Effects of Mass Change on Liquid–Liquid Phase Separation of the RNA-Binding Protein Fused in Sarcoma
Abstract
1. Introduction
2. Materials and Methods
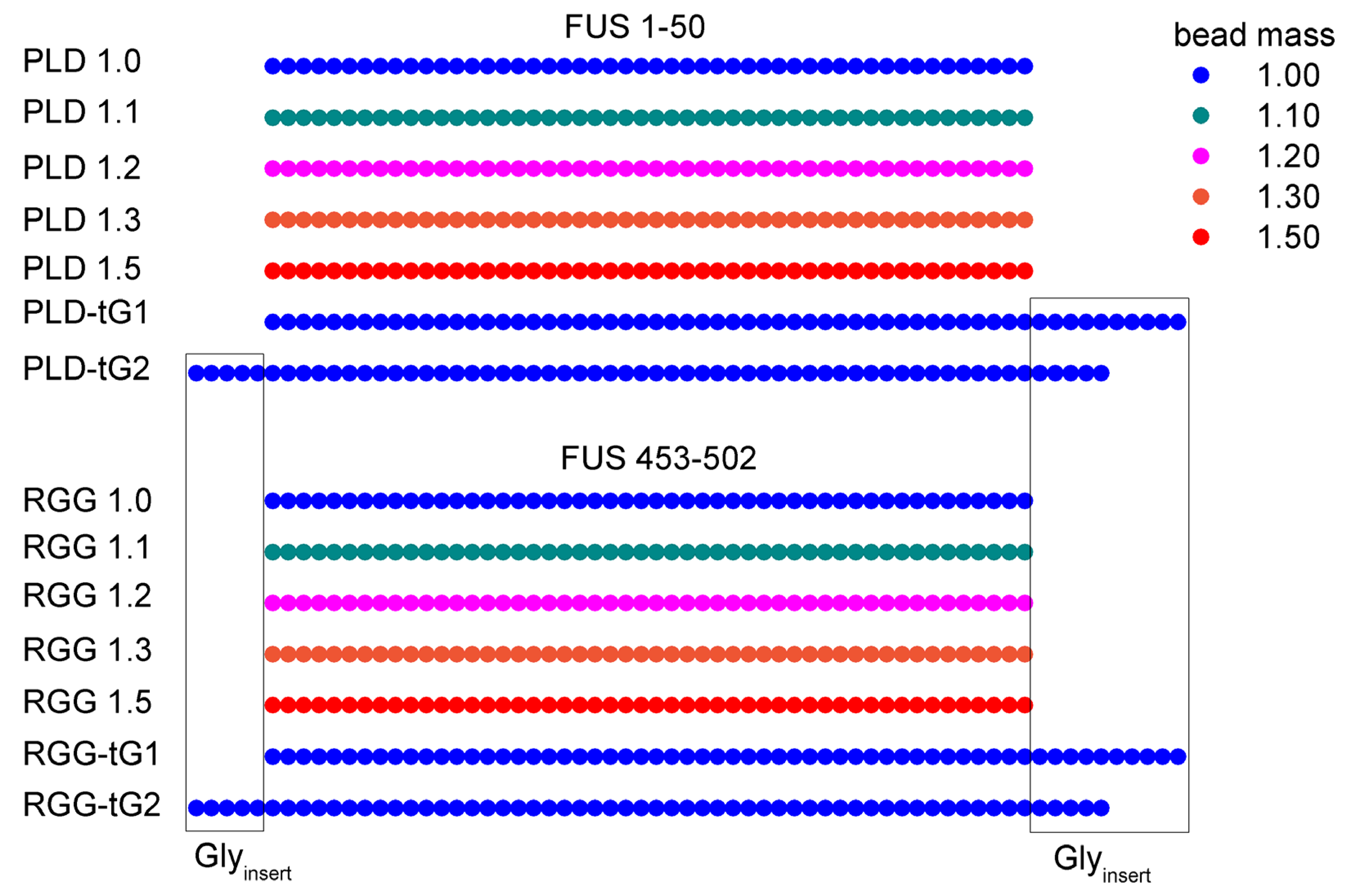
2.1. Simulation System
2.2. Data Analysis
3. Results and Discussion
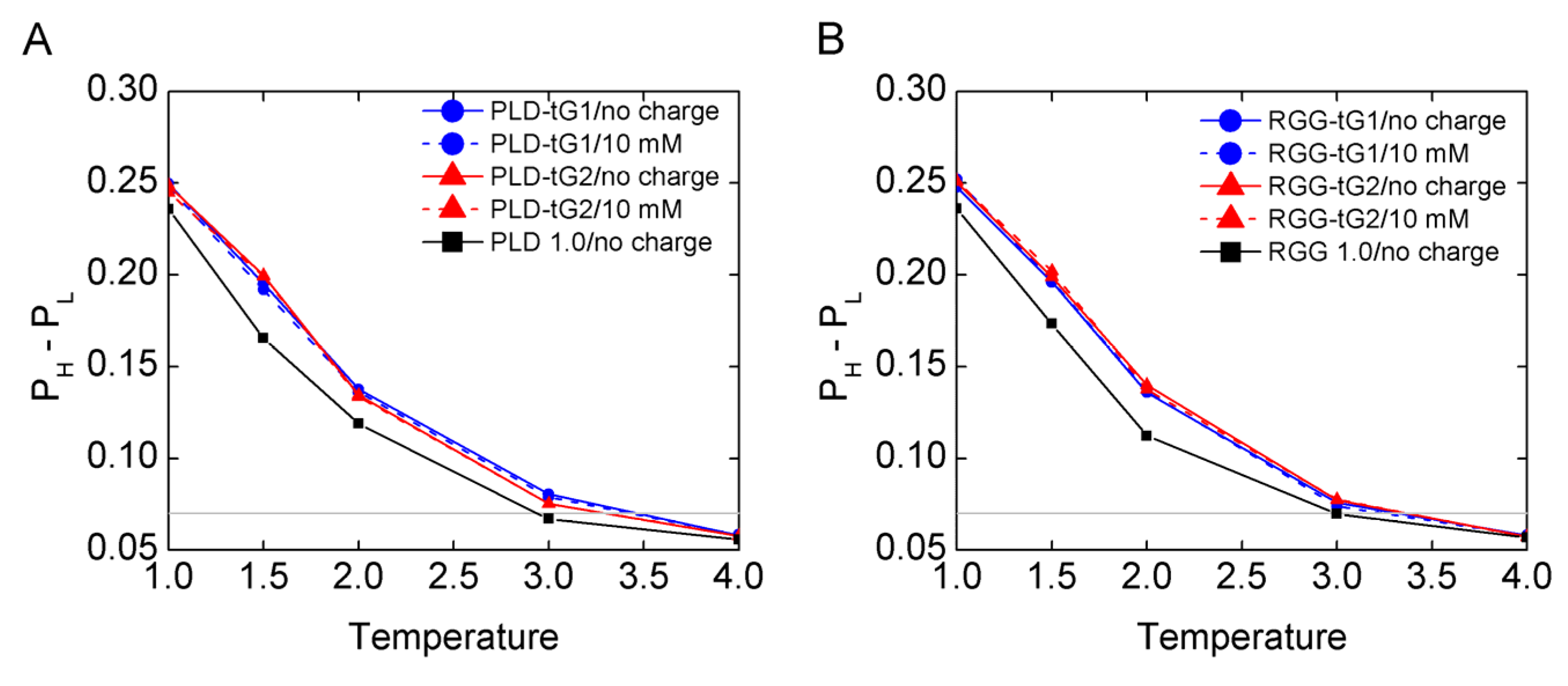
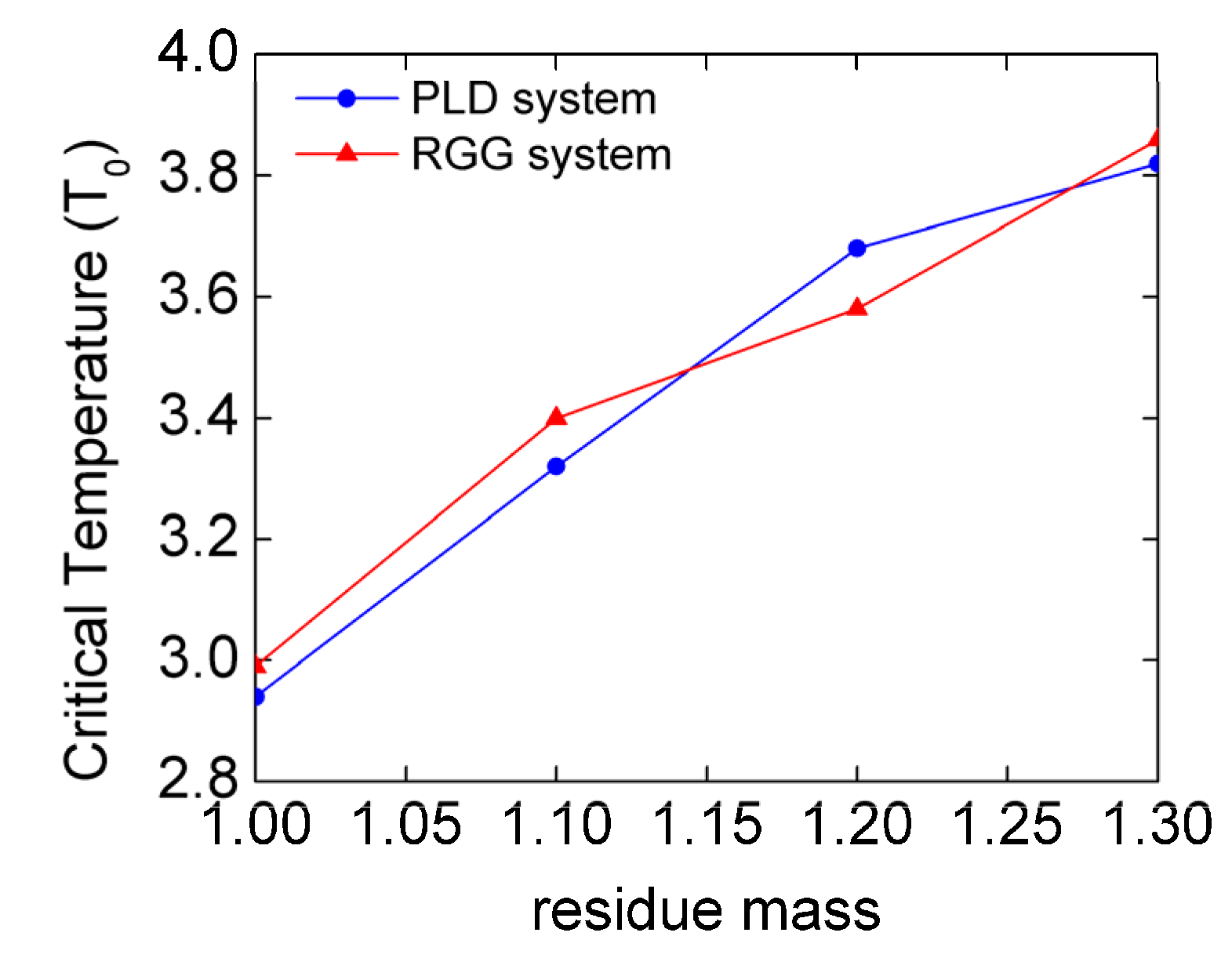
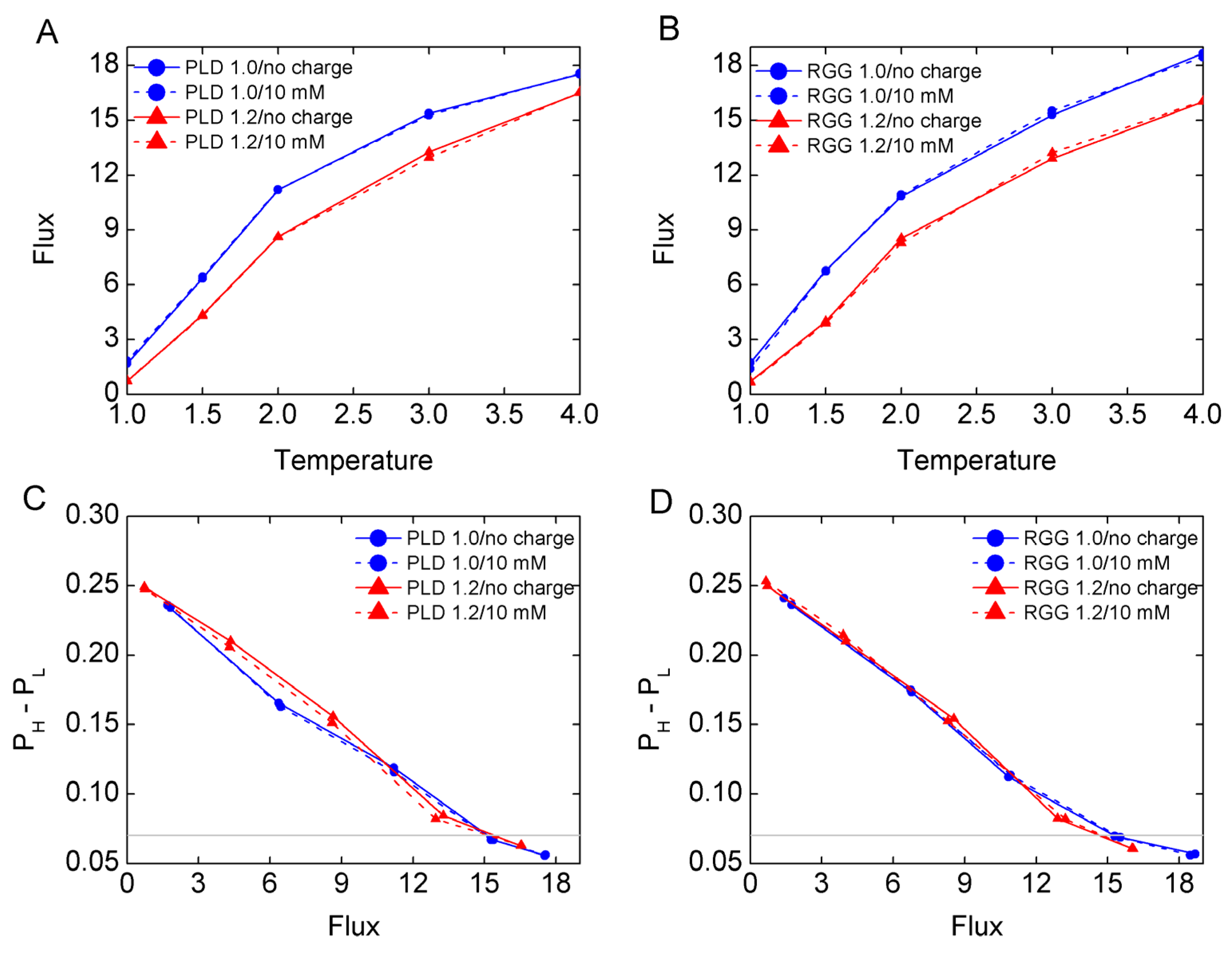
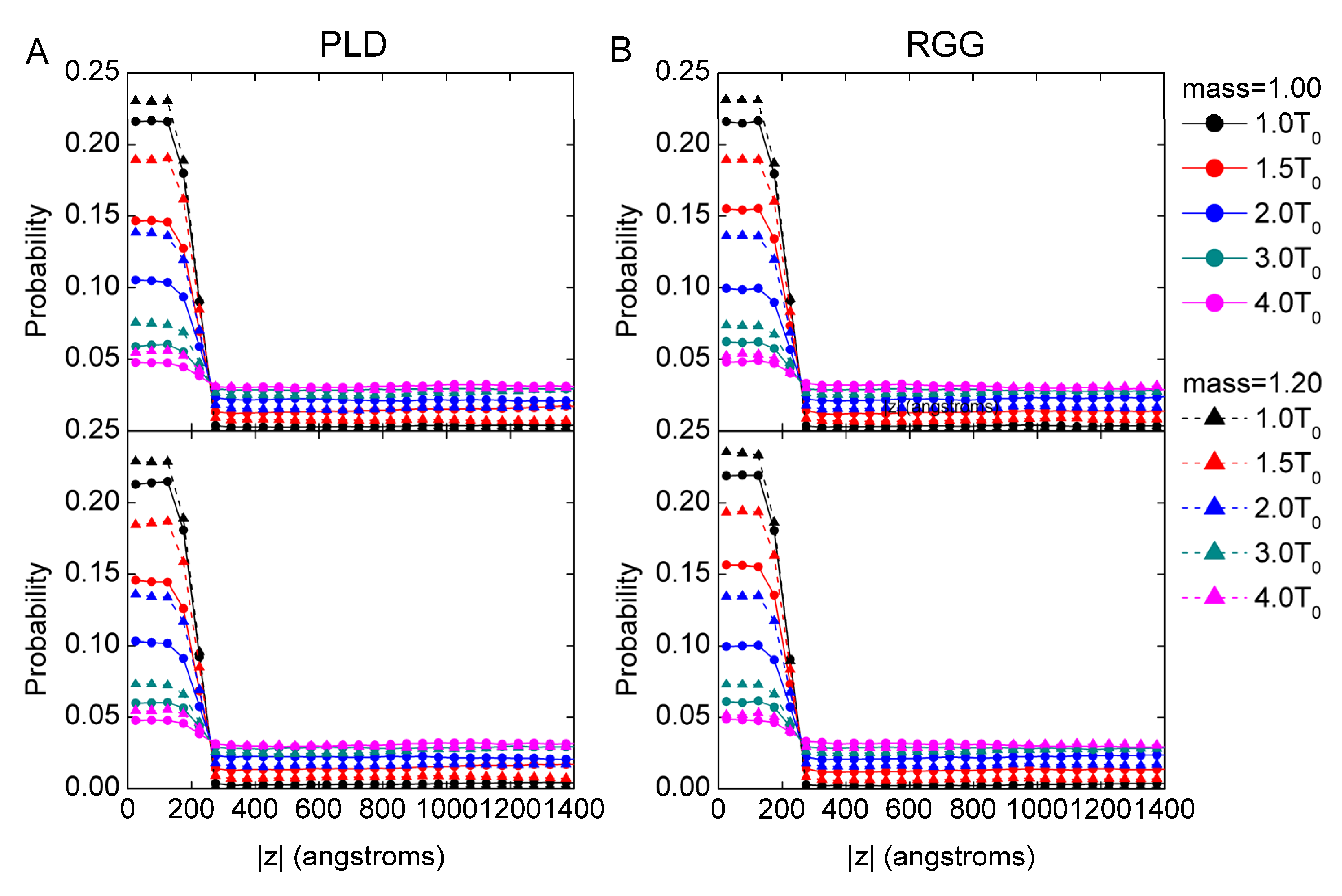
3.1. The Mass Effect on LLPS Stability
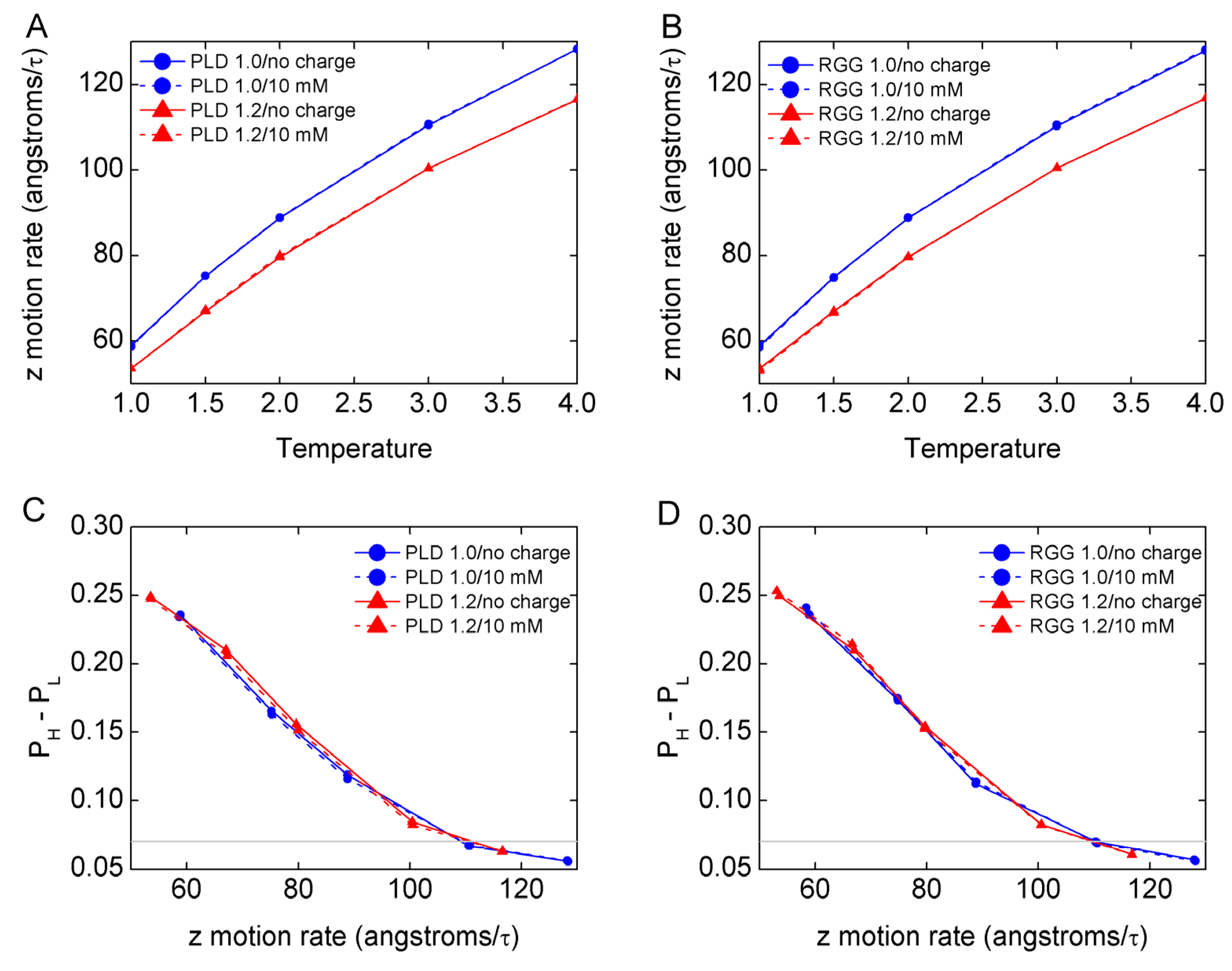
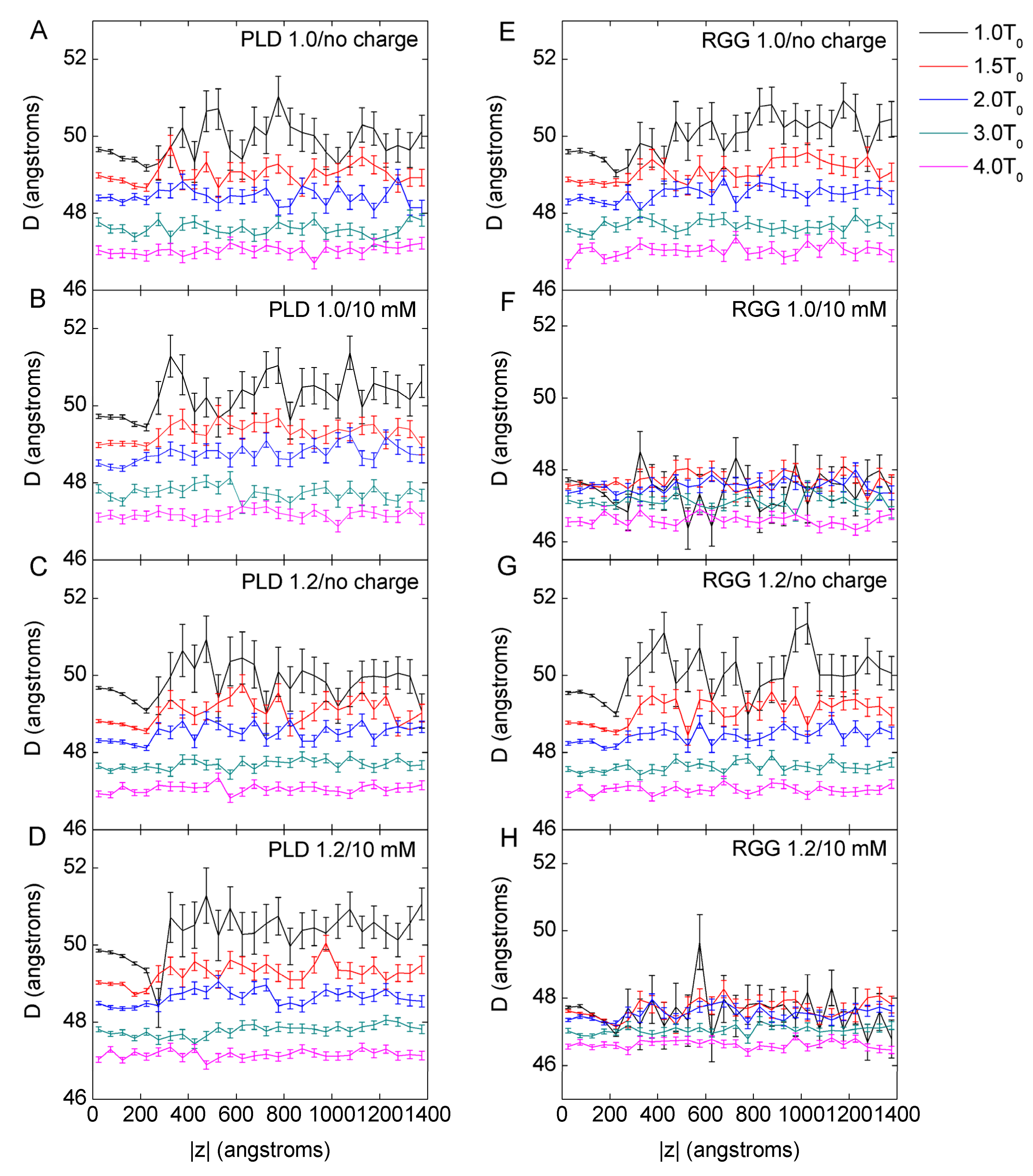
3.2. The Mechanism of Mass Effect on LLPS
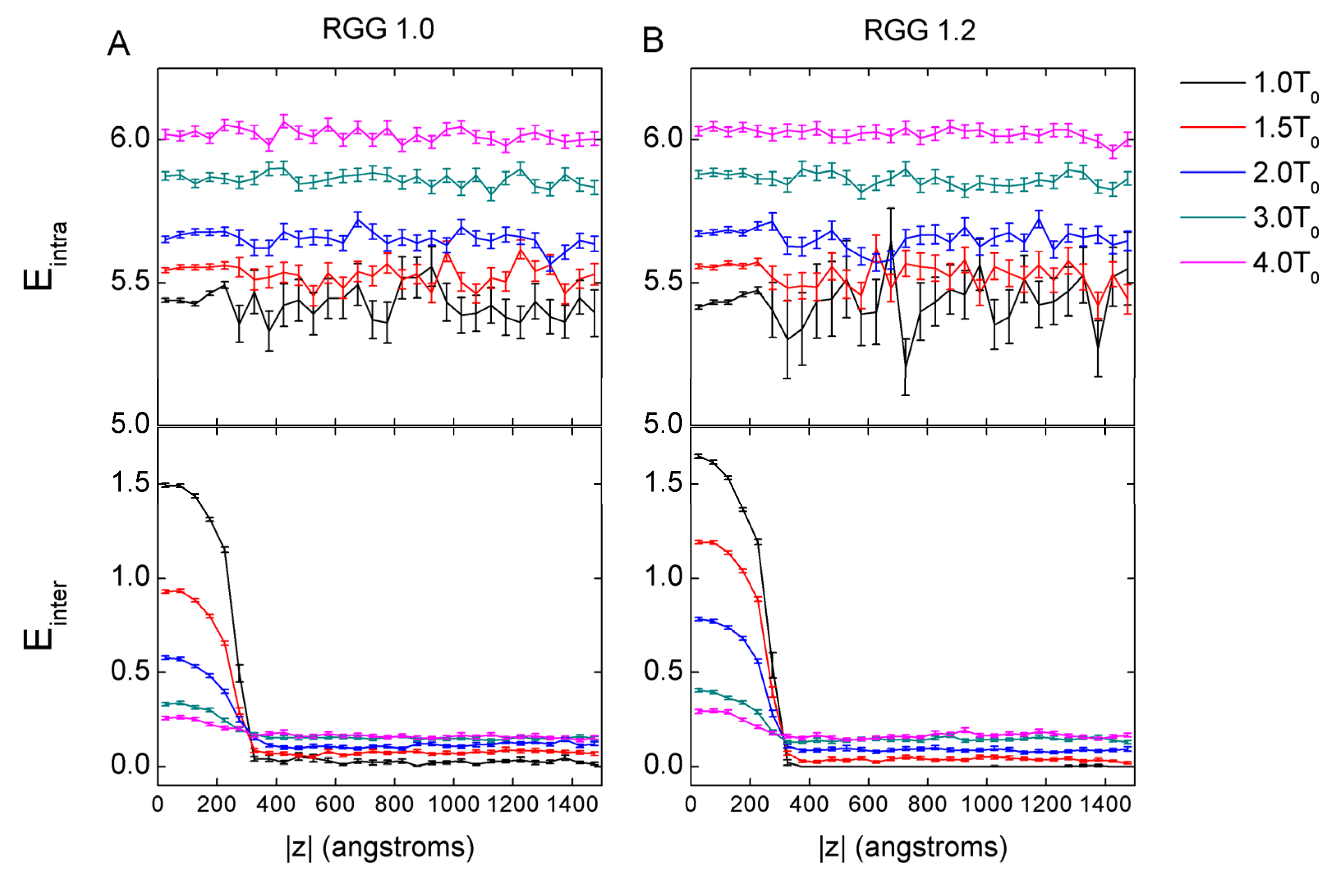
3.3. The Effect of Mass Increase on the Conformation of Chains and the Electrostatic Contact
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- An, H.; de Meritens, C.R.; Shelkovnikova, T.A. Connecting the “dots”: RNP granule network in health and disease. Biochim. Biophys. Acta-Mol. Cell Res. 2021, 1868, 119058. [Google Scholar] [CrossRef] [PubMed]
- Murthy, A.C.; Fawzi, N.L. The (un)structural biology of biomolecular liquid-liquid phase separation using NMR spectroscopy. J. Biol. Chem. 2020, 295, 2375–2384. [Google Scholar] [CrossRef] [PubMed]
- Alberti, S.; Gladfelter, A.; Mittag, T. Considerations and Challenges in Studying Liquid-Liquid Phase Separation and Biomolecular Condensates. Cell 2019, 176, 419–434. [Google Scholar] [CrossRef]
- Sawner, A.S.; Ray, S.; Yadav, P.; Mukherjee, S.; Panigrahi, R.; Poudyal, M.; Patel, K.; Ghosh, D.; Kummerant, E.; Kumar, A.; et al. Modulating alpha-Synuclein Liquid-Liquid Phase Separation. Biochemistry 2021, 60, 3676–3696. [Google Scholar] [CrossRef]
- Spegg, V.; Altmeyer, M. Biomolecular condensates at sites of DNA damage: More than just a phase. DNA Repair 2021, 106, 103179. [Google Scholar] [CrossRef] [PubMed]
- Mann, J.R.; Donnelly, C.J. RNA modulates physiological and neuropathological protein phase transitions. Neuron 2021, 109, 2663–2681. [Google Scholar] [CrossRef]
- Sehgal, P.B.; Westley, J.; Lerea, K.M.; DiSenso-Browne, S.; Etlinger, J.D. Biomolecular condensates in cell biology and virology: Phase-separated membraneless organelles (MLOs). Anal. Biochem. 2020, 597, 113691. [Google Scholar] [CrossRef]
- Garaizar, A.; Sanchez-Burgos, I.; Collepardo-Guevara, R.; Espinosa, J.R. Expansion of Intrinsically Disordered Proteins Increases the Range of Stability of Liquid-Liquid Phase Separation. Molecules 2020, 25, 4705. [Google Scholar] [CrossRef]
- Vodnala, M.; Choi, E.B.; Fong, Y.W. Low complexity domains, condensates, and stem cell pluripotency. World J. Stem Cells 2021, 13, 416–438. [Google Scholar] [CrossRef]
- Reber, S.; Jutzi, D.; Lindsay, H.; Devoy, A.; Mechtersheimer, J.; Levone, B.R.; Domanski, M.; Bentmann, E.; Dormann, D.; Muhlemann, O.; et al. The phase separation-dependent FUS interactome reveals nuclear and cytoplasmic function of liquid-liquid phase separation. Nucleic Acids Res. 2021, 49, 7713–7731. [Google Scholar] [CrossRef]
- Levone, B.R.; Lenzken, S.C.; Antonaci, M.; Maiser, A.; Rapp, A.; Conte, F.; Reber, S.; Mechtersheimer, J.; Ronchi, A.E.; Muhlemann, O.; et al. FUS-dependent liquid-liquid phase separation is important for DNA repair initiation. J. Cell Biol. 2021, 220, e202008030. [Google Scholar] [CrossRef] [PubMed]
- Su, J.M.; Wilson, M.Z.; Samuel, C.E.; Ma, D. Formation and Function of Liquid-Like Viral Factories in Negative-Sense Single-Stranded RNA Virus Infections. Viruses 2021, 13, 126. [Google Scholar] [CrossRef] [PubMed]
- Noda, N.N.; Wang, Z.; Zhang, H. Liquid-Liquid phase separation in autophagy. J. Cell Biol. 2020, 219, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Peng, Q.; Tan, S.M.; Xia, L.Z.; Wu, N.Y.; Oyang, L.; Tang, Y.Y.; Su, M.; Luo, X.; Wang, Y.; Sheng, X.W.; et al. Phase separation in Cancer: From the Impacts and Mechanisms to Treatment potentials. Int. J. Biol. Sci. 2022, 18, 5103–5122. [Google Scholar] [CrossRef] [PubMed]
- Milicevic, K.; Rankovic, B.; Andjus, P.R.; Bataveljic, D.; Milovanovic, D. Emerging Roles for Phase Separation of RNA-Binding Proteins in Cellular Pathology of ALS. Front. Cell Dev. Biol. 2022, 10, 840256. [Google Scholar] [CrossRef]
- Yoneda, R.; Ueda, N.; Kurokawa, R. m(6)A Modified Short RNA Fragments Inhibit Cytoplasmic TLS/FUS Aggregation Induced by Hyperosmotic Stress. Int. J. Mol. Sci. 2021, 22, 11014. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.L.; Shorter, J. Combating deleterious phase transitions in neurodegenerative disease. Biochim. Biophys. Acta-Mol. Cell Res. 2021, 1868, 118984. [Google Scholar] [CrossRef]
- Farina, S.; Esposito, F.; Battistoni, M.; Biamonti, G.; Francia, S. Post-Translational Modifications Modulate Proteinopathies of TDP-43, FUS and hnRNP-A/B in Amyotrophic Lateral Sclerosis. Front. Mol. Biosci. 2021, 8, 693325. [Google Scholar] [CrossRef]
- Hayashi, Y.; Ford, L.K.; Fioriti, L.; McGurk, L.; Zhang, M. Liquid-Liquid Phase Separation in Physiology and Pathophysiology of the Nervous System. J. Neurosci. 2021, 41, 834–844. [Google Scholar] [CrossRef]
- Lin, Y.-H.; Song, J.; Forman-Kay, J.D.; Chan, H.S. Random-phase-approximation theory for sequence-dependent, biologically functional liquid-liquid phase separation of intrinsically disordered proteins. J. Mol. Liq. 2017, 228, 176–193. [Google Scholar] [CrossRef]
- Uversky, V.N. Per aspera ad chaos: A personal journey to the wonderland of intrinsic disorder. Biochem. J. 2021, 478, 3015–3024. [Google Scholar] [CrossRef] [PubMed]
- Dunker, A.K.; Lawson, J.D.; Brown, C.J.; Williams, R.M.; Romero, P.; Oh, J.S.; Oldfield, C.J.; Campen, A.M.; Ratliff, C.M.; Hipps, K.W.; et al. Intrinsically disordered protein. J. Mol. Graph. Model. 2001, 19, 26–59. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Kuznetsova, I.M.; Turoverov, K.K.; Zaslavsky, B. Intrinsically disordered proteins as crucial constituents of cellular aqueous two phase systems and coacervates. FEBS Lett. 2015, 589, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Badaczewska-Dawid, A.E.; Uversky, V.N.; Potoyan, D.A. BIAPSS: A Comprehensive Physicochemical Analyzer of Proteins Undergoing Liquid-Liquid Phase Separation. Int. J. Mol. Sci. 2022, 23, 6204. [Google Scholar] [CrossRef] [PubMed]
- Portz, B.; Lee, B.L.; Shorter, J. FUS and TDP-43 Phases in Health and Disease. Trends Biochem. Sci. 2021, 46, 550–563. [Google Scholar] [CrossRef] [PubMed]
- Ishiguro, A.; Ishihama, A. Essential Roles and Risks of G-Quadruplex Regulation: Recognition Targets of ALS-Linked TDP-43 and FUS. Front. Mol. Biosci. 2022, 9, 957502. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Lim, L.Z.; Song, J.X. ATP enhances at low concentrations but dissolves at high concentrations liquid-liquid phase separation (LLPS) of ALS/FTD-causing FUS. Biochem. Biophys. Res. Commun. 2018, 504, 545–551. [Google Scholar] [CrossRef]
- Aida, H.; Shigeta, Y.; Harada, R. The role of ATP in solubilizing RNA-binding protein fused in sarcoma. Proteins 2022, 90, 1606–1612. [Google Scholar] [CrossRef]
- Lao, Z.; Dong, X.; Liu, X.; Li, F.; Chen, Y.; Tang, Y.; Wei, G. Insights into the Atomistic Mechanisms of Phosphorylation in Disrupting Liquid-Liquid Phase Separation and Aggregation of the FUS Low-Complexity Domain. J. Chem. Inf. Model 2022, 62, 3227–3238. [Google Scholar] [CrossRef]
- Bock, A.S.; Murthy, A.C.; Tang, W.S.; Jovic, N.; Shewmaker, F.; Mittal, J.; Fawzi, N.L. N-terminal acetylation modestly enhances phase separation and reduces aggregation of the low-complexity domain of RNA-binding protein fused in sarcoma. Protein Sci. 2021, 30, 1337–1349. [Google Scholar] [CrossRef]
- Yoshizawa, T.; Ali, R.; Jiou, J.; Fung, H.Y.J.; Burke, K.A.; Kim, S.J.; Lin, Y.; Peeples, W.B.; Saltzberg, D.; Soniat, M.; et al. Nuclear Import Receptor Inhibits Phase Separation of FUS through Binding to Multiple Sites. Cell 2018, 173, 693–705.e22. [Google Scholar] [CrossRef]
- Kaur, T.; Alshareedah, I.; Wang, W.; Ngo, J.; Moosa, M.M.; Banerjee, P.R. Molecular Crowding Tunes Material States of Ribonucleoprotein Condensates. Biomolecules 2019, 9, 71. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Bera, S.; Qiao, Q.; Tang, Y.; Lao, Z.; Luo, Y.; Gazit, E.; Wei, G. Liquid-Liquid Phase Separation of Tau Protein Is Encoded at the Monomeric Level. J. Phys. Chem. Lett. 2021, 12, 2576–2586. [Google Scholar] [CrossRef]
- Dignon, G.L.; Zheng, W.; Kim, Y.C.; Mittal, J. Temperature-Controlled Liquid-Liquid Phase Separation of Disordered Proteins. ACS Cent. Sci. 2019, 5, 821–830. [Google Scholar] [CrossRef] [PubMed]
- Maity, H.; Baidya, L.; Reddy, G. Salt-Induced Transitions in the Conformational Ensembles of Intrinsically Disordered Proteins. J. Phys. Chem. B 2022, 126, 5959–5971. [Google Scholar] [CrossRef] [PubMed]
- Li, S.J.; Yoshizawa, T.; Yamazaki, R.; Fujiwara, A.; Kameda, T.; Kitahara, R. Pressure and Temperature Phase Diagram for Liquid-Liquid Phase Separation of the RNA-Binding Protein Fused in Sarcoma. J. Phys. Chem. B 2021, 125, 6821–6829. [Google Scholar] [CrossRef]
- Kitahara, R.; Yamazaki, R.; Ide, F.; Li, S.J.; Shiramasa, Y.; Sasahara, N.; Yoshizawa, T. Pressure-Jump Kinetics of Liquid-Liquid Phase Separation: Comparison of Two Different Condensed Phases of the RNA-Binding Protein, Fused in Sarcoma. J. Am. Chem. Soc. 2021, 143, 19697–19702. [Google Scholar] [CrossRef]
- Burke, K.A.; Janke, A.M.; Rhine, C.L.; Fawzi, N.L. Residue-by-Residue View of In Vitro FUS Granules that Bind the C-Terminal Domain of RNA Polymerase II. Mol. Cell 2015, 60, 231–241. [Google Scholar] [CrossRef]
- Bari, K.J.; Prakashchand, D.D. Fundamental Challenges and Outlook in Simulating Liquid-Liquid Phase Separation of Intrinsically Disordered Proteins. J. Phys. Chem. Lett. 2021, 12, 1644–1656. [Google Scholar] [CrossRef]
- Zheng, W.; Dignon, G.L.; Jovic, N.; Xu, X.; Regy, R.M.; Fawzi, N.L.; Kim, Y.C.; Best, R.B.; Mittal, J. Molecular Details of Protein Condensates Probed by Microsecond Long Atomistic Simulations. J. Phys. Chem. B 2020, 124, 11671–11679. [Google Scholar] [CrossRef]
- Dignon, G.L.; Zheng, W.; Kim, Y.C.; Best, R.B.; Mittal, J. Sequence determinants of protein phase behavior from a coarse-grained model. PLoS Comput. Biol. 2018, 14, e1005941. [Google Scholar] [CrossRef] [PubMed]
- Dignon, G.L.; Zheng, W.; Best, R.B.; Kim, Y.C.; Mittal, J. Relation between single-molecule properties and phase behavior of intrinsically disordered proteins. Proc. Natl. Acad Sci. USA 2018, 115, 9929–9934. [Google Scholar] [CrossRef] [PubMed]
- Dannenhoffer-Lafage, T.; Best, R.B. A Data-Driven Hydrophobicity Scale for Predicting Liquid-Liquid Phase Separation of Proteins. J. Phys. Chem. B 2021, 125, 4046–4056. [Google Scholar] [CrossRef]
- Lin, X.; Kulkarni, P.; Bocci, F.; Schafer, N.P.; Roy, S.; Tsai, M.Y.; He, Y.; Chen, Y.; Rajagopalan, K.; Mooney, S.M.; et al. Structural and Dynamical Order of a Disordered Protein: Molecular Insights into Conformational Switching of PAGE4 at the Systems Level. Biomolecules 2019, 9, 77. [Google Scholar] [CrossRef] [PubMed]
- Lenard, A.J.; Zhou, Q.; Madreiter-Sokolowski, C.; Bourgeois, B.; Habacher, H.; Khanna, Y.; Madl, T. EGCG Promotes FUS Condensate Formation in a Methylation-Dependent Manner. Cells 2022, 11, 592. [Google Scholar] [CrossRef]
- Noel, J.K.; Whitford, P.C.; Sanbonmatsu, K.Y.; Onuchic, J.N. SMOG@ctbp: Simplified deployment of structure-based models in GROMACS. Nucleic Acids Res. 2010, 38, W657–W661. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Muthukumar, M. Microstructural Organization in α-Synuclein Solutions. Macromolecules 2022, 55, 4228–4236. [Google Scholar] [CrossRef]
- Chu, W.T.; Wang, J. Thermodynamic and sequential characteristics of phase separation and droplet formation for an intrinsically disordered region/protein ensemble. PLoS Comput. Biol. 2021, 17, e1008672. [Google Scholar] [CrossRef]
- Hazra, M.K.; Levy, Y. Charge pattern affects the structure and dynamics of polyampholyte condensates. Phys. Chem. Chem. Phys. 2020, 22, 19368–19375. [Google Scholar] [CrossRef]
- Silmore, K.S.; Howard, M.P.; Panagiotopoulos, A.Z. Vapour-liquid phase equilibrium and surface tension of fully flexible Lennard-Jones chains. Mol. Phys. 2017, 115, 320–327. [Google Scholar] [CrossRef]
- Kouza, M.; Li, M.S.; O’Brien, E.P.J.; Hu, C.K.; Thirumalai, D. Effect of finite size on cooperativity and rates of protein folding. J. Phys. Chem. A 2006, 110, 671–676. [Google Scholar] [CrossRef] [PubMed]
- Jackson, J.; Nguyen, K.; Whitford, P.C. Exploring the balance between folding and functional dynamics in proteins and RNA. Int. J. Mol. Sci. 2015, 16, 6868–6889. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, W.; Tang, C.; Chu, W.-T.; Wang, E.; Wang, J. Effects of Mass Change on Liquid–Liquid Phase Separation of the RNA-Binding Protein Fused in Sarcoma. Biomolecules 2023, 13, 625. https://doi.org/10.3390/biom13040625
Dong W, Tang C, Chu W-T, Wang E, Wang J. Effects of Mass Change on Liquid–Liquid Phase Separation of the RNA-Binding Protein Fused in Sarcoma. Biomolecules. 2023; 13(4):625. https://doi.org/10.3390/biom13040625
Chicago/Turabian StyleDong, Weiqian, Chun Tang, Wen-Ting Chu, Erkang Wang, and Jin Wang. 2023. "Effects of Mass Change on Liquid–Liquid Phase Separation of the RNA-Binding Protein Fused in Sarcoma" Biomolecules 13, no. 4: 625. https://doi.org/10.3390/biom13040625
APA StyleDong, W., Tang, C., Chu, W.-T., Wang, E., & Wang, J. (2023). Effects of Mass Change on Liquid–Liquid Phase Separation of the RNA-Binding Protein Fused in Sarcoma. Biomolecules, 13(4), 625. https://doi.org/10.3390/biom13040625







