Transcriptional Regulation Technology for Gene Perturbation in Fission Yeast
Abstract
1. Introduction
2. Transcriptional Regulation Technologies for S. pombe
2.1. Promoter Replacement
2.2. 3′UTR Disruption
2.3. RNA Interference
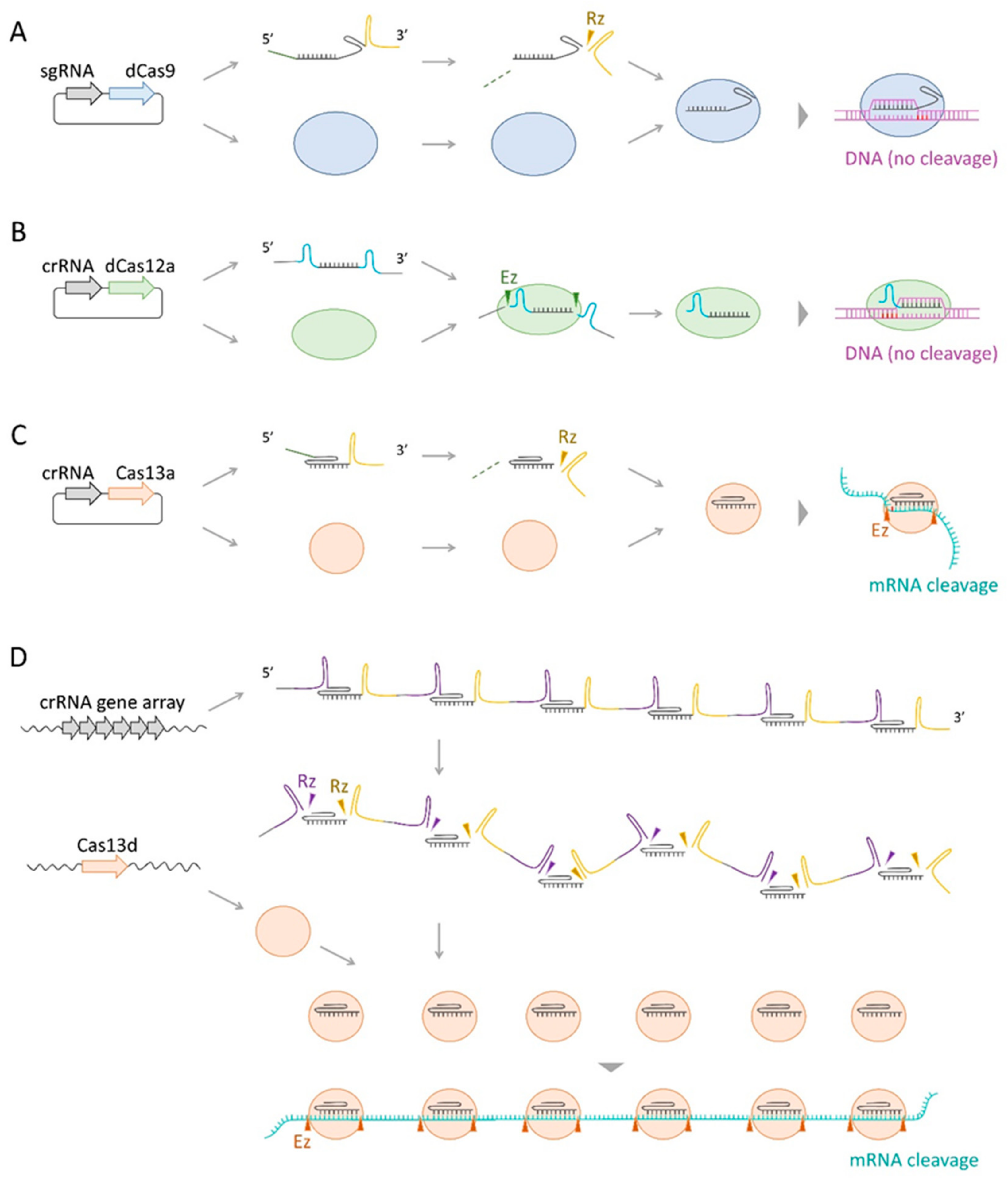
2.4. CRISPR–Cas Based Gene Knockdown Technology
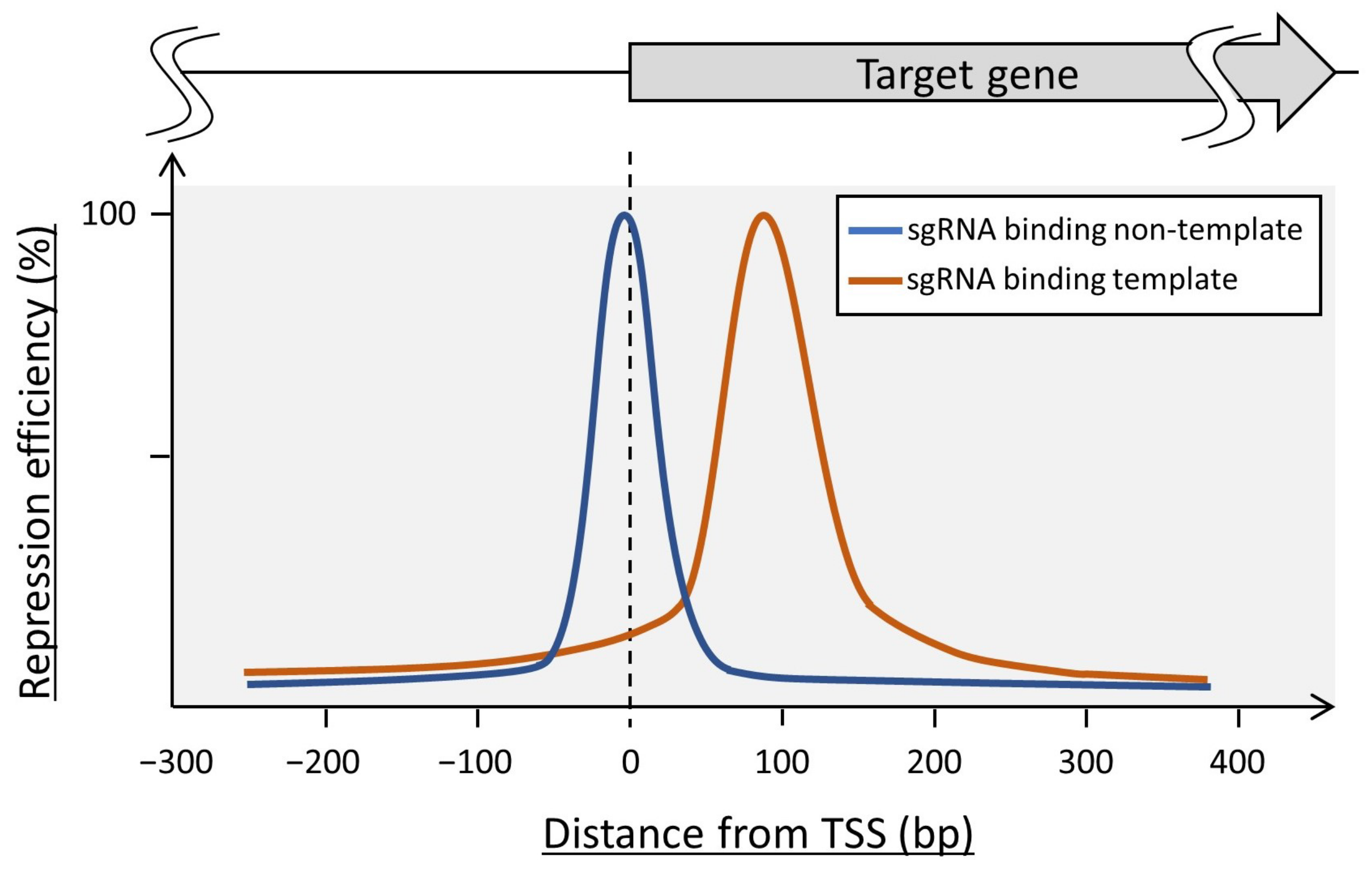
2.4.1. dCas9-Mediated CRISPR Interference
2.4.2. dCas12a-Mediated CRISPR Interference
2.4.3. Cas13-Mediated Gene Knockdown
3. Comparison of Gene Perturbation Technologies in S. pombe
4. Other CRISPR–Cas-Based Technologies on Transcriptional Manipulation
5. Applications of CRISPRi Resources
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 3'UTR | 3’-untranslated region |
| AID | auxin-inducible degron |
| Cas | CRISPR associated |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeat |
| CRISPRi | CRISPR interference |
| crRNA | CRISPR RNA |
| DAmP | Decreased Abundance by mRNA Perturbation |
| FKBP12 | FK506 binding protein |
| FRB domain | FKBP12-rapamycin-binding domain (of human mTOR) |
| KRAB | Krüppel associated box |
| Mxi1 | MAX-interacting protein 1 |
| PAM | protospacer adjacent motif |
| RNAi | RNA interference |
| sgRNA | small guide RNA |
| snRNA | small nuclear RNA |
| tracrRNA | transactivating crRNA |
References
- Hoffman, C.S.; Wood, V.; Fantes, P.A. An Ancient Yeast for Young Geneticists: A Primer on the Schizosaccharomyces pombe Model System. Genetics 2015, 201, 403–423. [Google Scholar] [CrossRef] [PubMed]
- Fantes, P.A.; Hoffman, C.S. A Brief History of Schizosaccharomyces pombe Research: A Perspective over the Past 70 Years. Genetics 2016, 203, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Wood, V.; Gwilliam, R.; Rajandream, M.A.; Lyne, M.; Lyne, R.; Stewart, A.; Sgouros, J.; Peat, N.; Hayles, J.; Baker, S.; et al. The genome sequence of Schizosaccharomyces pombe. Nature 2002, 415, 871–880. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.U.; Hayles, J.; Kim, D.; Wood, V.; Park, H.O.; Won, M.; Yoo, H.S.; Duhig, T.; Nam, M.; Palmer, G.; et al. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 2010, 28, 617–623. [Google Scholar] [CrossRef]
- Kanke, M.; Nishimura, K.; Kanemaki, M.; Kakimoto, T.; Takahashi, T.S.; Nakagawa, T.; Masukata, H. Auxin-inducible protein depletion system in fission yeast. BMC Cell Biol. 2011, 12, 8. [Google Scholar] [CrossRef]
- Zhang, X.R.; Zhao, L.; Suo, F.; Gao, Y.; Wu, Q.; Qi, X.; Du, L.L. An improved auxin-inducible degron system for fission yeast. G3 2022, 12, jkab393. [Google Scholar] [CrossRef]
- Haruki, H.; Nishikawa, J.; Laemmli, U.K. The anchor-away technique: Rapid, conditional establishment of yeast mutant phenotypes. Mol. Cell 2008, 31, 925–932. [Google Scholar] [CrossRef]
- Ding, L.; Laor, D.; Weisman, R.; Forsburg, S.L. Rapid regulation of nuclear proteins by rapamycin-induced translocation in fission yeast. Yeast 2014, 31, 253–264. [Google Scholar] [CrossRef]
- Kim, H.; Kim, M.; Im, S.K.; Fang, S. Mouse Cre-LoxP system: General principles to determine tissue-specific roles of target genes. Lab. Anim. Res. 2018, 34, 147–159. [Google Scholar] [CrossRef]
- Iwaki, T.; Takegawa, K. A set of loxP marker cassettes for Cre-mediated multiple gene disruption in Schizosaccharomyces pombe. Biosci. Biotechnol. Biochem. 2004, 68, 545–550. [Google Scholar] [CrossRef]
- Hirashima, K.; Iwaki, T.; Takegawa, K.; Giga-Hama, Y.; Tohda, H. A simple and effective chromosome modification method for large-scale deletion of genome sequences and identification of essential genes in fission yeast. Nucleic Acids Res. 2006, 34, e11. [Google Scholar] [CrossRef]
- Ishii, K.; Ogiyama, Y.; Chikashige, Y.; Soejima, S.; Masuda, F.; Kakuma, T.; Hiraoka, Y.; Takahashi, K. Heterochromatin integrity affects chromosome reorganization after centromere dysfunction. Science 2008, 321, 1088–1091. [Google Scholar] [CrossRef]
- Bahler, J.; Wu, J.Q.; Longtine, M.S.; Shah, N.G.; McKenzie, A., 3rd; Steever, A.B.; Wach, A.; Philippsen, P.; Pringle, J.R. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 1998, 14, 943–951. [Google Scholar] [CrossRef]
- Schuldiner, M.; Collins, S.R.; Thompson, N.J.; Denic, V.; Bhamidipati, A.; Punna, T.; Ihmels, J.; Andrews, B.; Boone, C.; Greenblatt, J.F.; et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell 2005, 123, 507–519. [Google Scholar] [CrossRef]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef]
- Housden, B.E.; Muhar, M.; Gemberling, M.; Gersbach, C.A.; Stainier, D.Y.; Seydoux, G.; Mohr, S.E.; Zuber, J.; Perrimon, N. Loss-of-function genetic tools for animal models: Cross-species and cross-platform differences. Nat. Rev. Genet. 2017, 18, 24–40. [Google Scholar] [CrossRef]
- Daga, R.R.; Jimenez, J. Translational control of the cdc25 cell cycle phosphatase: A molecular mechanism coupling mitosis to cell growth. J. Cell Sci. 1999, 112 Pt 18, 3137–3146. [Google Scholar] [CrossRef]
- Hou, M.C.; Wiley, D.J.; Verde, F.; McCollum, D. Mob2p interacts with the protein kinase Orb6p to promote coordination of cell polarity with cell cycle progression. J. Cell Sci. 2003, 116, 125–135. [Google Scholar] [CrossRef]
- Shams-Eldin, H.; Azzouz, N.; Eckert, V.; Blaschke, T.; Kedees, M.H.; Hubel, A.; Schwarz, R.T. The Schizosaccharomyces pombe GPI8 gene complements a Saccharomyces cerevisiae GPI8 anchoring mutant. Yeast 2001, 18, 33–39. [Google Scholar] [CrossRef]
- Forsburg, S.L. Comparison of Schizosaccharomyces pombe expression systems. Nucleic Acids Res. 1993, 21, 2955–2956. [Google Scholar] [CrossRef] [PubMed]
- Roguev, A.; Bandyopadhyay, S.; Zofall, M.; Zhang, K.; Fischer, T.; Collins, S.R.; Qu, H.; Shales, M.; Park, H.O.; Hayles, J.; et al. Conservation and rewiring of functional modules revealed by an epistasis map in fission yeast. Science 2008, 322, 405–410. [Google Scholar] [CrossRef]
- Muhlrad, D.; Parker, R. Aberrant mRNAs with extended 3’ UTRs are substrates for rapid degradation by mRNA surveillance. RNA 1999, 5, 1299–1307. [Google Scholar] [CrossRef] [PubMed]
- Breslow, D.K.; Cameron, D.M.; Collins, S.R.; Schuldiner, M.; Stewart-Ornstein, J.; Newman, H.W.; Braun, S.; Madhani, H.D.; Krogan, N.J.; Weissman, J.S. A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat. Methods 2008, 5, 711–718. [Google Scholar] [CrossRef] [PubMed]
- Grewal, S.I.; Jia, S. Heterochromatin revisited. Nat. Rev. Genet. 2007, 8, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Volpe, T.A.; Kidner, C.; Hall, I.M.; Teng, G.; Grewal, S.I.; Martienssen, R.A. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 2002, 297, 1833–1837. [Google Scholar] [CrossRef]
- Hall, I.M.; Shankaranarayana, G.D.; Noma, K.; Ayoub, N.; Cohen, A.; Grewal, S.I. Establishment and maintenance of a heterochromatin domain. Science 2002, 297, 2232–2237. [Google Scholar] [CrossRef]
- Raponi, M.; Arndt, G.M. Double-stranded RNA-mediated gene silencing in fission yeast. Nucleic Acids Res. 2003, 31, 4481–4489. [Google Scholar] [CrossRef]
- Sigova, A.; Rhind, N.; Zamore, P.D. A single Argonaute protein mediates both transcriptional and posttranscriptional silencing in Schizosaccharomyces pombe. Genes Dev. 2004, 18, 2359–2367. [Google Scholar] [CrossRef]
- Iida, T.; Nakayama, J.; Moazed, D. siRNA-mediated heterochromatin establishment requires HP1 and is associated with antisense transcription. Mol. Cell 2008, 31, 178–189. [Google Scholar] [CrossRef]
- Simmer, F.; Buscaino, A.; Kos-Braun, I.C.; Kagansky, A.; Boukaba, A.; Urano, T.; Kerr, A.R.; Allshire, R.C. Hairpin RNA induces secondary small interfering RNA synthesis and silencing in trans in fission yeast. EMBO Rep. 2010, 11, 112–118. [Google Scholar] [CrossRef]
- Makarova, K.S.; Wolf, Y.I.; Iranzo, J.; Shmakov, S.A.; Alkhnbashi, O.S.; Brouns, S.J.J.; Charpentier, E.; Cheng, D.; Haft, D.H.; Horvath, P.; et al. Evolutionary classification of CRISPR-Cas systems: A burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020, 18, 67–83. [Google Scholar] [CrossRef]
- Zhao, Y.; Boeke, J.D. CRISPR-Cas12a system in fission yeast for multiplex genomic editing and CRISPR interference. Nucleic Acids Res. 2020, 48, 5788–5798. [Google Scholar] [CrossRef]
- Ishikawa, K.; Soejima, S.; Masuda, F.; Saitoh, S. Implementation of dCas9-mediated CRISPRi in the fission yeast Schizosaccharomyces pombe. G3 2021, 11, jkab051. [Google Scholar] [CrossRef]
- Chen, Z.; Zheng, S.; Fu, C. Shotgun knockdown of RNA by CRISPR-Cas13d in fission yeast. J. Cell Sci. 2023, 136, jcs260769. [Google Scholar] [CrossRef]
- Jing, X.; Xie, B.; Chen, L.; Zhang, N.; Jiang, Y.; Qin, H.; Wang, H.; Hao, P.; Yang, S.; Li, X. Implementation of the CRISPR-Cas13a system in fission yeast and its repurposing for precise RNA editing. Nucleic Acids Res. 2018, 46, e90. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Sternberg, S.H.; Redding, S.; Jinek, M.; Greene, E.C.; Doudna, J.A. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 2014, 507, 62–67. [Google Scholar] [CrossRef]
- Hayashi, A.; Tanaka, K. Short-Homology-Mediated CRISPR/Cas9-Based Method for Genome Editing in Fission Yeast. G3 2019, 9, 1153–1163. [Google Scholar] [CrossRef]
- Jacobs, J.Z.; Ciccaglione, K.M.; Tournier, V.; Zaratiegui, M. Implementation of the CRISPR-Cas9 system in fission yeast. Nat. Commun. 2014, 5, 5344. [Google Scholar] [CrossRef]
- Rodriguez-Lopez, M.; Cotobal, C.; Fernandez-Sanchez, O.; Borbaran Bravo, N.; Oktriani, R.; Abendroth, H.; Uka, D.; Hoti, M.; Wang, J.; Zaratiegui, M.; et al. A CRISPR/Cas9-based method and primer design tool for seamless genome editing in fission yeast. Wellcome Open Res. 2016, 1, 19. [Google Scholar] [CrossRef]
- Zhang, X.R.; He, J.B.; Wang, Y.Z.; Du, L.L. A Cloning-Free Method for CRISPR/Cas9-Mediated Genome Editing in Fission Yeast. G3 2018, 8, 2067–2077. [Google Scholar] [CrossRef] [PubMed]
- Gasiunas, G.; Barrangou, R.; Horvath, P.; Siksnys, V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. USA 2012, 109, E2579–E2586. [Google Scholar] [CrossRef] [PubMed]
- Bikard, D.; Jiang, W.; Samai, P.; Hochschild, A.; Zhang, F.; Marraffini, L.A. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013, 41, 7429–7437. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, L.A.; Horlbeck, M.A.; Adamson, B.; Villalta, J.E.; Chen, Y.; Whitehead, E.H.; Guimaraes, C.; Panning, B.; Ploegh, H.L.; Bassik, M.C.; et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell 2014, 159, 647–661. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, K.; Soejima, S.; Saitoh, S. Genetic knockdown of obscure, conserved, essential genes using CRISPR interference methods in the fission yeast S. pombe. J. Cell Sci. 2023, in press. [Google Scholar]
- Horlbeck, M.A.; Witkowsky, L.B.; Guglielmi, B.; Replogle, J.M.; Gilbert, L.A.; Villalta, J.E.; Torigoe, S.E.; Tjian, R.; Weissman, J.S. Nucleosomes impede Cas9 access to DNA in vivo and in vitro. Elife 2016, 5, e12677. [Google Scholar] [CrossRef]
- Smith, J.D.; Suresh, S.; Schlecht, U.; Wu, M.; Wagih, O.; Peltz, G.; Davis, R.W.; Steinmetz, L.M.; Parts, L.; St Onge, R.P. Quantitative CRISPR interference screens in yeast identify chemical-genetic interactions and new rules for guide RNA design. Genome Biol. 2016, 17, 45. [Google Scholar] [CrossRef]
- Givens, R.M.; Lai, W.K.; Rizzo, J.M.; Bard, J.E.; Mieczkowski, P.A.; Leatherwood, J.; Huberman, J.A.; Buck, M.J. Chromatin architectures at fission yeast transcriptional promoters and replication origins. Nucleic Acids Res. 2012, 40, 7176–7189. [Google Scholar] [CrossRef]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; van der Oost, J.; Regev, A.; et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef]
- Fonfara, I.; Richter, H.; Bratovic, M.; Le Rhun, A.; Charpentier, E. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 2016, 532, 517–521. [Google Scholar] [CrossRef]
- Zetsche, B.; Heidenreich, M.; Mohanraju, P.; Fedorova, I.; Kneppers, J.; DeGennaro, E.M.; Winblad, N.; Choudhury, S.R.; Abudayyeh, O.O.; Gootenberg, J.S.; et al. Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat. Biotechnol. 2017, 35, 31–34. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, J.; Cheng, Q.; Zheng, X.; Zhao, G.; Wang, J. Multiplex gene regulation by CRISPR-ddCpf1. Cell Discov. 2017, 3, 17018. [Google Scholar] [CrossRef]
- Abudayyeh, O.O.; Gootenberg, J.S.; Konermann, S.; Joung, J.; Slaymaker, I.M.; Cox, D.B.; Shmakov, S.; Makarova, K.S.; Semenova, E.; Minakhin, L.; et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016, 353, aaf5573. [Google Scholar] [CrossRef]
- Abudayyeh, O.O.; Gootenberg, J.S.; Essletzbichler, P.; Han, S.; Joung, J.; Belanto, J.J.; Verdine, V.; Cox, D.B.T.; Kellner, M.J.; Regev, A.; et al. RNA targeting with CRISPR-Cas13. Nature 2017, 550, 280–284. [Google Scholar] [CrossRef]
- Konermann, S.; Lotfy, P.; Brideau, N.J.; Oki, J.; Shokhirev, M.N.; Hsu, P.D. Transcriptome Engineering with RNA-Targeting Type VI-D CRISPR Effectors. Cell 2018, 173, 665–676.e614. [Google Scholar] [CrossRef]
- Yan, W.X.; Chong, S.; Zhang, H.; Makarova, K.S.; Koonin, E.V.; Cheng, D.R.; Scott, D.A. Cas13d Is a Compact RNA-Targeting Type VI CRISPR Effector Positively Modulated by a WYL-Domain-Containing Accessory Protein. Mol. Cell 2018, 70, 327–339.e325. [Google Scholar] [CrossRef]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef]
- Li, Y.; Xu, J.; Guo, X.; Li, Z.; Cao, L.; Liu, S.; Guo, Y.; Wang, G.; Luo, Y.; Zhang, Z.; et al. The collateral activity of RfxCas13d can induce lethality in a RfxCas13d knock-in mouse model. Genome Biol. 2023, 24, 20. [Google Scholar] [CrossRef]
- Guo, L.Y.; Bian, J.; Davis, A.E.; Liu, P.; Kempton, H.R.; Zhang, X.; Chemparathy, A.; Gu, B.; Lin, X.; Rane, D.A.; et al. Multiplexed genome regulation in vivo with hyper-efficient Cas12a. Nat. Cell Biol. 2022, 24, 590–600. [Google Scholar] [CrossRef]
- Evers, B.; Jastrzebski, K.; Heijmans, J.P.; Grernrum, W.; Beijersbergen, R.L.; Bernards, R. CRISPR knockout screening outperforms shRNA and CRISPRi in identifying essential genes. Nat. Biotechnol. 2016, 34, 631–633. [Google Scholar] [CrossRef] [PubMed]
- Smith, I.; Greenside, P.G.; Natoli, T.; Lahr, D.L.; Wadden, D.; Tirosh, I.; Narayan, R.; Root, D.E.; Golub, T.R.; Subramanian, A.; et al. Evaluation of RNAi and CRISPR technologies by large-scale gene expression profiling in the Connectivity Map. PLoS Biol. 2017, 15, e2003213. [Google Scholar] [CrossRef] [PubMed]
- Stojic, L.; Lun, A.T.L.; Mangei, J.; Mascalchi, P.; Quarantotti, V.; Barr, A.R.; Bakal, C.; Marioni, J.C.; Gergely, F.; Odom, D.T. Specificity of RNAi, LNA and CRISPRi as loss-of-function methods in transcriptional analysis. Nucleic Acids Res. 2018, 46, 5950–5966. [Google Scholar] [CrossRef] [PubMed]
- Larson, M.H.; Gilbert, L.A.; Wang, X.; Lim, W.A.; Weissman, J.S.; Qi, L.S. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc. 2013, 8, 2180–2196. [Google Scholar] [CrossRef]
- Gilbert, L.A.; Larson, M.H.; Morsut, L.; Liu, Z.; Brar, G.A.; Torres, S.E.; Stern-Ginossar, N.; Brandman, O.; Whitehead, E.H.; Doudna, J.A.; et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 2013, 154, 442–451. [Google Scholar] [CrossRef]
- Hu, J.H.; Miller, S.M.; Geurts, M.H.; Tang, W.; Chen, L.; Sun, N.; Zeina, C.M.; Gao, X.; Rees, H.A.; Lin, Z.; et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 2018, 556, 57–63. [Google Scholar] [CrossRef]
- Nishimasu, H.; Shi, X.; Ishiguro, S.; Gao, L.; Hirano, S.; Okazaki, S.; Noda, T.; Abudayyeh, O.O.; Gootenberg, J.S.; Mori, H.; et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science 2018, 361, 1259–1262. [Google Scholar] [CrossRef]
- Walton, R.T.; Christie, K.A.; Whittaker, M.N.; Kleinstiver, B.P. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science 2020, 368, 290–296. [Google Scholar] [CrossRef]
- Momen-Roknabadi, A.; Oikonomou, P.; Zegans, M.; Tavazoie, S. An inducible CRISPR interference library for genetic interrogation of Saccharomyces cerevisiae biology. Commun. Biol. 2020, 3, 723. [Google Scholar] [CrossRef]
- McGlincy, N.J.; Meacham, Z.A.; Reynaud, K.K.; Muller, R.; Baum, R.; Ingolia, N.T. A genome-scale CRISPR interference guide library enables comprehensive phenotypic profiling in yeast. BMC Genom. 2021, 22, 205. [Google Scholar] [CrossRef]
- Muller, R.; Meacham, Z.A.; Ferguson, L.; Ingolia, N.T. CiBER-seq dissects genetic networks by quantitative CRISPRi profiling of expression phenotypes. Science 2020, 370, eabb9662. [Google Scholar] [CrossRef]
| Method | Material Preparation | Conditional Induction | Systematic Design | Relative Throughput | Relative Specificity |
|---|---|---|---|---|---|
| genetic mutation | genetic screening | Yes a | No | + | +++ |
| AID | degron tag insertion | Yes | Yes | ++ | +++ |
| anchor-away | FRB tag insertion | Yes | Yes | ++ | +++ |
| Cre–loxP | loxP sites insertion | Yes | Yes | ++ | +++ |
| promoter replacement | promoter replacement | Yes | Yes | ++ | +++ |
| DAmP | marker gene insertion | No | Yes | ++ | +++ |
| RNAi | long hairpin RNA (chromosome or plasmid) | Yes | No | ++ | + |
| CRISPRi (dCas9) | short oligo DNA cloning (plasmid) | Yes | Yes | +++ | ++ |
| CRISPRi (dCas12a) | short oligo DNA cloning (plasmid) | No b | No c | ++ c | ++ |
| Cas13a | short oligo DNA cloning (plasmid) | Yes | No c | ++ c | ++ d |
| Cas13d | multiplex crRNA gene cloning (chromosome) | Yes | Yes | ++ | ++ d |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ishikawa, K.; Saitoh, S. Transcriptional Regulation Technology for Gene Perturbation in Fission Yeast. Biomolecules 2023, 13, 716. https://doi.org/10.3390/biom13040716
Ishikawa K, Saitoh S. Transcriptional Regulation Technology for Gene Perturbation in Fission Yeast. Biomolecules. 2023; 13(4):716. https://doi.org/10.3390/biom13040716
Chicago/Turabian StyleIshikawa, Ken, and Shigeaki Saitoh. 2023. "Transcriptional Regulation Technology for Gene Perturbation in Fission Yeast" Biomolecules 13, no. 4: 716. https://doi.org/10.3390/biom13040716
APA StyleIshikawa, K., & Saitoh, S. (2023). Transcriptional Regulation Technology for Gene Perturbation in Fission Yeast. Biomolecules, 13(4), 716. https://doi.org/10.3390/biom13040716





