pharmacological studies. Psychopharmacology 2001, 156, 234–258. [Google Scholar] [CrossRef] [PubMed]
- Geyer, M.A.; Krebs-Thomson, K.; Braff, D.L.; Swerdlow, N.R. Pharmacological studies of prepulse inhibition models of sensorimotor gating deficits in schizophrenia: A decade in review. Psychopharmacology 2001, 156, 117–154. [Google Scholar] [CrossRef] [PubMed]
- De Lau, L.M.L.; Breteler, M.M.B. Epidemiology of Parkinson’s disease. Lancet Neurol. 2006, 5, 525–535. [Google Scholar] [CrossRef] [PubMed]
- Arioka, Y.; Shishido, E.; Kushima, I.; Suzuki, T.; Saito, R.; Aiba, A.; Mori, D.; Ozaki, N. Chromosome 22q11.2 deletion causes PERK-dependent vulnerability in dopaminergic neurons. Ebiomedicine 2021, 63, 103138. [Google Scholar] [CrossRef] [PubMed]
- Männistö, P.T.; Kaakkola, S. Catechol-O-methyltransferase (COMT): Biochemistry, molecular biology, pharmacology, and clinical efficacy of the new selective COMT inhibitors. Pharmacol. Rev. 1999, 51, 593–628. [Google Scholar]
- Chen, J.; Lipska, B.K.; Halim, N.; Ma, Q.D.; Matsumoto, M.; Melhem, S.; Kolachana, B.S.; Hyde, T.M.; Herman, M.M.; Apud, J.; et al. Functional Analysis of Genetic Variation in Catechol-O-Methyltransferase (COMT): Effects on mRNA, Protein, and Enzyme Activity in Postmortem Human Brain. Am. J. Hum. Genet. 2004, 75, 807–821. [Google Scholar] [CrossRef]
- Kikinis, Z.; Cho, K.I.K.; Coman, I.L.; Radoeva, P.D.; Bouix, S.; Tang, Y.; Eckbo, R.; Makris, N.; Kwon, J.S.; Kubicki, M.; et al. Abnormalities in brain white matter in adolescents with 22q11.2 deletion syndrome and psychotic symptoms. Brain Imaging Behav. 2017, 11, 1353–1364. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tabata, H.; Mori, D.; Matsuki, T.; Yoshizaki, K.; Asai, M.; Nakayama, A.; Ozaki, N.; Nagata, K.-i. Histological Analysis of a Mouse Model of the 22q11.2 Microdeletion Syndrome. Biomolecules 2023, 13, 763. https://doi.org/10.3390/biom13050763
Tabata H, Mori D, Matsuki T, Yoshizaki K, Asai M, Nakayama A, Ozaki N, Nagata K-i. Histological Analysis of a Mouse Model of the 22q11.2 Microdeletion Syndrome. Biomolecules. 2023; 13(5):763. https://doi.org/10.3390/biom13050763
Chicago/Turabian StyleTabata, Hidenori, Daisuke Mori, Tohru Matsuki, Kaichi Yoshizaki, Masato Asai, Atsuo Nakayama, Norio Ozaki, and Koh-ichi Nagata. 2023. "Histological Analysis of a Mouse Model of the 22q11.2 Microdeletion Syndrome" Biomolecules 13, no. 5: 763. https://doi.org/10.3390/biom13050763
APA StyleTabata, H., Mori, D., Matsuki, T., Yoshizaki, K., Asai, M., Nakayama, A., Ozaki, N., & Nagata, K.-i. (2023). Histological Analysis of a Mouse Model of the 22q11.2 Microdeletion Syndrome. Biomolecules, 13(5), 763. https://doi.org/10.3390/biom13050763







