Integrative Analysis and Experimental Validation of Competing Endogenous RNAs in Obstructive Sleep Apnea
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection and Preprocessing
2.2. Differential Expression Analysis
2.3. Weighted Gene Co-Expression Network Analysis
2.4. Functional Enrichment Analysis
2.5. Feature Selection, Modeling and Validation
2.6. Construction of the lncRNA-miRNA-mRNA Regulatory Networks
2.7. Correlation Analysis between Hub mRNAs and Immune Characteristics
2.8. Validation the Expression of the ceRNA In Vitro
2.8.1. Cell Culture
2.8.2. Chronic Intermittent Hypoxia Protocols
2.8.3. RNA Extraction and qRT-PCR
2.9. Statistical Analysis
3. Results
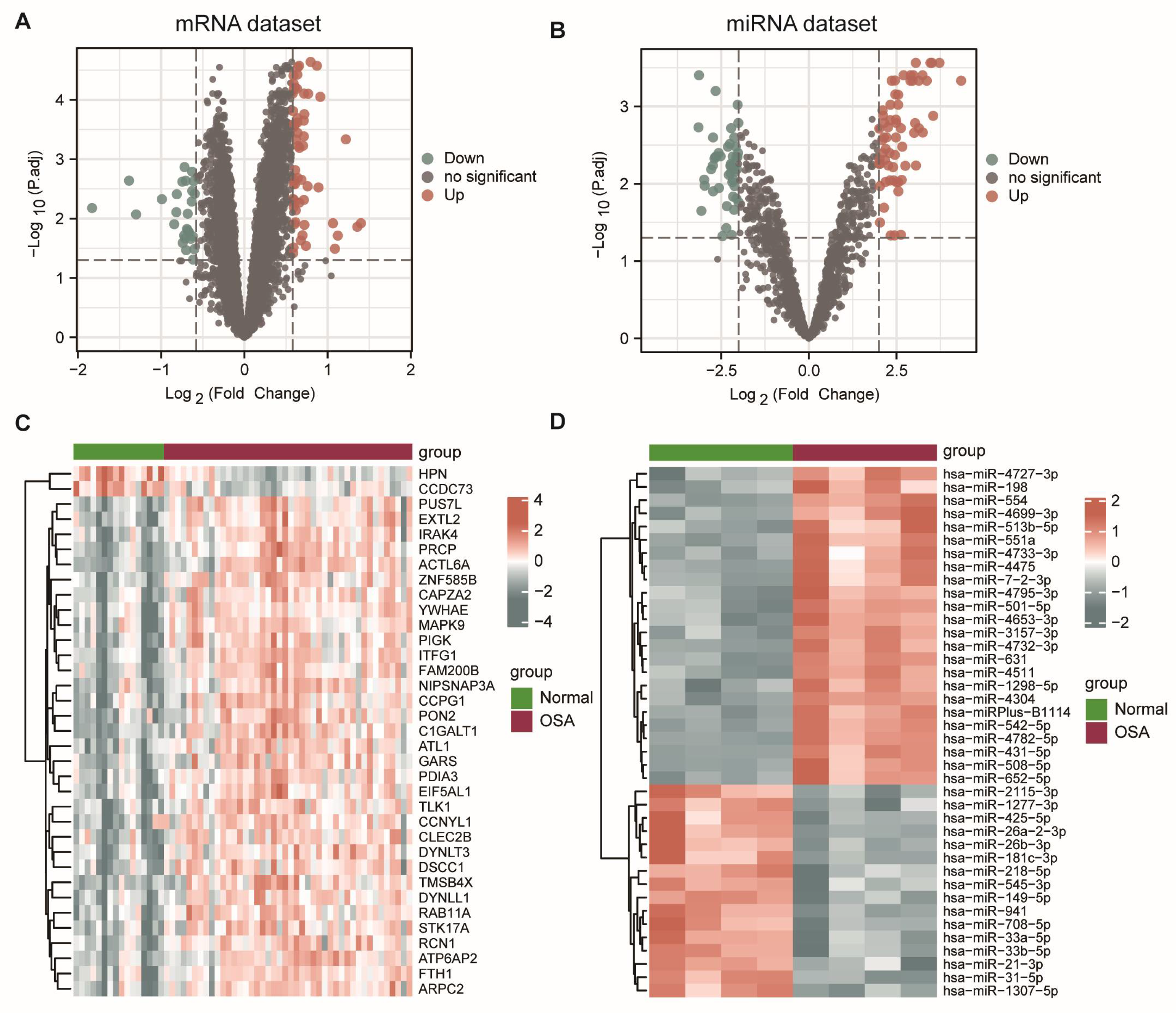
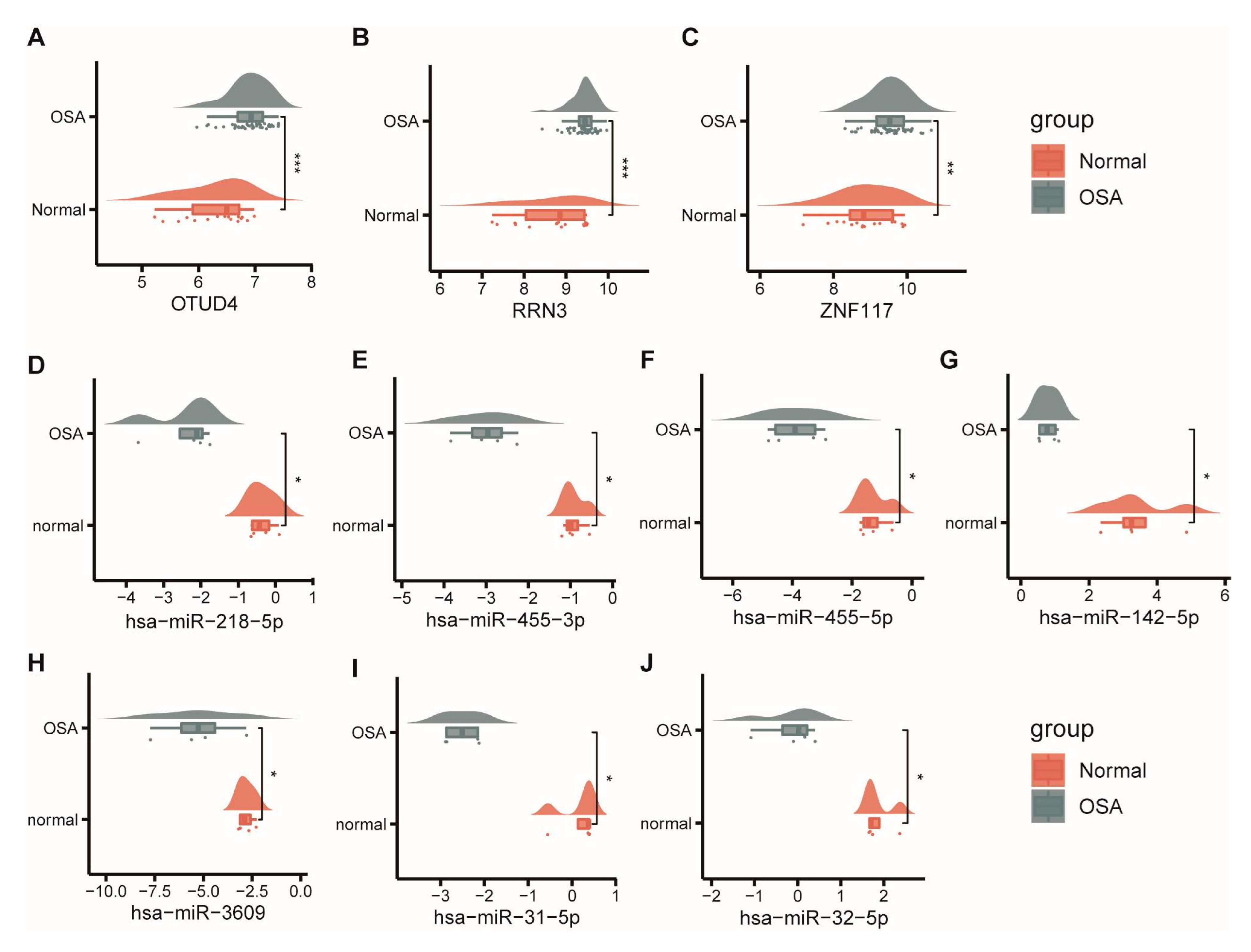
3.1. Differential Expression Analysis between Controls and OSA Patients
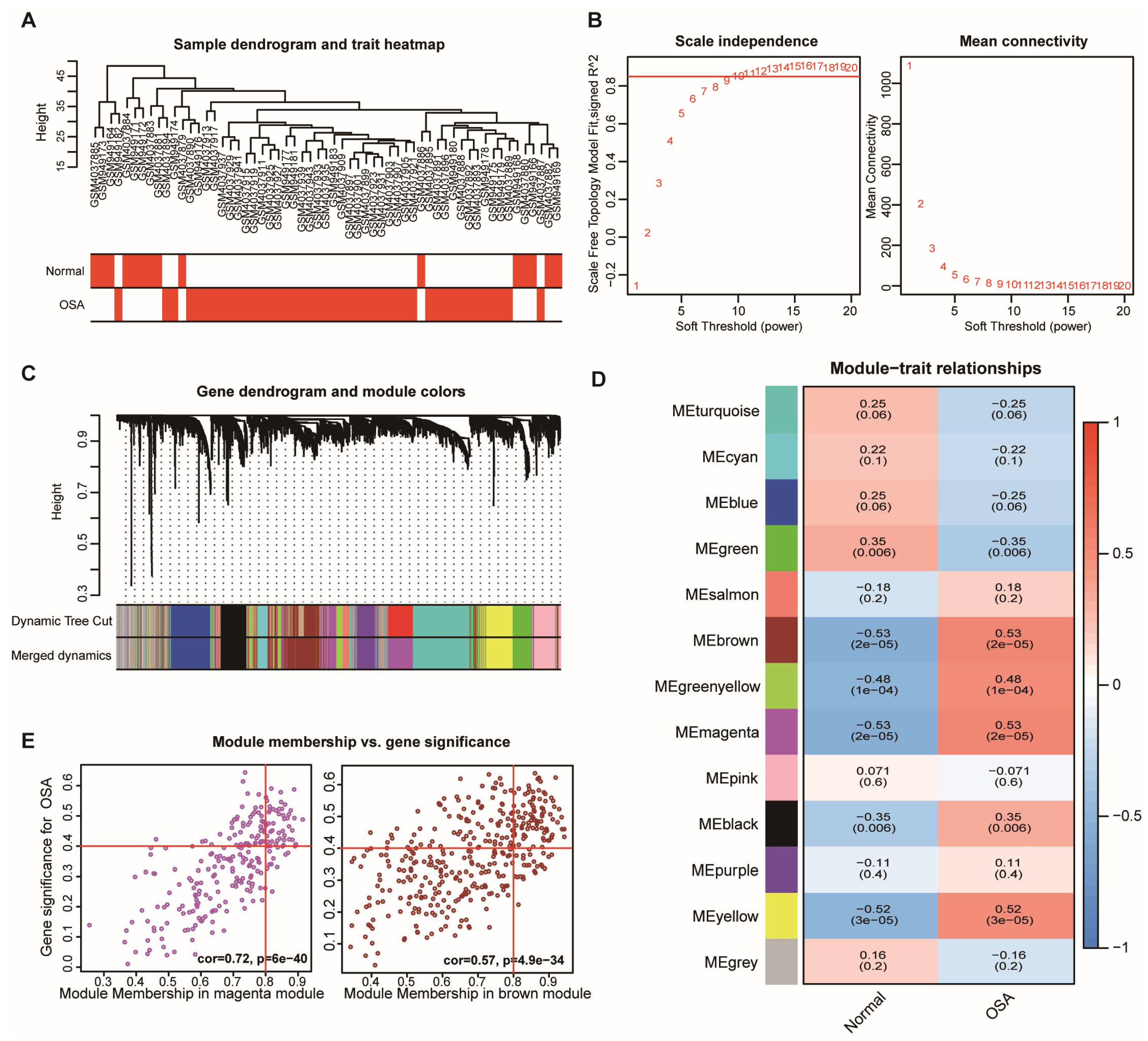
3.2. Identification of the Key Modules in OSA
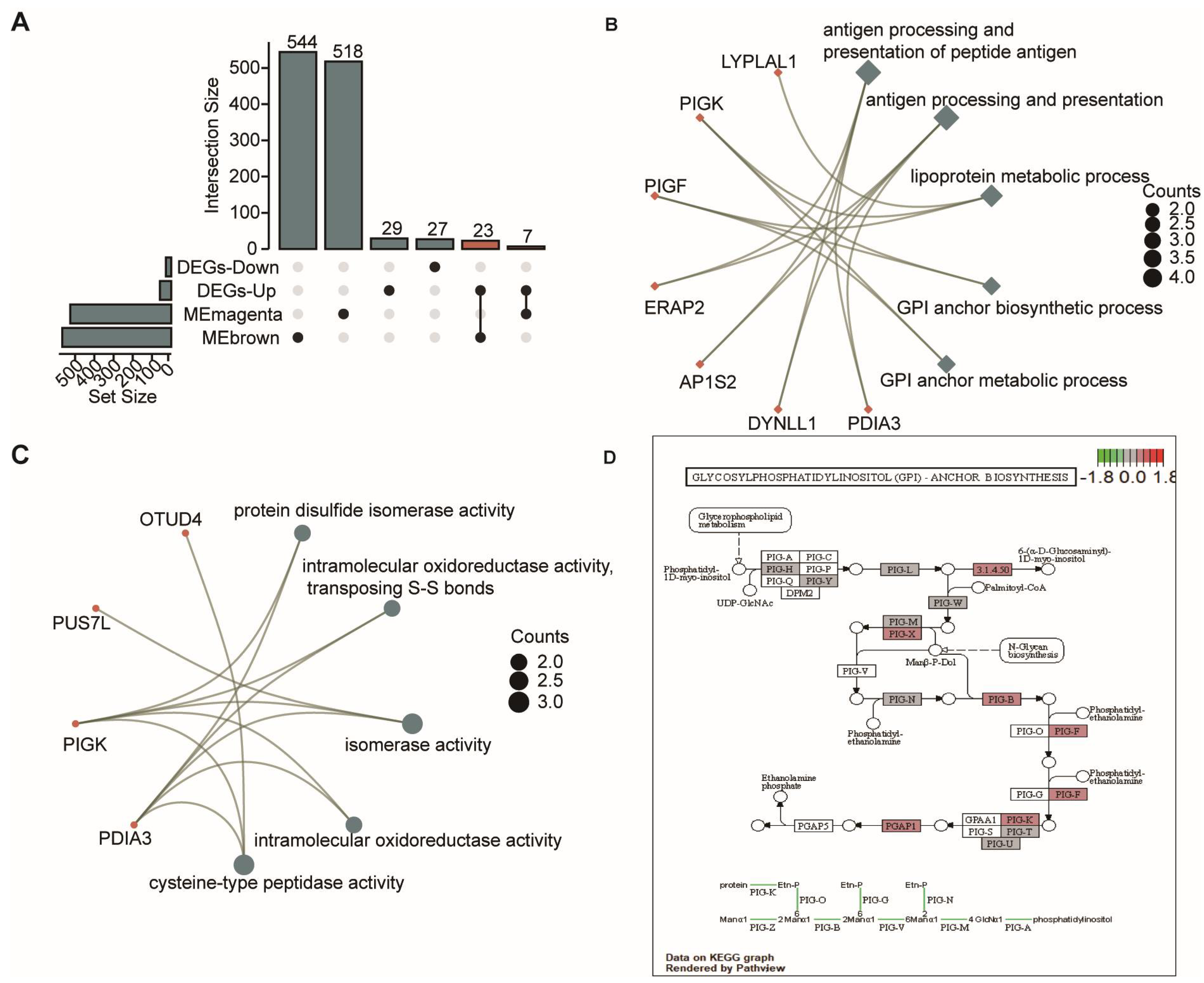
3.3. Functional Enrichment Analysis for OSA-Specific mRNAs
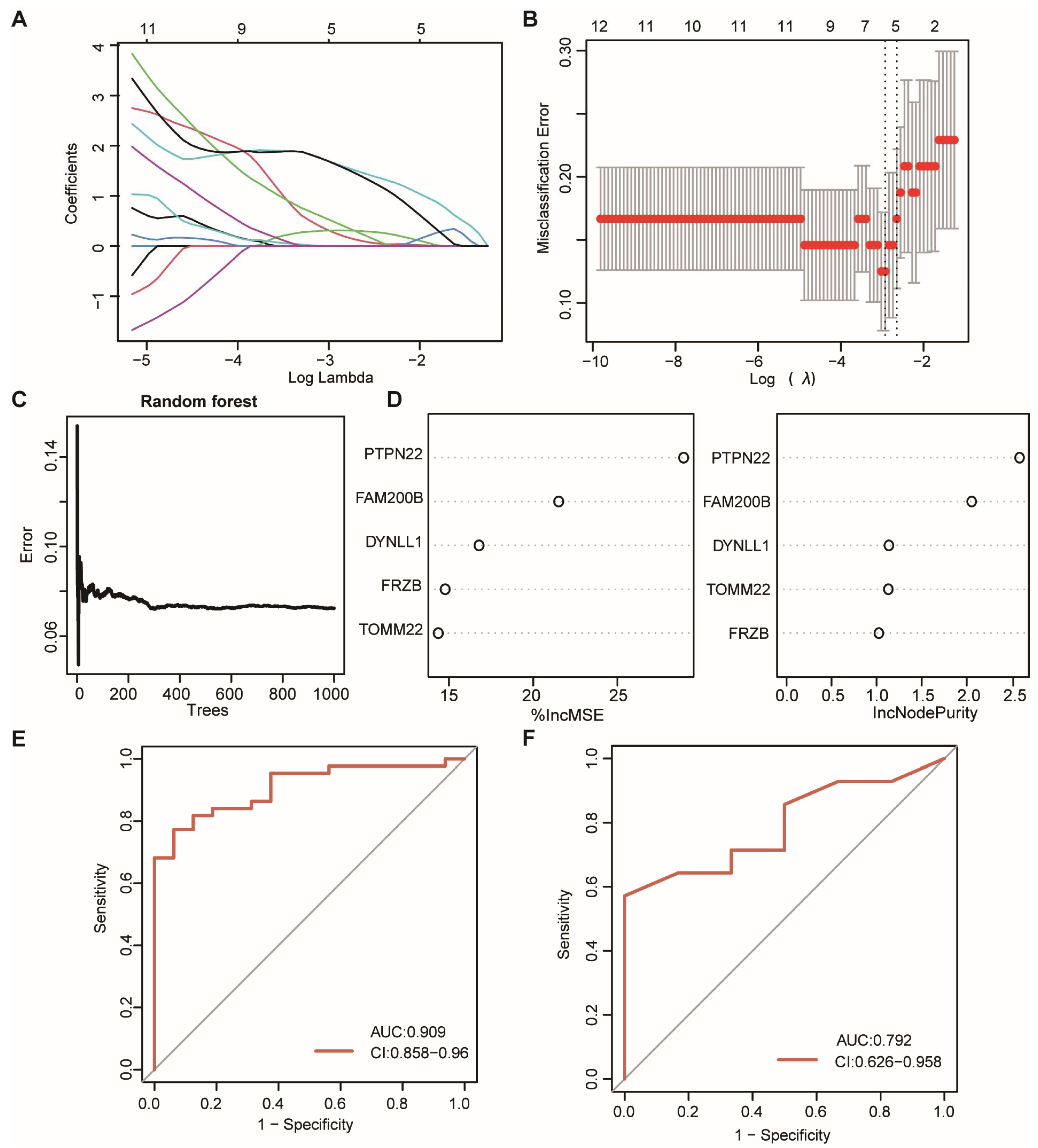
3.4. Construction of the mRNA Signature and Validation
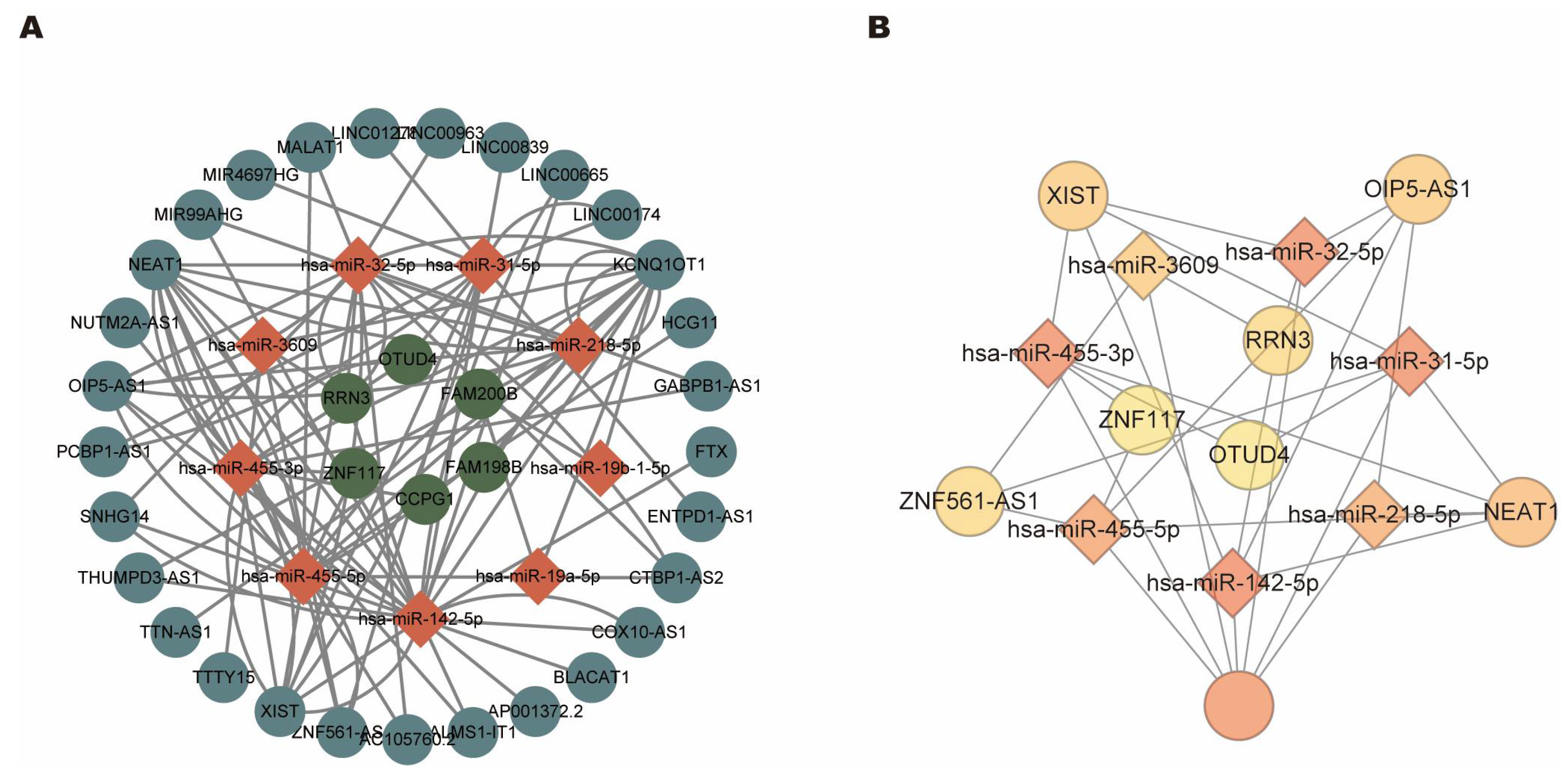
3.5. Construct CeRNA Regulatory Networks
3.6. Identification of Hub CeRNA Networks
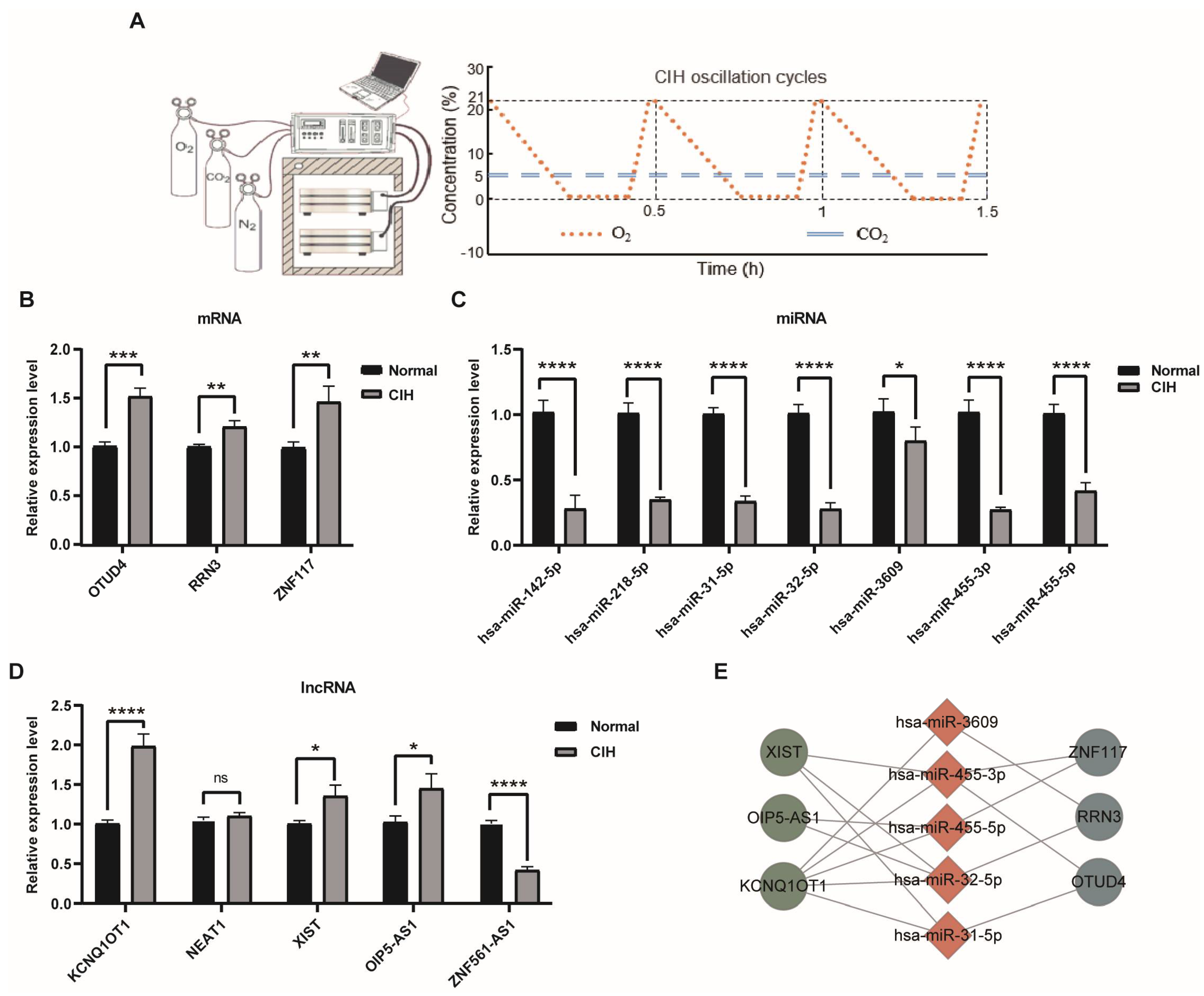
3.7. Validation of Expression Levels of Hub CeRNA Networks In Vitro
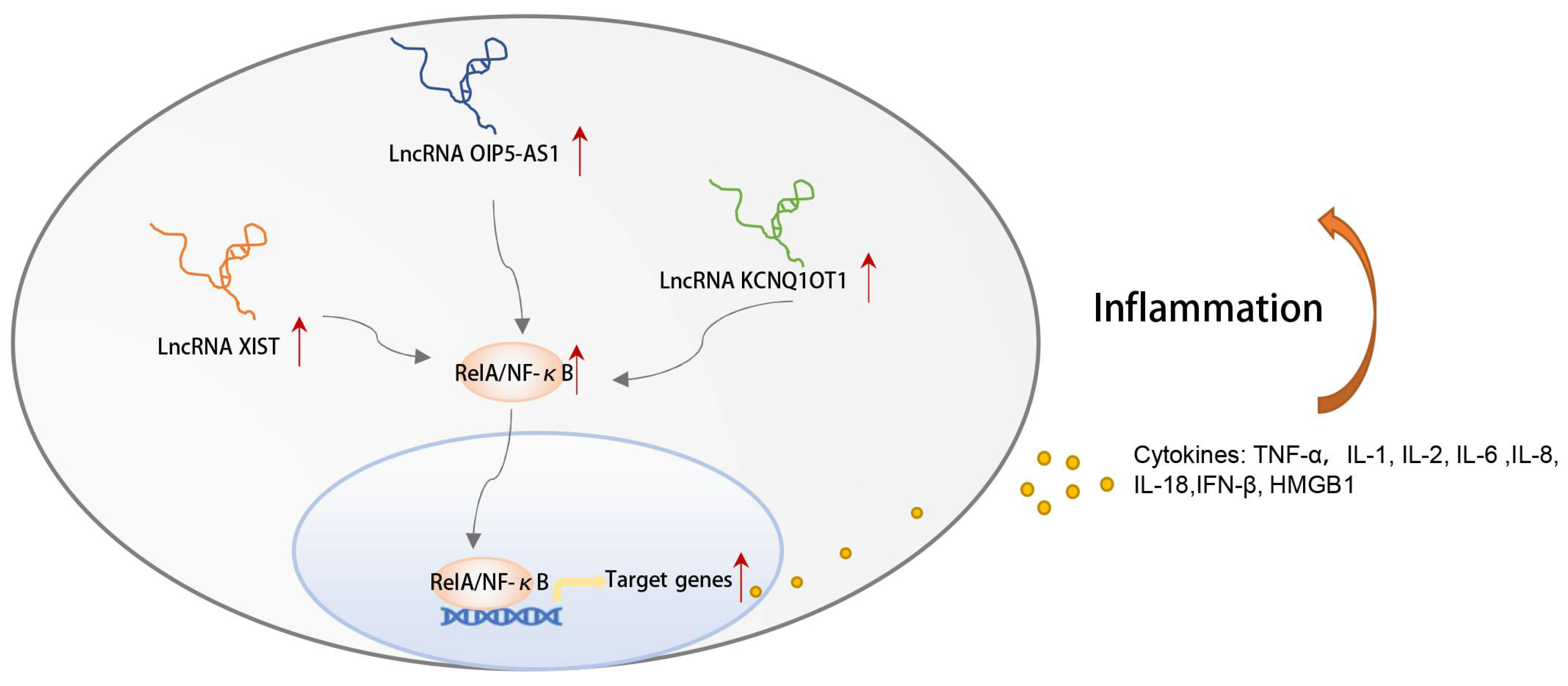
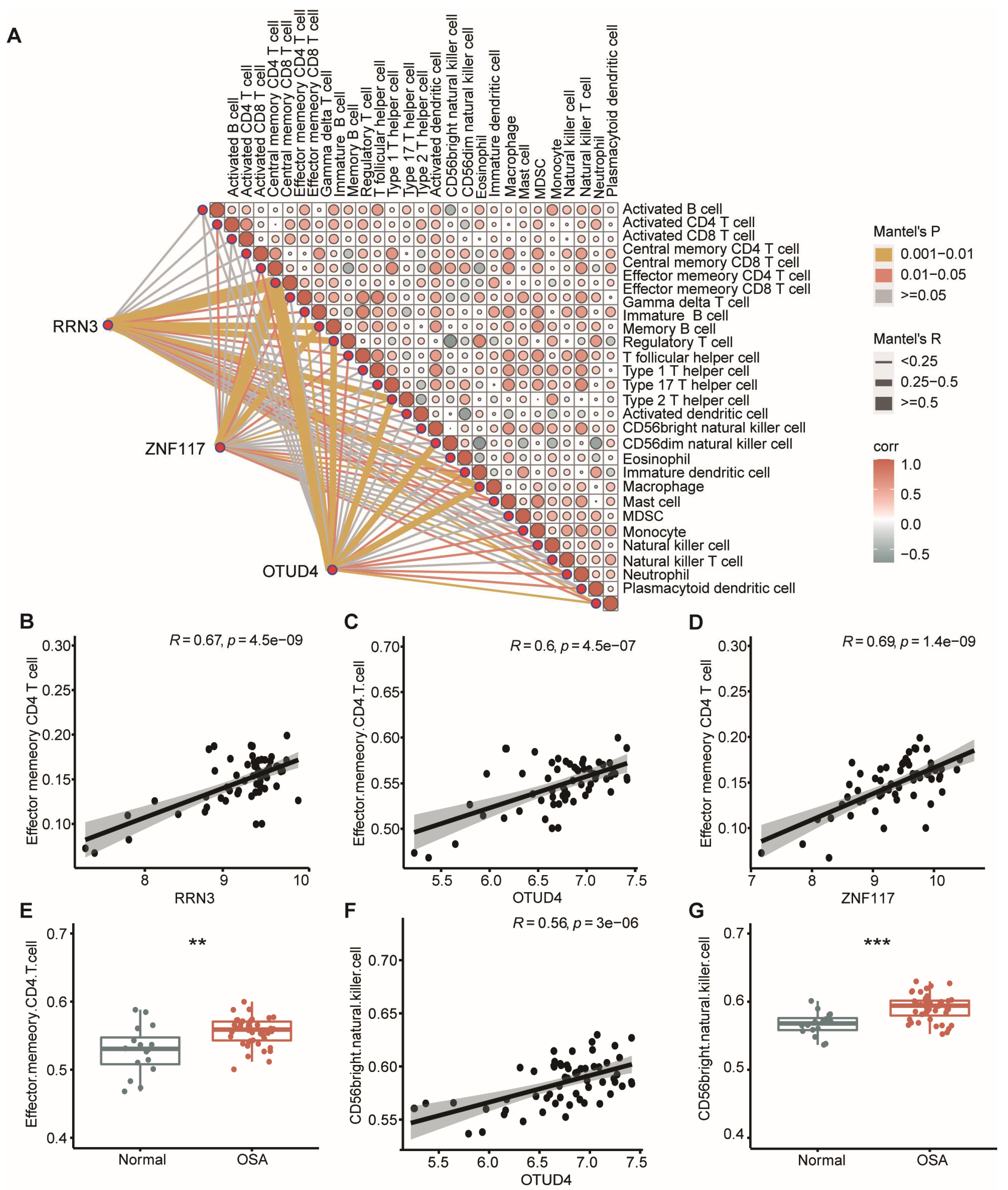
3.8. CeRNA Networks Related to Inflammation and Immune Characteristics of OSA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Peppard, P.E.; Young, T.; Barnet, J.H.; Palta, M.; Hagen, E.W.; Hla, K.M. Increased Prevalence of Sleep-Disordered Breathing in Adults. Am. J. Epidemiol. 2013, 177, 1006–1014. [Google Scholar] [CrossRef]
- Benjafield, A.V.; Ayas, N.T.; Eastwood, P.R.; Heinzer, R.; Ip, M.S.M.; Morrell, M.J.; Nunez, C.M.; Patel, S.R.; Penzel, T.; Pépin, J.L.; et al. Estimation of the global prevalence and burden of obstructive sleep apnoea: A literature-based analysis. Lancet Respir. Med. 2019, 7, 687–698. [Google Scholar] [CrossRef] [PubMed]
- Baldanzi, G.; Sayols-Baixeras, S.; Theorell-Haglöw, J.; Dekkers, K.F.; Hammar, U.; Nguyen, D.; Lin, Y.T.; Ahmad, S.; Holm, J.B.; Nielsen, H.B.; et al. Obstructive sleep apnea was associated with the human gut microbiota composition and functional potential in the population-based Swedish CardioPulmonary bioImage Study (SCAPIS). Chest 2023, in press. [Google Scholar] [CrossRef]
- Mangione, C.M.; Barry, M.J.; Nicholson, W.K.; Cabana, M.; Chelmow, D.; Rucker Coker, T.; Davidson, K.W.; Davis, E.M.; Donahue, K.E.; Jaén, C.R.; et al. Screening for Obstructive Sleep Apnea in Adults: US Preventive Services Task Force Recommendation Statement. Jama 2022, 328, 1945–1950. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Cerón, E.; Casitas, R.; Galera, R.; Sánchez-Sánchez, B.; Zamarrón, E.; Garcia-Sanchez, A.; Jaureguizar, A.; Cubillos-Zapata, C.; Garcia-Rio, F. Contribution of sleep characteristics to the association between obstructive sleep apnea and dyslipidemia. Sleep Med. 2021, 84, 63–72. [Google Scholar] [CrossRef]
- Kawano, Y.; Tamura, A.; Kadota, J. Association between the severity of obstructive sleep apnea and the ratio of low-density lipoprotein cholesterol to high-density lipoprotein cholesterol. Metab. Clin. Exp. 2012, 61, 186–192. [Google Scholar] [CrossRef]
- Kent, B.D.; Garvey, J.F.; Ryan, S.; Nolan, G.; Dodd, J.D.; McNicholas, W.T. Severity of obstructive sleep apnoea predicts coronary artery plaque burden: A coronary computed tomographic angiography study. Eur. Respir. J. 2013, 42, 1263–1270. [Google Scholar] [CrossRef]
- Yaggi, H.K.; Concato, J.; Kernan, W.N.; Lichtman, J.H.; Brass, L.M.; Mohsenin, V. Obstructive sleep apnea as a risk factor for stroke and death. New. Engl. J. Med. 2005, 353, 2034–2041. [Google Scholar] [CrossRef] [PubMed]
- Kuo, C.Y.; Hsiao, H.T.; Lo, I.H.; Nikolai, T. Association Between Obstructive Sleep Apnea, Its Treatment, and Alzheimer’s Disease: Systematic Mini-Review. Front. Aging Neurosci. 2020, 12, 591737. [Google Scholar] [CrossRef]
- Turkiewicz, S.; Ditmer, M.; Sochal, M.; Białasiewicz, P.; Strzelecki, D.; Gabryelska, A. Obstructive Sleep Apnea as an Acceleration Trigger of Cellular Senescence Processes through Telomere Shortening. Int. J. Mol. Sci. 2021, 22, 12536. [Google Scholar] [CrossRef]
- Díaz-García, E.; García-Tovar, S.; Alfaro, E.; Jaureguizar, A.; Casitas, R.; Sánchez-Sánchez, B.; Zamarrón, E.; Fernández-Lahera, J.; López-Collazo, E.; Cubillos-Zapata, C.; et al. Inflammasome Activation: A Keystone of Proinflammatory Response in Obstructive Sleep Apnea. Am. J. Respir. Crit. Care Med. 2022, 205, 1337–1348. [Google Scholar] [CrossRef] [PubMed]
- Drager, L.F.; Li, J.; Shin, M.K.; Reinke, C.; Aggarwal, N.R.; Jun, J.C.; Bevans-Fonti, S.; Sztalryd, C.; O’Byrne, S.M.; Kroupa, O.; et al. Intermittent hypoxia inhibits clearance of triglyceride-rich lipoproteins and inactivates adipose lipoprotein lipase in a mouse model of sleep apnoea. Eur. Heart, J. 2012, 33, 783–790. [Google Scholar] [CrossRef]
- Xu, H.; Liu, F.; Li, Z.; Li, X.; Liu, Y.; Li, N.; Zhang, X.; Gao, Z.; Zhang, X.; Liu, Y.; et al. Genome-Wide Association Study of Obstructive Sleep Apnea and Objective Sleep-Related Traits Identifies Novel Risk Loci in Han Chinese Individuals. Am. J. Respir. Crit. Care Med. 2022, 206, 1534–1545. [Google Scholar] [CrossRef] [PubMed]
- Mohit; Shrivastava, A.; Chand, P. Molecular determinants of obstructive sleep apnea. Sleep Med. 2021, 80, 105–112. [Google Scholar] [CrossRef]
- Palmer, L.J.; Buxbaum, S.G.; Larkin, E.; Patel, S.R.; Elston, R.C.; Tishler, P.V.; Redline, S. A whole-genome scan for obstructive sleep apnea and obesity. Am. J. Hum. Genet. 2003, 72, 340–350. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.Y.; Lin, Y.C.; Cui, S.; Huang, Y.; Tang, Y.; Xu, J.; Bao, J.; Li, Y.; Wen, J.; Zuo, H.; et al. miRTarBase update 2022: An informative resource for experimentally validated miRNA-target interactions. Nucleic. Acids Res. 2022, 50, D222–D230. [Google Scholar] [CrossRef]
- Yildirim, O.; Baloglu, U.B.; Acharya, U.R. A Deep Learning Model for Automated Sleep Stages Classification Using PSG Signals. Int. J. Environ. Res. Public Health 2019, 16, 599. [Google Scholar] [CrossRef]
- Ho-Xuan, H.; Glažar, P.; Latini, C.; Heizler, K.; Haase, J.; Hett, R.; Anders, M.; Weichmann, F.; Bruckmann, A.; Van den Berg, D.; et al. Comprehensive analysis of translation from overexpressed circular RNAs reveals pervasive translation from linear transcripts. Nucleic Acids Res. 2020, 48, 10368–10382. [Google Scholar] [CrossRef]
- Chan, J.J.; Kwok, Z.H.; Chew, X.H.; Zhang, B.; Liu, C.; Soong, T.W.; Yang, H.; Tay, Y. A FTH1 gene:pseudogene:microRNA network regulates tumorigenesis in prostate cancer. Nucleic Acids Res. 2018, 46, 1998–2011. [Google Scholar] [CrossRef]
- Song, W.; Ren, J.; Xiang, R.; Yuan, W.; Fu, T. Cross-Talk Between m(6)A- and m(5)C-Related lncRNAs to Construct a Novel Signature and Predict the Immune Landscape of Colorectal Cancer Patients. Front. Immunol. 2022, 13, 740960. [Google Scholar] [CrossRef]
- Zhou, Z.; Ni, H.; Li, Y.; Jiang, B. LncRNA XIST promotes inflammation by downregulating GRα expression in the adenoids of children with OSAHS. Exp. Ther. Med. 2021, 21, 500. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, Z.; Wang, Y.; Wei, L.; Chen, H. Overexpressed long noncoding RNA CPS1-IT alleviates pulmonary arterial hypertension in obstructive sleep apnea by reducing interleukin-1β expression via HIF1 transcriptional activity. J. Cell. Physiol. 2019, 234, 19715–19727. [Google Scholar] [CrossRef]
- Chen, Q.; Lin, G.; Lin, L.; Huang, J.; Chen, L.; Lian, N.; Chen, M.; Zeng, A.; Lin, Q. LncRNA XR_596701 protects H9c2 cells against intermittent hypoxia-induced injury through regulation of the miR-344b-5p/FAIM3 axis. Cell Death Discov. 2022, 8, 42. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, Z.; Du, Y.; Xing, Y.; Guo, Y.; Zhang, Y.; Guo, R.; Gong, W.; Nie, S.; Wang, X. lncRNA Mirt1: A Critical Regulatory Factor in Chronic Intermittent Hypoxia Exaggerated Post-MI Cardiac Remodeling. Front. Genet. 2022, 13, 818823. [Google Scholar] [CrossRef]
- Chen, Y.C.; Hsu, P.Y.; Su, M.C.; Chin, C.H.; Liou, C.W.; Wang, T.Y.; Lin, Y.Y.; Lee, C.P.; Lin, M.C.; Hsiao, C.C. miR-21-5p Under-Expression in Patients with Obstructive Sleep Apnea Modulates Intermittent Hypoxia with Re-Oxygenation-Induced-Cell Apoptosis and Cytotoxicity by Targeting Pro-Inflammatory TNF-α-TLR4 Signaling. Int. J. Mol. Sci. 2020, 21, 999. [Google Scholar] [CrossRef]
- Santamaria-Martos, F.; Benítez, I.; Ortega, F.; Zapater, A.; Giron, C.; Pinilla, L.; Pascual, L.; Cortijo, A.; Dalmases, M.; Fernandez-Real, J.M.; et al. Circulating microRNA profile as a potential biomarker for obstructive sleep apnea diagnosis. Sci. Rep. 2019, 9, 13456. [Google Scholar] [CrossRef]
- Zhang, R.; Hao, Y.; Zhang, J. The lncRNA DANCR promotes development of atherosclerosis by regulating the miR-214-5p/COX20 signaling pathway. Cell. Mol. Biol. Lett. 2022, 27, 15. [Google Scholar] [CrossRef] [PubMed]
- Xue, S.T.; Zheng, B.; Cao, S.Q.; Ding, J.C.; Hu, G.S.; Liu, W.; Chen, C. Long non-coding RNA LINC00680 functions as a ceRNA to promote esophageal squamous cell carcinoma progression through the miR-423-5p/PAK6 axis. Mol. Cancer 2022, 21, 69. [Google Scholar] [CrossRef]
- Liu, B.; Zhou, X.; Wu, D.; Zhang, X.; Shen, X.; Mi, K.; Qu, Z.; Jiang, Y.; Shang, D. Comprehensive characterization of a drug-resistance-related ceRNA network across 15 anti-cancer drug categories. Mol. Ther. Nucleic Acids 2021, 24, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Glenfield, C.; McLysaght, A. Pseudogenes Provide Evolutionary Evidence for the Competitive Endogenous RNA Hypothesis. Mol. Biol. Evol. 2018, 35, 2886–2899. [Google Scholar] [CrossRef]
- Bosson, A.D.; Zamudio, J.R.; Sharp, P.A. Endogenous miRNA and target concentrations determine susceptibility to potential ceRNA competition. Mol. Cell 2014, 56, 347–359. [Google Scholar] [CrossRef] [PubMed]
- Gharib, S.A.; Hurley, A.L.; Rosen, M.J.; Spilsbury, J.C.; Schell, A.E.; Mehra, R.; Patel, S.R. Obstructive sleep apnea and CPAP therapy alter distinct transcriptional programs in subcutaneous fat tissue. Sleep 2020, 43, zsz314. [Google Scholar] [CrossRef]
- Gharib, S.A.; Hayes, A.L.; Rosen, M.J.; Patel, S.R. A pathway-based analysis on the effects of obstructive sleep apnea in modulating visceral fat transcriptome. Sleep 2013, 36, 23–30. [Google Scholar] [CrossRef]
- Chen, Y.C.; Chen, K.D.; Su, M.C.; Chin, C.H.; Chen, C.J.; Liou, C.W.; Chen, T.W.; Chang, Y.C.; Huang, K.T.; Wang, C.C.; et al. Genome-wide gene expression array identifies novel genes related to disease severity and excessive daytime sleepiness in patients with obstructive sleep apnea. PLoS ONE 2017, 12, e0176575. [Google Scholar] [CrossRef] [PubMed]
- Davis, S.; Meltzer, P.S. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007, 23, 1846–1847. [Google Scholar] [CrossRef] [PubMed]
- Cederberg, K.L.J.; Hanif, U.; Peris Sempere, V.; Hédou, J.; Leary, E.B.; Schneider, L.D.; Lin, L.; Zhang, J.; Morse, A.M.; Blackman, A.; et al. Proteomic Biomarkers of the Apnea Hypopnea Index and Obstructive Sleep Apnea: Insights into the Pathophysiology of Presence, Severity, and Treatment Response. Int. J. Mol. Sci. 2022, 23, 7983. [Google Scholar] [CrossRef]
- Gaspar, L.S.; Hesse, J.; Yalçin, M.; Santos, B.; Carvalhas-Almeida, C.; Ferreira, M.; Moita, J.; Relógio, A.; Cavadas, C.; Álvaro, A.R. Long-term continuous positive airway pressure treatment ameliorates biological clock disruptions in obstructive sleep apnea. EBioMedicine 2021, 65, 103248. [Google Scholar] [CrossRef]
- Sofer, T.; Li, R.; Joehanes, R.; Lin, H.; Gower, A.C.; Wang, H.; Kurniansyah, N.; Cade, B.E.; Lee, J.; Williams, S.; et al. Low oxygen saturation during sleep reduces CD1D and RAB20 expressions that are reversed by CPAP therapy. EBioMedicine 2020, 56, 102803. [Google Scholar] [CrossRef] [PubMed]
- Christensson, E.; Mkrtchian, S.; Ebberyd, A.; Österlund Modalen, Å.; Franklin, K.A.; Eriksson, L.I.; Jonsson Fagerlund, M. Whole blood gene expression signature in patients with obstructive sleep apnea and effect of continuous positive airway pressure treatment. Respir. Physiol. Neurobiol. 2021, 294, 103746. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic. Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Hou, J.; Zhao, L.; Yan, J.; Ren, X.; Zhu, K.; Gao, T.; Du, X.; Luo, H.; Li, Z.; Xu, M. MicroRNA expression profile is altered in the upper airway skeletal muscle tissue of patients with obstructive sleep apnea-hypopnea syndrome. J. Int. Med. Res. 2019, 47, 4163–4182. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant graphics for data analysis. Springer-Verlag: New York, NY, USA, 2016; pp. 189–201. ISBN 978-3-319-24277-4. [Google Scholar]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. Bmc Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Brouwer, C. Pathview: An R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 2013, 29, 1830–1831. [Google Scholar] [CrossRef] [PubMed]
- Friedman, J.; Hastie, T.; Tibshirani, R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J. Stat. Softw. 2010, 33, 1–22. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.C.; Müller, M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. Bmc Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef] [PubMed]
- Li, J.H.; Liu, S.; Zhou, H.; Qu, L.H.; Yang, J.H. starBase v2. 0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein–RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014, 42, 92–97. [Google Scholar] [CrossRef] [PubMed]
- Paraskevopoulou, M.D.; Vlachos, I.S.; Karagkouni, D.; Georgakilas, G.; Kanellos, I.; Vergoulis, T.; Zagganas, K.; Tsanakas, P.; Floros, E.; Dalamagas, T.; et al. DIANA-LncBase v2: Indexing microRNA targets on non-coding transcripts. Nucleic Acids Res. 2016, 44, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.H.; Chen, S.H.; Wu, H.H.; Ho, C.W.; Ko, M.T.; Lin, C.Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. Bmc Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Zhou, L.; Chen, J.; Wei, T. ggcor: Extended tools for correlation analysis and visualization. R package version 0.9. 2020; 7. [Google Scholar]
- Xu, M.; Kong, Y.; Chen, N.; Peng, W.; Zi, R.; Jiang, M.; Zhu, J.; Wang, Y.; Yue, J.; Lv, J.; et al. Identification of Immune-Related Gene Signature and Prediction of CeRNA Network in Active Ulcerative Colitis. Front. Immunol. 2022, 13, 855645. [Google Scholar] [CrossRef]
- Zheng, Z.; Zhang, G.; Liang, X.; Li, T. LncRNA OIP5-AS1 facilitates ox-LDL-induced endothelial cell injury through the miR-98-5p/HMGB1 axis. Mol. Cell. Biochem. 2021, 476, 443–455. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Sun, H.; Li, X.; Cao, W.; Peng, A.; Dong, S.; Yu, Z. Downregulation of lncRNA KCNQ1OT1 relieves traumatic brain injury induced neurological deficits via promoting "M2" microglia polarization. Brain Res. Bull 2021, 171, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Wang, T.; Zhao, Z.; Wei, W.; Yang, X.; Wang, X.; Xin, W. Novel Insights into the Emerging Role of Neat1 and Its Effects Downstream in the Regulation of Inflammation. J. Inflamm. Res. 2022, 15, 557–571. [Google Scholar] [CrossRef]
- Swanson, L.M.; Arnedt, J.T.; Rosekind, M.R.; Belenky, G.; Balkin, T.J.; Drake, C. Sleep disorders and work performance: Findings from the 2008 National Sleep Foundation Sleep in America poll. J. Sleep Res. 2011, 20, 487–494. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, C.E.; Issa, F.G.; Berthon-Jones, M.; Eves, L. Reversal of obstructive sleep apnoea by continuous positive airway pressure applied through the nares. Lancet 1981, 1, 862–865. [Google Scholar] [CrossRef]
- Jackson, M.L.; McEvoy, R.D.; Banks, S.; Barnes, M. Neurobehavioral Impairment and CPAP Treatment Response in Mild-Moderate Obstructive Sleep Apneas. J. Clin. Sleep Med. JCSM Off. Publ. Am. Acad. Sleep Med. 2018, 14, 47–56. [Google Scholar] [CrossRef]
- Kylstra, W.A.; Aaronson, J.A.; Hofman, W.F.; Schmand, B.A. Neuropsychological functioning after CPAP treatment in obstructive sleep apnea: A meta-analysis. Sleep Med. Rev. 2013, 17, 341–347. [Google Scholar] [CrossRef]
- Almendros, I.; Gileles-Hillel, A.; Khalyfa, A.; Wang, Y.; Zhang, S.X.; Carreras, A.; Farré, R.; Gozal, D. Adipose tissue macrophage polarization by intermittent hypoxia in a mouse model of OSA: Effect of tumor microenvironment. Cancer Lett. 2015, 361, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Akbarpour, M.; Khalyfa, A.; Qiao, Z.; Gileles-Hillel, A.; Almendros, I.; Farré, R.; Gozal, D. Altered CD8+ T-Cell Lymphocyte Function and TC1 Cell Stemness Contribute to Enhanced Malignant Tumor Properties in Murine Models of Sleep Apnea. Sleep 2017, 40, zsw040. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Jiménez, E.; Cubillos-Zapata, C.; Toledano, V.; Pérez de Diego, R.; Fernández-Navarro, I.; Casitas, R.; Carpio, C.; Casas-Martín, J.; Valentín, J.; Varela-Serrano, A.; et al. Monocytes inhibit NK activity via TGF-β in patients with obstructive sleep apnoea. Eur. Respir. J. 2017, 49, 1602456. [Google Scholar] [CrossRef]
- Sun, M.; Liang, C.; Lin, H.; Meng, Y.; Tang, Q.; Shi, X.; Zhang, E.; Tang, Q. Monocyte to HDL cholesterol ratio as a marker of the presence and severity of obstructive sleep apnea in hypertensive patients. Sci. Rep. 2021, 11, 15821. [Google Scholar] [CrossRef]
- Johns, E.C.; Halligan, D.L.; Tammsalu, T.; Hill, E.A.; Riha, R.L.; Denison, F.C.; Reynolds, R.M. Gene expression profiling of placentae from women with obesity and obstructive sleep apnoea. Placenta 2022, 121, 53–60. [Google Scholar] [CrossRef]
- Deng, J.; Deng, H.; Liu, C.; Liang, Y.; Wang, S. Long non-coding RNA OIP5-AS1 functions as an oncogene in lung adenocarcinoma through targeting miR-448/Bcl-2. Biomed. Pharmacother. Biomed. Pharmacother. 2018, 98, 102–110. [Google Scholar] [CrossRef]
- Kun-Peng, Z.; Chun-Lin, Z.; Xiao-Long, M.; Lei, Z. Fibronectin-1 modulated by the long noncoding RNA OIP5-AS1/miR-200b-3p axis contributes to doxorubicin resistance of osteosarcoma cells. J. Cell. Physiol. 2019, 234, 6927–6939. [Google Scholar] [CrossRef]
- Li, Y.; Han, X.; Feng, H.; Han, J. Long noncoding RNA OIP5-AS1 in cancer. Clin. Chim. Acta. Int. J. Clin. Chem. 2019, 499, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Che, Z.; Xueqin, J.; Zhang, Z. LncRNA OIP5-AS1 accelerates intervertebral disc degeneration by targeting miR-25-3p. Bioengineered 2021, 12, 11201–11212. [Google Scholar] [CrossRef]
- Ren, M.; Wang, T.; Han, Z.; Fu, P.; Liu, Z.; Ouyang, C. Long Noncoding RNA OIP5-AS1 Contributes to the Progression of Atherosclerosis by Targeting miR-26a-5p Through the AKT/NF-κB Pathway. J. Cardiovasc. Pharmacol. 2020, 76, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.; Liu, R.; Wu, X.; Ma, K.; Luo, W.; Nie, K.; Zhang, C.; Meng, X.; Tong, T.; Chen, X.; et al. LncRNA NEAT1 mediates intestinal inflammation by regulating TNFRSF1B. Ann. Transl. Med. 2021, 9, 773. [Google Scholar] [CrossRef]
- Bangert, C.; Villazala-Merino, S.; Fahrenberger, M.; Krausgruber, T.; Bauer, W.M.; Stanek, V.; Campion, N.J.; Bartosik, T.; Quint, T.; Regelsberger, G.; et al. Comprehensive Analysis of Nasal Polyps Reveals a More Pronounced Type 2 Transcriptomic Profile of Epithelial Cells and Mast Cells in Aspirin-Exacerbated Respiratory Disease. Front. Immunol. 2022, 13, 850494. [Google Scholar] [CrossRef]
- Wang, J.; Jiao, P.; Wei, X.; Zhou, Y. Silencing Long Non-coding RNA Kcnq1ot1 Limits Acute Kidney Injury by Promoting miR-204-5p and Blocking the Activation of NLRP3 Inflammasome. Front. Physiol. 2021, 12, 721524. [Google Scholar] [CrossRef]
- Wen, W.W.; Sun, H.L.; Yang, Y.X.; Jia, Y.F.; Huang, M.L.; Du, Y.H.; Qin, Y.W.; Fang, F.; Zhang, M.; Wei, Y.X. The association between circulating APRIL levels and severity of obstructive sleep apnea in Chinese adults. Clin. Chim. Acta. Int. J. Clin. Chem. 2020, 508, 161–169. [Google Scholar] [CrossRef]
- Ryan, S.; Taylor, C.T.; McNicholas, W.T. Systemic inflammation: A key factor in the pathogenesis of cardiovascular complications in obstructive sleep apnoea syndrome? Postgrad. Med. J. 2009, 85, 693–698. [Google Scholar] [CrossRef] [PubMed]
- Dempsey, J.A.; Veasey, S.C.; Morgan, B.J.; O’Donnell, C.P. Pathophysiology of sleep apnea. Physiol. Rev. 2010, 90, 47–112. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.L.; Gozal, D.; Wang, Y.; Bandla, H.P.; Bhattacharjee, R.; Kulkarni, R.; Kheirandish-Gozal, L. Alterations in circulating T-cell lymphocyte populations in children with obstructive sleep apnea. Sleep 2013, 36, 913–922. [Google Scholar] [CrossRef]
- Vicente, E.; Marin, J.M.; Carrizo, S.J.; Osuna, C.S.; González, R.; Marin-Oto, M.; Forner, M.; Vicente, P.; Cubero, P.; Gil, A.V.; et al. Upper airway and systemic inflammation in obstructive sleep apnoea. Eur. Respir. J. 2016, 48, 1108–1117. [Google Scholar] [CrossRef]
- Domagała-Kulawik, J.; Osińska, I.; Piechuta, A.; Bielicki, P.; Skirecki, T. T, B, and NKT Cells in Systemic Inflammation in Obstructive Sleep Apnoea. Mediat. Inflamm. 2015, 2015, 161579. [Google Scholar] [CrossRef]
- Said, E.A.; Al-Abri, M.A.; Al-Saidi, I.; Al-Balushi, M.S.; Al-Busaidi, J.Z.; Al-Reesi, I.; Koh, C.Y.; Hasson, S.S.; Idris, M.A.; Al-Jabri, A.A.; et al. Altered blood cytokines, CD4 T cells, NK and neutrophils in patients with obstructive sleep apnea. Immunol. Lett. 2017, 190, 272–278. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, N.; Zhu, Y.; Liu, F.; Zhang, X.; Liu, Y.; Wang, X.; Gao, Z.; Guan, J.; Yin, S. Integrative Analysis and Experimental Validation of Competing Endogenous RNAs in Obstructive Sleep Apnea. Biomolecules 2023, 13, 639. https://doi.org/10.3390/biom13040639
Li N, Zhu Y, Liu F, Zhang X, Liu Y, Wang X, Gao Z, Guan J, Yin S. Integrative Analysis and Experimental Validation of Competing Endogenous RNAs in Obstructive Sleep Apnea. Biomolecules. 2023; 13(4):639. https://doi.org/10.3390/biom13040639
Chicago/Turabian StyleLi, Niannian, Yaxin Zhu, Feng Liu, Xiaoman Zhang, Yuenan Liu, Xiaoting Wang, Zhenfei Gao, Jian Guan, and Shankai Yin. 2023. "Integrative Analysis and Experimental Validation of Competing Endogenous RNAs in Obstructive Sleep Apnea" Biomolecules 13, no. 4: 639. https://doi.org/10.3390/biom13040639
APA StyleLi, N., Zhu, Y., Liu, F., Zhang, X., Liu, Y., Wang, X., Gao, Z., Guan, J., & Yin, S. (2023). Integrative Analysis and Experimental Validation of Competing Endogenous RNAs in Obstructive Sleep Apnea. Biomolecules, 13(4), 639. https://doi.org/10.3390/biom13040639



