Purification and Biological Properties of Raniseptins-3 and -6, Two Antimicrobial Peptides from Boana raniceps (Cope, 1862) Skin Secretion
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Skin Secretion
2.2. Fractionation of Crude Secretion by Reverse-Phase High-Performance Liquid Chromatography (RP-HPLC)
2.3. Purification of Antimicrobial Peptides
2.4. Mass Spectrometry
2.5. Chemical Sequencing
2.6. Circular Dichroism
2.7. Bioinformatic Analysis
2.8. Solid-Phase Synthesis and Purification
2.9. Quantification of Peptides
2.10. Antibacterial Activity
2.11. Antifungical Activity
2.12. Antiproliferative Activity
2.13. Human Hemolysis Activity
2.14. Scanning Electron Microscopy (SEM)
2.15. Infection and Viability Tests Protocol
3. Results
3.1. Purification of Antimicrobial Peptides from B. raniceps
3.2. Structural Characterization of Antimicrobial Peptides from B. raniceps
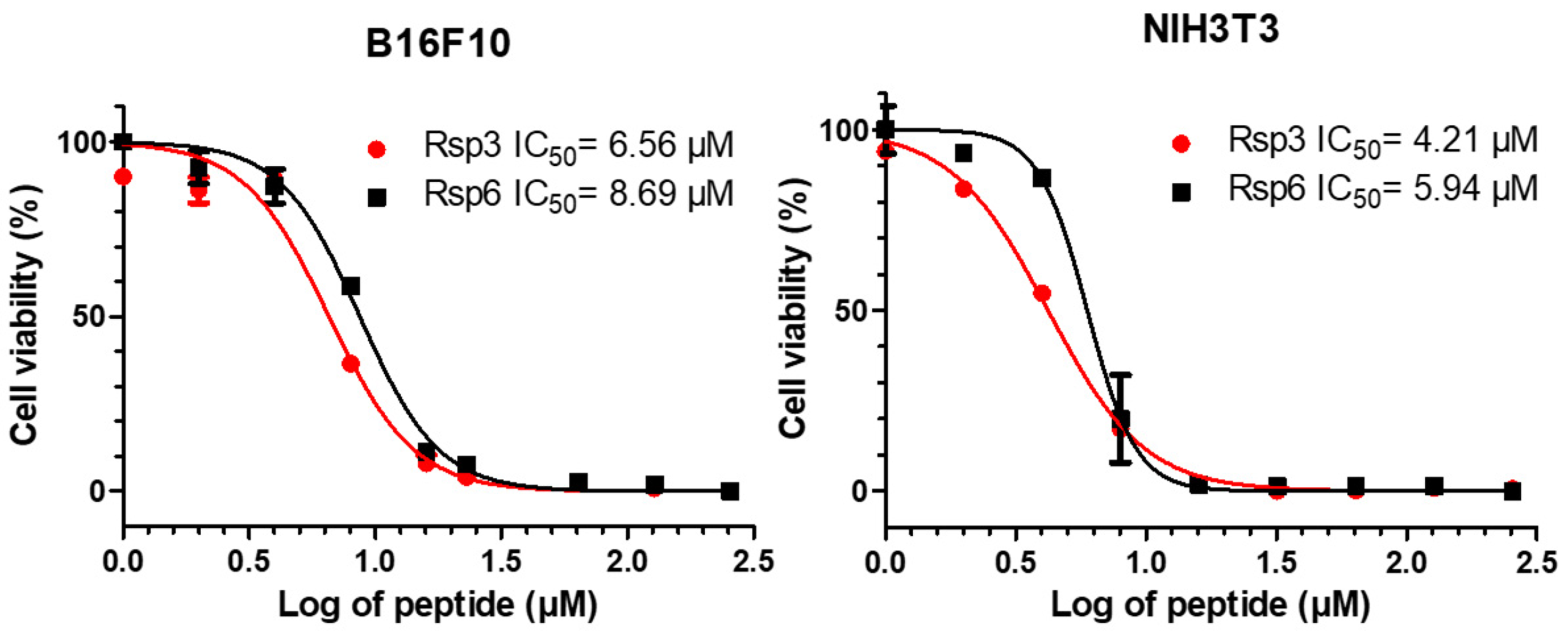
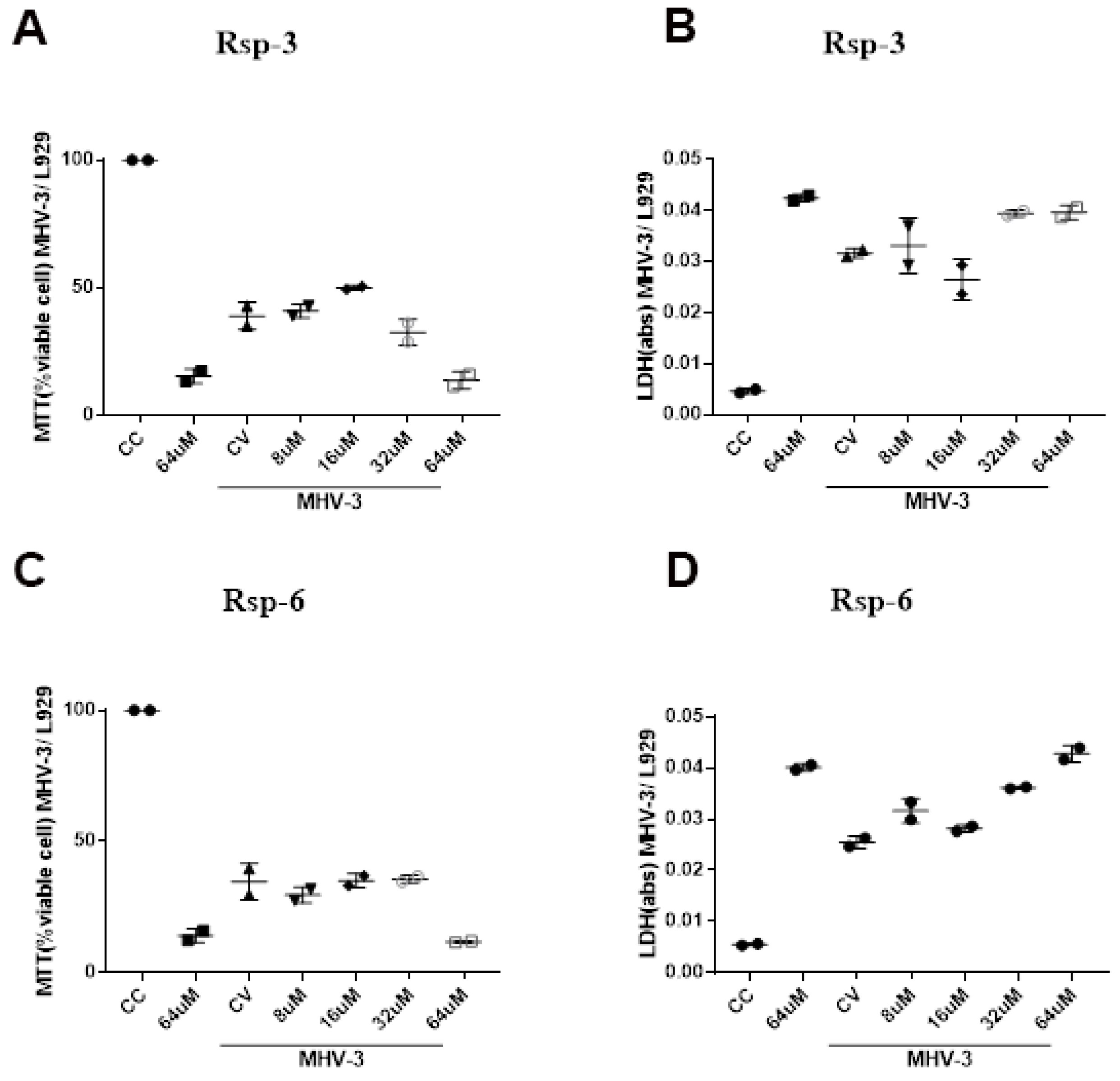
3.3. Biological Characterization of Antimicrobial Peptides from B. raniceps
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial Resistance: A Global Multifaceted Phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef]
- Roca, I.; Akova, M.; Baquero, F.; Carlet, J.; Cavaleri, M.; Coenen, S.; Cohen, J.; Findlay, D.; Gyssens, I.; Heure, O.E.; et al. The Global Threat of Antimicrobial Resistance: Science for Intervention. New Microbes New Infect. 2015, 6, 22–29. [Google Scholar] [CrossRef]
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Iskandar, K.; Murugaiyan, J.; Halat, D.H.; El Hage, S.; Chibabhai, V.; Adukkadukkam, S.; Roques, C.; Molinier, L.; Salameh, P.; Van Dongen, M. Antibiotic Discovery and Resistance: The Chase and the Race. Antibiotics 2022, 11, 182. [Google Scholar] [CrossRef] [PubMed]
- Ventola, C.L. The Antibiotic Resistance Crisis: Part 1: Causes and Threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- Laxminarayan, R.; Matsoso, P.; Pant, S.; Brower, C.; Røttingen, J.-A.; Klugman, K.; Davies, S. Access to Effective Antimicrobials: A Worldwide Challenge. Lancet 2016, 387, 168–175. [Google Scholar] [CrossRef] [PubMed]
- Le, C.-F.; Fang, C.-M.; Sekaran, S.D. Intracellular Targeting Mechanisms by Antimicrobial Peptides. Antimicrob. Agents Chemother. 2017, 61, e02340-16. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, A.C.; Fontes, W.; Sebben, A.; Castro, M. Antimicrobial Peptides from Anurans Skin Secretions. Protein Pept. Lett. 2003, 10, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Ladram, A.; Nicolas, P. Antimicrobial Peptides from Frog Skin: Biodiversity and Therapeutic Promises. Front. Biosci. 2016, 21, 1341–1371. [Google Scholar] [CrossRef]
- Patocka, J.; Nepovimova, E.; Klimova, B.; Wu, Q.; Kuca, K. Antimicrobial Peptides: Amphibian Host Defense Peptides. Curr. Med. Chem. 2019, 26, 5924–5946. [Google Scholar] [CrossRef] [PubMed]
- Conlon, J.M.; Mechkarska, M.; Leprince, J. Peptidomic Analysis in the Discovery of Therapeutically Valuable Peptides in Amphibian Skin Secretions. Expert Rev. Proteom. 2019, 16, 897–908. [Google Scholar] [CrossRef]
- Romero, S.M.; Cardillo, A.B.; Martínez Ceron, M.C.; Camperi, S.A.; Giudicessi, S.L. Temporins: An Approach of Potential Pharmaceutic Candidates. Surg. Infect. 2020, 21, 309–322. [Google Scholar] [CrossRef] [PubMed]
- Conlon, J.M. Structural Diversity and Species Distribution of Host-Defense Peptides in Frog Skin Secretions. Cell. Mol. Life Sci. 2011, 68, 2303–2315. [Google Scholar] [CrossRef]
- Mojsoska, B.; Jenssen, H. Peptides and Peptidomimetics for Antimicrobial Drug Design. Pharmaceuticals 2015, 8, 366–415. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Cha, H.J. Disperse Distribution of Cationic Amino Acids on Hydrophilic Surface of Helical Wheel Enhances Antimicrobial Peptide Activity. Biotechnol. Bioeng. 2010, 107, 216–223. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.-J.; Gallo, R.L. Antimicrobial Peptides. Curr. Biol. 2016, 26, R14–R19. [Google Scholar] [CrossRef]
- Almeida, R.A.; Gordo, M.; Da Silva, F.M.A.; De Araújo, R.C.; Ramada, M.H.S.; Abrão, F.Y.; Costa, T.O.G.; Koolen, H.H.F.; De Souza, A.D.; Bloch, C., Jr. Cinerascetins, New Peptides from Hypsiboas cinerascens: MALDI LIFT-TOF-MS/MS de novo Sequence and Imaging Analysis. J. Braz. Chem. Soc. 2015, 26, 2290–2297. [Google Scholar] [CrossRef]
- Nacif-Marçal, L.; Pereira, G.R.; Abranches, M.V.; Costa, N.C.; Cardoso, S.A.; Honda, E.R.; de Paula, S.O.; Feio, R.N.; Oliveira, L.L. Identification and Characterization of an Antimicrobial Peptide of Hypsiboas semilineatus (Spix, 1824) (Amphibia, Hylidae). Toxicon 2015, 99, 16–22. [Google Scholar] [CrossRef]
- Vaz-Silva, W.; Maciel, N.M.; Nomura, F.; de Morais, A.R.; Batista, V.G.; Santos, D.L.; Andrade, S.P.; de Oliveira, A.Â.B.; Brandão, R.A.; Bastos, R.P. Guia de Identificação das Espécies de Anfíbios (Anura e Gymnophiona) do Estado de Goiás e do Distrito Federal, Brasil Central; Sociedade Brasileira de Zoologia: Curitiba, Brazil, 2020; ISBN 9786587590011. [Google Scholar]
- Frost, D.R. Amphibian Species of the World: An Online Reference. Version 6.1. Available online: https://amphibiansoftheworld.amnh.org/index.php (accessed on 7 June 2022).
- La Marca, E.; Azevedodo-Ramos, C.; Silvano, D.; Scott, N.; Aquino, L.; Faivovich, J. Hypsiboas raniceps. The IUCN Red List of Threatened Species. Available online: https://dx.doi.org/10.2305/IUCN.UK.2004.RLTS.T55622A11341908.en (accessed on 8 June 2022).
- Fouquet, A.; Gilles, A.; Vences, M.; Marty, C.; Blanc, M.; Gemmell, N. Underestimation of Species Richness in Neotropical Frogs Revealed by MtDNA Analyses. PLoS ONE 2007, 2, e1109. [Google Scholar] [CrossRef]
- Greenfield, N.J.; Fasman, G.D. Computed Circular Dichroism Spectra for the Evaluation of Protein Conformation. Biochemistry 1969, 8, 4108–4116. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Li, X.; Wang, Z. APD3: The Antimicrobial Peptide Database as a Tool for Research and Education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef] [PubMed]
- Needleman, S.B.; Wunsch, C.D. A General Method Applicable to the Search for Similarities in the Amino Acid Sequence of Two Proteins. J. Mol. Biol. 1970, 48, 443–453. [Google Scholar] [CrossRef] [PubMed]
- Mol, A.R.; Castro, M.S.; Fontes, W. NetWheels: A Web Application to Create High Quality Peptide Helical Wheel and Net Projections. BioRxiv 2018, 416347. [Google Scholar]
- Geourjon, C.; Deléage, G. SOPMA: Significant Improvements in Protein Secondary Structure Prediction by Consensus Prediction from Multiple Alignments. Bioinformatics 1995, 11, 681–684. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Chan, W.; White, P. (Eds.) Fmoc Solid Phase Peptide Synthesis; Oxford University Press: Oxford, UK, 1999; ISBN 9780199637256. [Google Scholar]
- Silva, L.P.; Leite, J.R.; Brand, G.D.; Regis, W.; Tedesco, A.; Azevedo, R.B.; Freitas, S.M.; Bloch, C. Dermaseptins from Phyllomedusa oreades and Phyllomedusa distincta: Liposomes Fusion and/or Lysis Investigated by Fluorescence and Atomic Force Microscopy. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2008, 151, 329–335. [Google Scholar] [CrossRef]
- Aitken, A.; Learmonth, M.P. Protein Determination by UV Absorption. In The Protein Protocols Handbook; Humana Press: Totowa, NJ, USA, 1996; pp. 3–6. [Google Scholar]
- CLSI Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Document M7-A6; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2003.
- CLSI Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts, 3rd ed.; Document M27-A3; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2008.
- Libério, M.S.; Joanitti, G.; Azevedo, R.B.; Cilli, E.M.; Zanotta, L.C.; Nascimento, A.C.; de Sousa, M.V.; Júnior, O.R.P.; Fontes, W.; Castro, M.S. Anti-Proliferative and Cytotoxic Activity of Pentadactylin Isolated from Leptodactylus labyrinthicus on Melanoma Cells. Amino Acids 2011, 40, 51–59. [Google Scholar] [CrossRef]
- Santana, C.J.C.; Magalhães, A.C.M.; Prías-Márquez, C.A.; Falico, D.A.; Júnior, A.C.M.D.S.; Lima, B.D.; Ricart, C.A.O.; De Pilger, D.R.B.; Bonotto, R.M.; Moraes, C.B.; et al. Biological Properties of a Novel Multifunctional Host Defense Peptide from the Skin Secretion of the Chaco Tree Frog, Boana raniceps. Biomolecules 2020, 10, 790. [Google Scholar] [CrossRef]
- Garcia, A.B.; de Moraes, A.P.; Rodrigues, D.M.; Gilioli, R.; de Oliveira-Filho, E.F.; Durães-Carvalho, R.; Arns, C.W. Coding-Complete Genome Sequence of Murine Hepatitis Virus Strain 3 from Brazil. Microbiol. Resour. Announc. 2021, 10, e00248-21. [Google Scholar] [CrossRef]
- Magalhães, B.S.; Melo, J.A.; Leite, J.R.S.; Silva, L.P.; Prates, M.V.; Vinecky, F.; Barbosa, E.A.; Verly, R.M.; Mehta, A.; Nicoli, J.R.; et al. Post-Secretory Events Alter the Peptide Content of the Skin Secretion of Hypsiboas raniceps. Biochem. Biophys. Res. Commun. 2008, 377, 1057–1061. [Google Scholar] [CrossRef]
- Barbosa, E. Avaliação Da Transcrição, Expressão e Indução de Genes Que Codificam Peptídeos Antimicrobianos Em Hypsiboas raniceps Por Ferramentas de Biologia Molecular e Espectrometria de Massa. Master’s Thesis, University of Brasilia, Brasilia, Brazil, 2010. [Google Scholar]
- Santana, C.J.C.; Magalhães, A.C.M.; Júnior, A.C.M.D.S.; Ricart, C.A.O.; Lima, B.D.; Álvares, A.D.C.M.; De Freitas, S.M.; Pires, J.O.R.; Fontes, W.; Castro, M.S. Figainin 1, a Novel Amphibian Skin Peptide with Antimicrobial and Antiproliferative Properties. Antibiotics 2020, 9, 625. [Google Scholar] [CrossRef] [PubMed]
- Prates, M.V.; Sforça, M.L.; Regis, W.C.B.; Leite, J.R.S.A.; Silva, L.P.; Pertinhez, T.A.; Araújo, A.L.T.; Azevedo, R.B.; Spisni, A.; Bloch, C. The NMR-derived Solution Structure of a New Cationic Antimicrobial Peptide from the Skin Secretion of the Anuran Hyla punctata. J. Biol. Chem. 2004, 279, 13018–13026. [Google Scholar] [CrossRef]
- Castro, M.S.; Ferreira, T.C.G.; Cilli, E.M.; Crusca, E.; Mendes-Giannini, M.J.S.; Sebben, A.; Ricart, C.A.O.; Sousa, M.V.; Fontes, W. Hylin a1, the First Cytolytic Peptide Isolated from the Arboreal South American Frog Hypsiboas albopunctatus (“spotted treefrog”). Peptides 2009, 30, 291–296. [Google Scholar] [CrossRef]
- Bechinger, B.; Gorr, S.-U. Antimicrobial Peptides: Mechanisms of Action and Resistance. J. Dent. Res. 2017, 96, 254–260. [Google Scholar] [CrossRef] [PubMed]
- Kyte, J.; Doolittle, R.F. A Simple Method for Displaying the Hydropathic Character of a Protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef] [PubMed]
- Neelay, O.P.; Peterson, C.A.; Snavely, M.E.; Brown, T.C.; TecleMariam, A.F.; Campbell, J.A.; Blake, A.M.; Schneider, S.C.; Cremeens, M.E. Antimicrobial Peptides Interact with Peptidoglycan. J. Mol. Struct. 2017, 1146, 329–336. [Google Scholar] [CrossRef]
- Li, J.; Koh, J.J.; Liu, S.; Lakshminarayanan, R.; Verma, C.S.; Beuerman, R.W. Membrane Active Antimicrobial Peptides: Translating Mechanistic Insights to Design. Front Neurosci. 2017, 11, 73. [Google Scholar] [CrossRef]
- Wang, G. (Ed.) Antimicrobial Peptides: Discovery, Design and Novel Therapeutic Strategies; CABI: Wallingford, UK, 2010; ISBN 9781845936570. [Google Scholar]
- Wang, Y.; Zhang, Y.; Lee, W.-H.; Yang, X.; Zhang, Y. Novel Peptides from Skins of Amphibians Showed Broad-Spectrum Antimicrobial Activities. Chem. Biol. Drug Des. 2016, 87, 419–424. [Google Scholar] [CrossRef]
- Blondelle, S.E.; Lohner, K.; Aguilar, M.-I. Lipid-Induced Conformation and Lipid-Binding Properties of Cytolytic and Antimicrobial Peptides: Determination and Biological Specificity. Biochim. Biophys. Acta (BBA) Biomembr. 1999, 1462, 89–108. [Google Scholar] [CrossRef]
- Dennison, S.R.; Harris, F.; Mura, M.; Phoenix, D.A. An Atlas of Anionic Antimicrobial Peptides from Amphibians. Curr. Protein Pept. Sci. 2018, 19, 823–838. [Google Scholar] [CrossRef]
- Oelkrug, C.; Hartke, M.; Schubert, A. Mode of Action of Anticancer Peptides (ACPs) from Amphibian Origin. Anticancer Res. 2015, 35, 635–643. [Google Scholar]
- Hoskin, D.W.; Ramamoorthy, A. Studies on Anticancer Activities of Antimicrobial Peptides. Biochim. Biophys. Acta (BBA) Biomembr. 2008, 1778, 357–375. [Google Scholar] [CrossRef]
- Conlon, J.M.; Woodhams, D.C.; Raza, H.; Coquet, L.; Leprince, J.; Jouenne, T.; Vaudry, H.; Rollins-Smith, L.A. Peptides with Differential Cytolytic Activity from Skin Secretions of the Lemur Leaf Frog Hylomantis lemur (Hylidae: Phyllomedusinae). Toxicon 2007, 50, 498–506. [Google Scholar] [CrossRef] [PubMed]
- Elnagdy, S.; AlKhazindar, M. The Potential of Antimicrobial Peptides as an Antiviral Therapy against COVID-19. ACS Pharmacol. Transl. Sci. 2020, 3, 780–782. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.; Siman-Tov, G.; Hall, G.; Bhalla, N.; Narayanan, A. Human Antimicrobial Peptides as Therapeutics for Viral Infections. Viruses 2019, 11, 704. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Guan, W.J.; Ni, Z.Y.; Hu, Y.; Liang, W.H.; Qu, C.Q.; He, J.X.; Liu, L.; Shan, H.; Lei, C.L.; Hui, D.S.C.; et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.-L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021, 19, 141–154. [Google Scholar] [CrossRef]
- Oliveira, G.P.; Kroon, E.G. Mouse Hepatitis Virus: A Betacoronavirus Model to Study the Virucidal Activity of Air Disinfection Equipment on Surface Contamination. J. Virol. Methods 2021, 297, 114274. [Google Scholar] [CrossRef]
- Makowski, M.; Silva, Í.C.; Pais do Amaral, C.; Gonçalves, S.; Santos, N.C. Advances in Lipid and Metal Nanoparticles for Antimicrobial Peptide Delivery. Pharmaceutics 2019, 11, 588. [Google Scholar] [CrossRef]
- Thakur, A.; Sharma, A.; Alajangi, H.K.; Jaiswal, P.K.; Lim, Y.-B.; Singh, G.; Barnwal, R.P. In Pursuit of Next-Generation Therapeutics: Antimicrobial Peptides against Superbugs, their Sources, Mechanism of Action, Nanotechnology-Based Delivery, and Clinical Applications. Int. J. Biol. Macromol. 2022, 218, 135–156. [Google Scholar] [CrossRef] [PubMed]




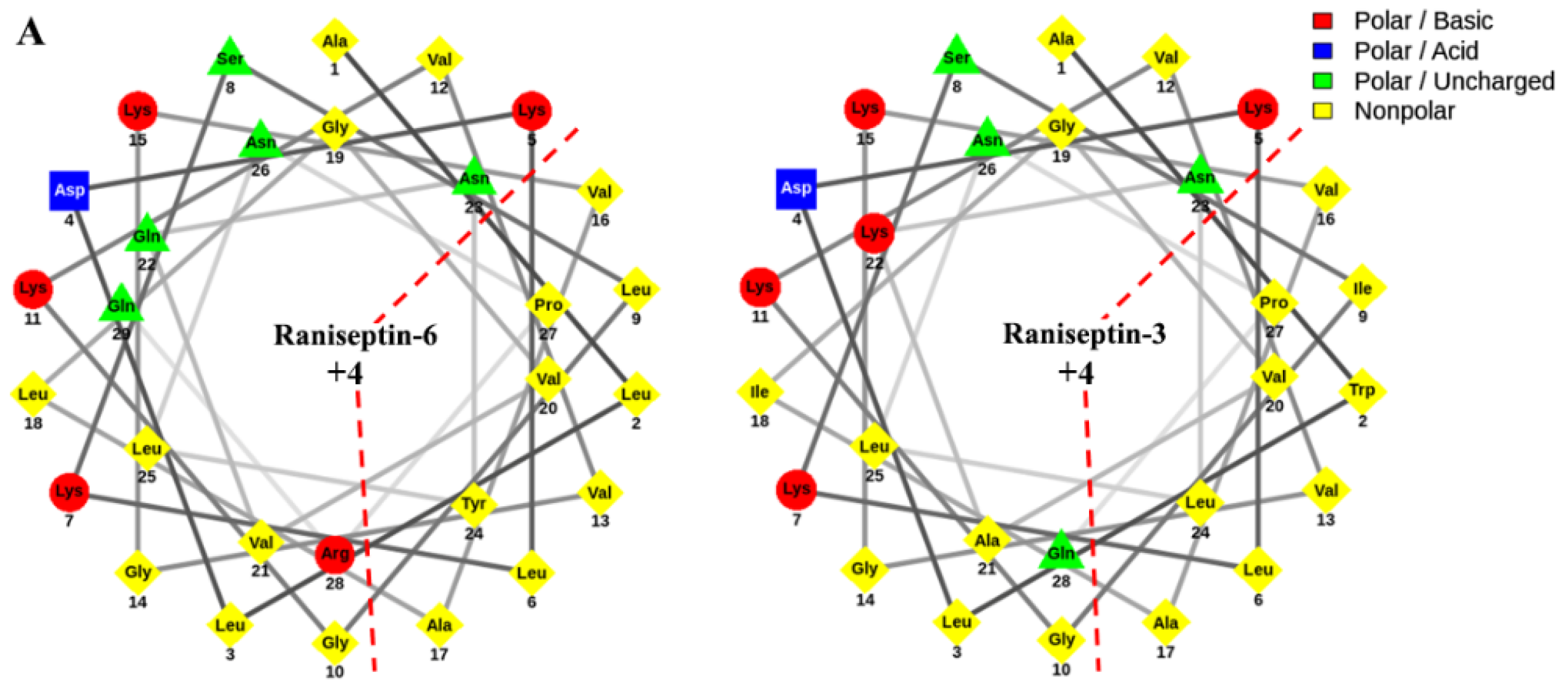
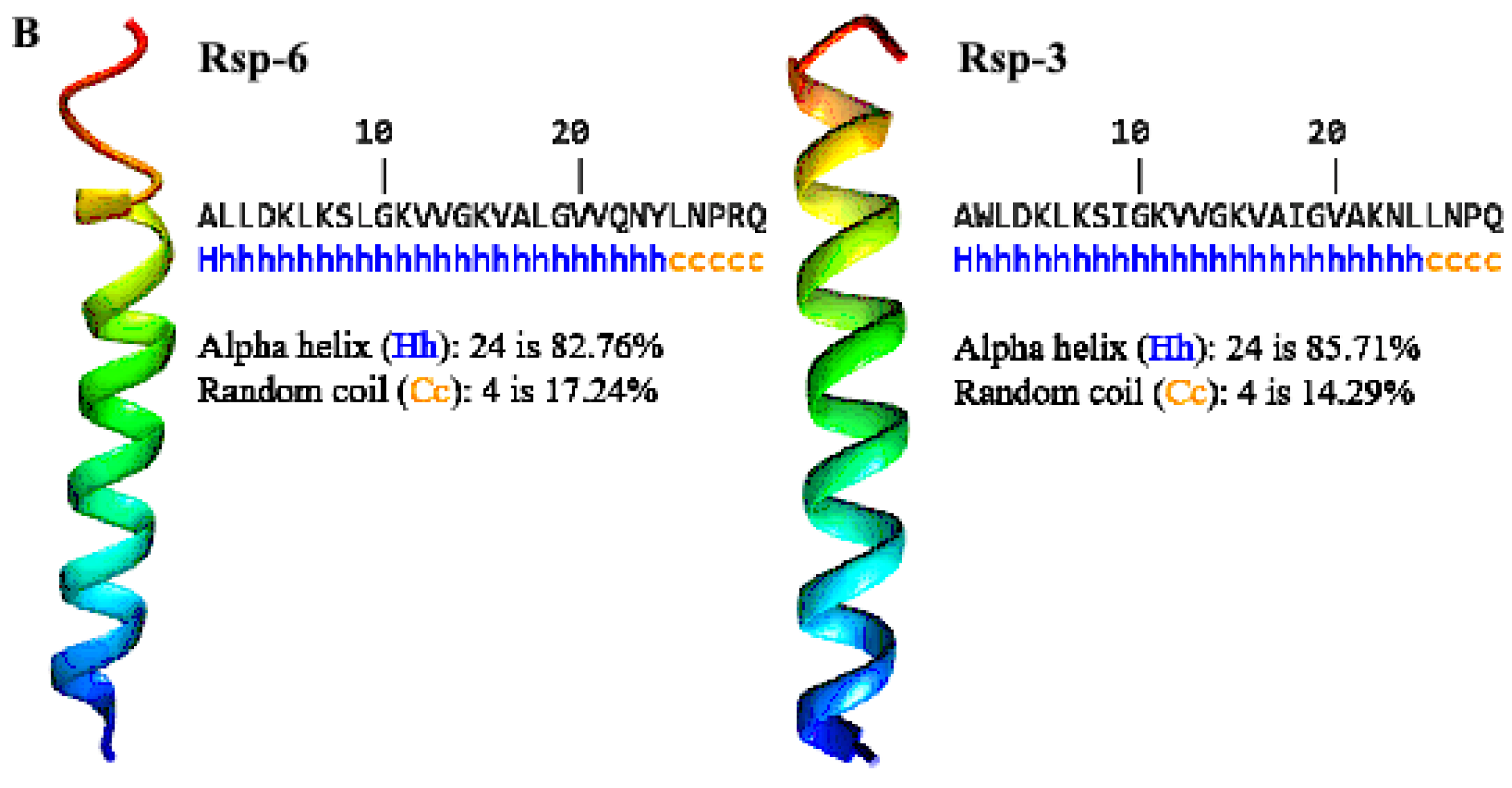
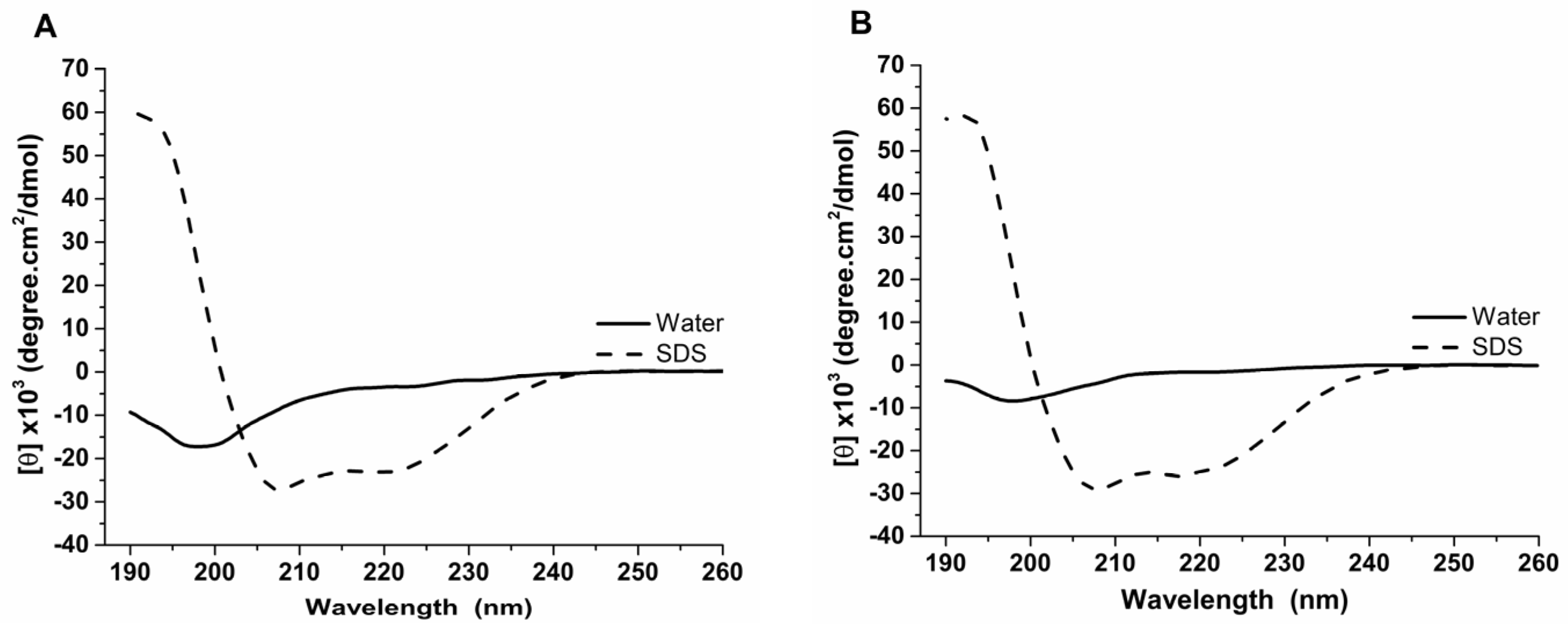
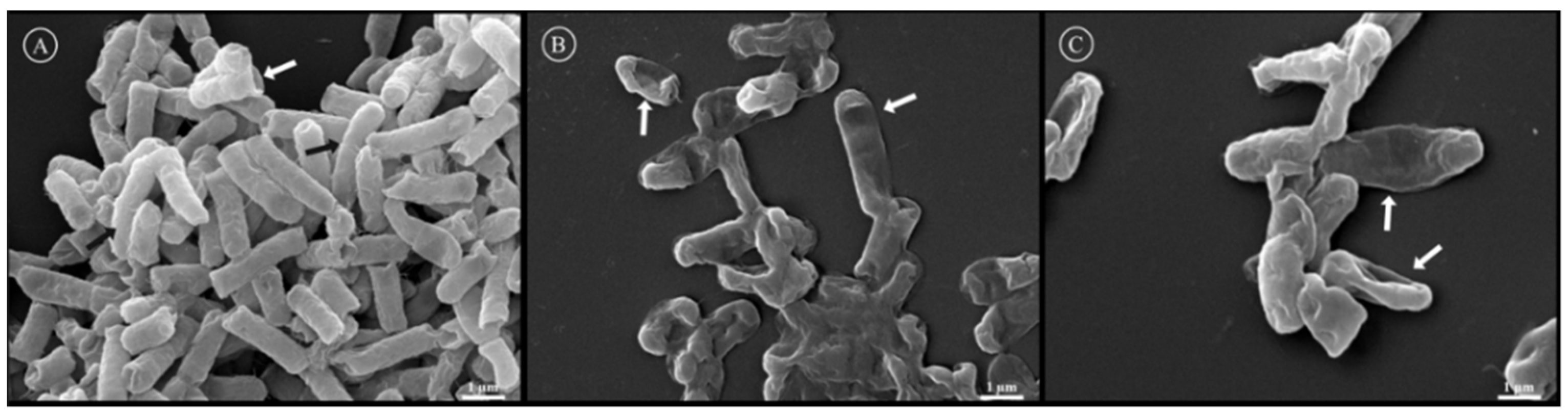

| Peptide | Mass calc. (Da) | Mass obs. (Da) | Net Charge | Hydrophobic Face | Hydrophobicity <H> | GRAVY |
|---|---|---|---|---|---|---|
| Raniseptin-3 | 2958.77 | 2958.7 | +4 | VIPWVVLLA | 43.77 | 0.300 |
| Raniseptin-6 | 3119.85 | 3119.5 | +4 | VLPLVVLYA | 43.60 | 0.169 |
| Microorganisms | Rsp-3 | Rsp-6 |
|---|---|---|
| Gram-negative bacteria | ||
| E. coli (ATCC 25922) | 2 | 2 |
| K. pneumoniae (ATCC 13883) | 1 | 1 |
| K. pneumoniae carbapanemase (KPC CAPB053) | 4 | 4 |
| Gram-positive bacteria | ||
| S. aureus (ATCC 25923) | 4 | 32 |
| S. epidermidis (ATCC 12228) | 8 | 8 |
| Yeast | ||
| C. albicans (ATCC 14053) | >128 | >128 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Freitas, G.G.d.; Barbosa, J.M.; Santana, C.J.C.d.; Magalhães, A.C.M.; Macedo, K.W.R.; Souza, J.O.d.; Castro, J.S.d.; Vasconcelos, I.A.d.; Souza, A.A.; Freitas, S.M.d.; et al. Purification and Biological Properties of Raniseptins-3 and -6, Two Antimicrobial Peptides from Boana raniceps (Cope, 1862) Skin Secretion. Biomolecules 2023, 13, 576. https://doi.org/10.3390/biom13030576
Freitas GGd, Barbosa JM, Santana CJCd, Magalhães ACM, Macedo KWR, Souza JOd, Castro JSd, Vasconcelos IAd, Souza AA, Freitas SMd, et al. Purification and Biological Properties of Raniseptins-3 and -6, Two Antimicrobial Peptides from Boana raniceps (Cope, 1862) Skin Secretion. Biomolecules. 2023; 13(3):576. https://doi.org/10.3390/biom13030576
Chicago/Turabian StyleFreitas, Gabriel Gonçalves de, João Martins Barbosa, Carlos José Correia de Santana, Ana Carolina Martins Magalhães, Keven Wender Rodrigues Macedo, Jéssica Oliveira de Souza, Jessica Schneider de Castro, Isadora Alves de Vasconcelos, Amanda Araújo Souza, Sonia Maria de Freitas, and et al. 2023. "Purification and Biological Properties of Raniseptins-3 and -6, Two Antimicrobial Peptides from Boana raniceps (Cope, 1862) Skin Secretion" Biomolecules 13, no. 3: 576. https://doi.org/10.3390/biom13030576
APA StyleFreitas, G. G. d., Barbosa, J. M., Santana, C. J. C. d., Magalhães, A. C. M., Macedo, K. W. R., Souza, J. O. d., Castro, J. S. d., Vasconcelos, I. A. d., Souza, A. A., Freitas, S. M. d., Báo, S. N., Costa, S. R., Brand, G. D., Chaves, I. d. M., Costa, V. V., Fontes, W., Pires Júnior, O. R., & Castro, M. S. (2023). Purification and Biological Properties of Raniseptins-3 and -6, Two Antimicrobial Peptides from Boana raniceps (Cope, 1862) Skin Secretion. Biomolecules, 13(3), 576. https://doi.org/10.3390/biom13030576








