Genome-Wide Identification and Expression Analysis of Rosa roxburghii Autophagy-Related Genes in Response to Top-Rot Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome-Wide Identification of ATG Family Genes in R. roxburghii
2.2. Bioinformatics Analysis of RrATGs
2.3. Plant Materials and Treatments
2.4. RNA-Seq Analysis of RrATGs Tissue-Specific Expression
2.5. RNA Extraction, cDNA Synthesis, and qRT-PCR Analysis
2.6. Total Calcium Content Detection
2.7. Statistical Analysis
3. Results
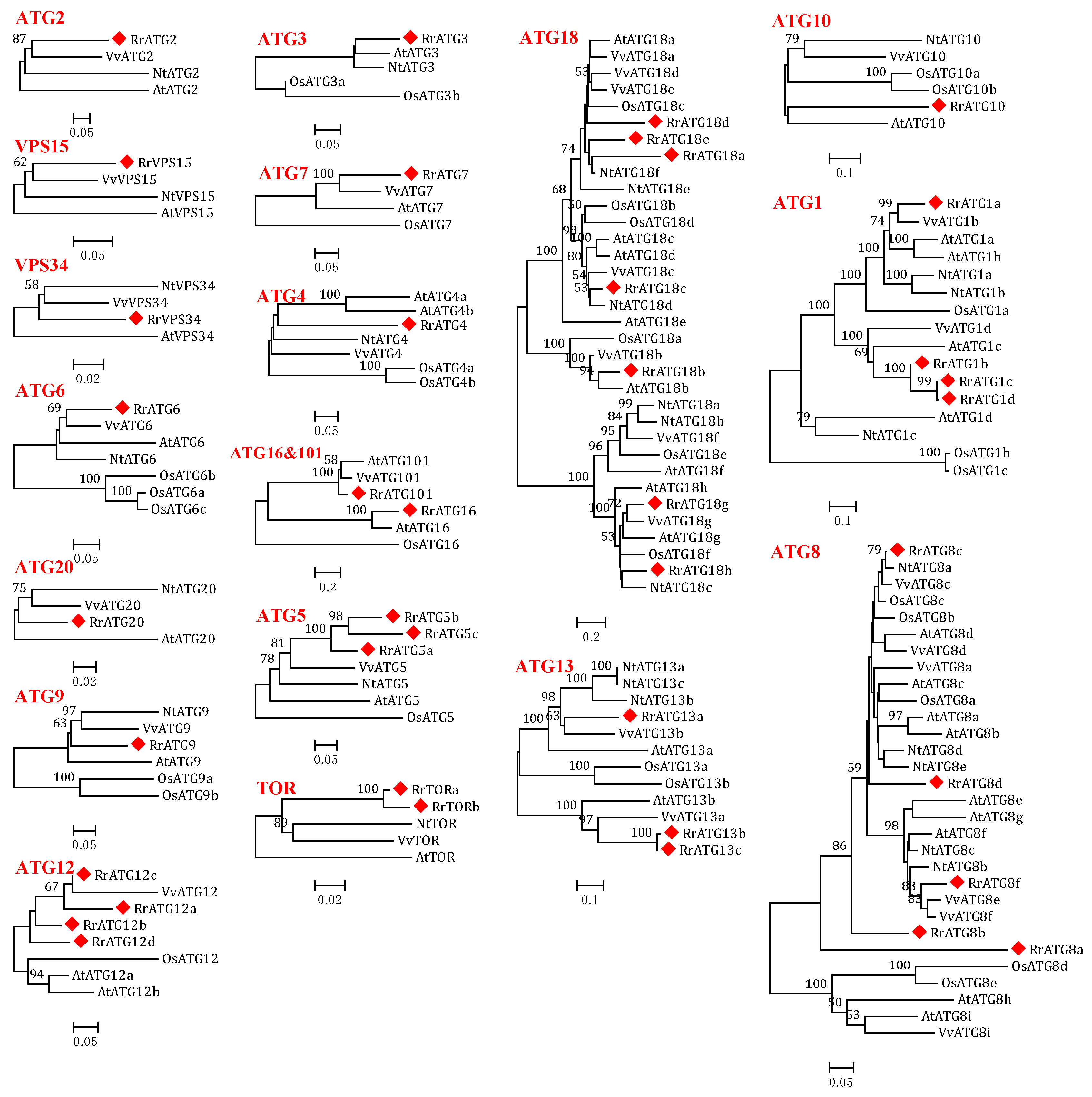
3.1. Identification of 40 ATGs in R. roxburghii
3.2. Bioinformatics Analysis of RrATGs
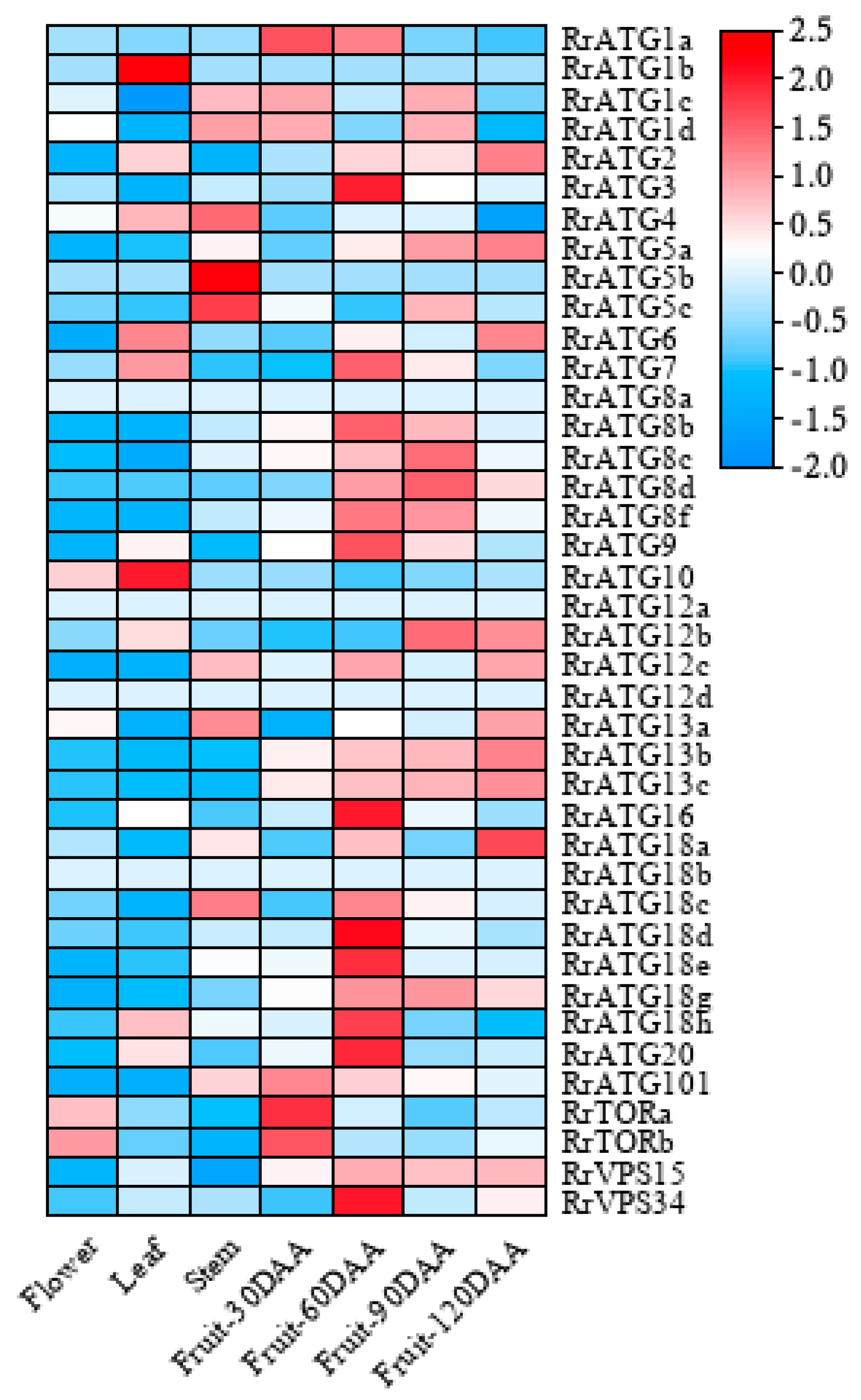
3.3. RNA-Seq Analyses of RrATGs in Different Tissues and Developmental Stage-Specific Expressions
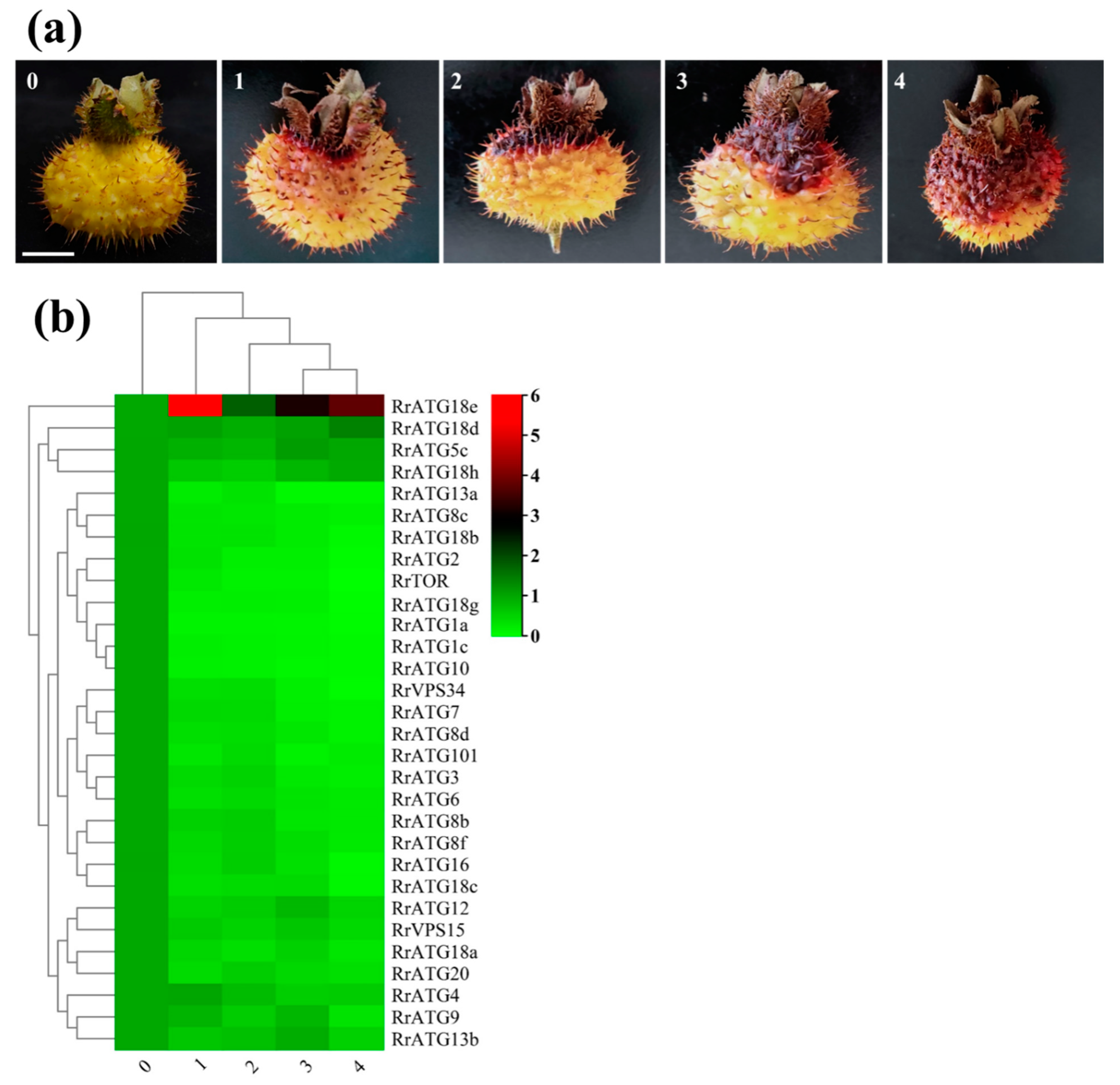
3.4. Expression Profiles of RrATGs with Different TRD Grades of R. roxburghii Fruits
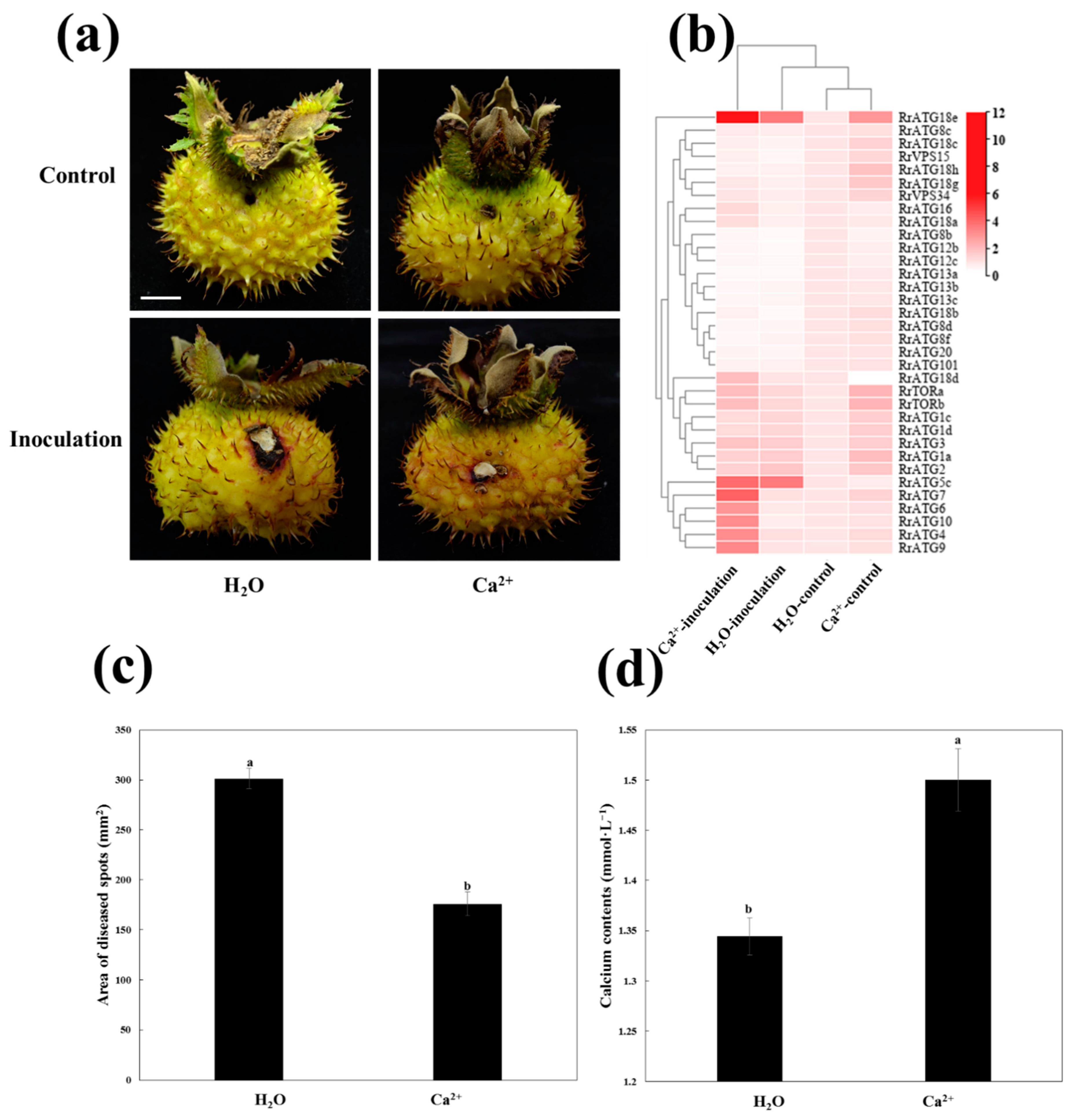
3.5. Field Control Effect of 2% Calcium Acetate (Ca2+) against TRD in R. roxburghii Fruits
3.6. Expression Profiles of RrATGs under C. fructicola Infection after Ca2+ Treatment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, L.; Wei, T.; Zheng, L.; Jiang, F.; Ma, W.; Lu, M.; Wu, X.; An, H. Recent advances on main active ingredients, pharmacological activities of Rosa roxbughii and its development and utilization. Foods 2023, 12, 1051. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Luo, Y.; Lu, M.; Wu, X.; An, H. The pathogen of top rot disease in Rosa roxburghii and its effective control fungicides. Horticulturae 2022, 8, 1036. [Google Scholar] [CrossRef]
- Jones, J.D.G.; Dangl, J. The plant immune system. Nature 2006, 444, 323. [Google Scholar] [CrossRef] [PubMed]
- Van Loon, L.C. The intelligent behavior of plants. Trends Plant Sci. 2016, 21, 286–294. [Google Scholar] [CrossRef]
- Hacquard, S.; Spaepen, S.; Garrido-Oter, R.; Schulze-Lefert, P. Interplay between innate immunity and the plant microbiota. Annu. Rev. Phytopathol. 2017, 55, 565–589. [Google Scholar] [CrossRef]
- Dangl, J.L.; Horvath, D.M.; Staskawicz, B.J. Pivoting the plant immune system from dissection to deployment. Science 2013, 341, 746–751. [Google Scholar] [CrossRef] [PubMed]
- Kroemer, G.; Eldeiry, W.S.; Golstein, P.; Peter, M.E.; Vaux, D.; Vandenabeele, P.; Zhivotovsky, B.; Blagosklonny, M.V.; Malorni, W.; Knight, R.A. Classification of cell death: Recommendations of the nomenclature committee on cell death 2009. Cell Death Differ. 2009, 16, 3–11. [Google Scholar] [CrossRef]
- Künstler, A.; Bacsó, R.; Gullner, G.; Hafez, Y.M.; Király, L. Staying alive-is cell death dispensable for plant disease resistance during the hypersensitive response? Physiol. Mol. Plant Pathol. 2016, 93, 75–84. [Google Scholar] [CrossRef]
- Liu, Y.; Bassham, D.C. Autophagy: Pathways for self-eating in plant cells. Annu. Rev. Plant Biol. 2012, 63, 215–237. [Google Scholar] [CrossRef] [PubMed]
- Shibutani, S.T.; Yoshimori, T. A current perspective of autophagosome biogenesis. Cell Res. 2014, 24, 58–68. [Google Scholar] [CrossRef] [PubMed]
- Mehrpour, M.; Esclatine, A.; Beau, I.; Codogno, P. Overview of macroautophagy regulation in mammalian cells. Cell Res. 2010, 20, 748–762. [Google Scholar] [CrossRef]
- Han, S.; Yu, B.; Wang, Y.; Liu, Y. Role of plant autophagy in stress response. Protein Cell. 2011, 2, 784–791. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Zhang, B.; Chen, W. Research progress of ATGs involved in plant immunity and NPR1 metabolism. Int. J. Mol. Sci. 2021, 22, 12093. [Google Scholar] [CrossRef]
- Yang, Y.; Xiang, Y.; Niu, Y. An overview of the molecular mechanisms and functions of autophagic pathways in plants. Plant Signal Behav. 2021, 16, 1977527. [Google Scholar] [CrossRef] [PubMed]
- Avila-Ospina, L.; Moison, M.; Yoshimoto, K.; Masclaux-Daubresse, C. Autophagy, plant senescence, and nutrient recycling. J. Exp. Bot. 2014, 65, 3799–3811. [Google Scholar] [CrossRef] [PubMed]
- Lenz, H.D.; Haller, E.; Melzer, E.; Kober, K.; Wurster, K.; Stahl, M.; Bassham, D.C.; Vierstra, R.D.; Parker, J.E.; Bautor, J.; et al. Autophagy differentially controls plant basal immunity to biotrophic and necrotrophic pathogens. Plant J. 2011, 66, 818–830. [Google Scholar] [CrossRef]
- Xia, K.; Liu, T.; Ouyang, J.; Wang, R.; Fan, T.; Zhang, M. Genome-wide identification, classification, and expression analysis of autophagy-associated gene homologues in rice (Oryza sativa L.). DNA Res. 2011, 18, 363–377. [Google Scholar] [CrossRef]
- Zhou, X.; Zhao, P.; Wang, W.; Zou, J.; Cheng, T.; Peng, X.; Sun, M. A comprehensive, genome-wide analysis of autophagy-related genes identified in tobacco suggests a central role of autophagy in plant response to various environmental cues. DNA Res. 2015, 22, 245–257. [Google Scholar] [CrossRef]
- Li, F.; Chung, T.; Pennington, J.G.; Federico, M.L.; Kaeppler, H.F.; Kaeppler, S.M.; Otegui, M.S.; Vierstra, R.D. Autophagic recycling plays a central role in maize nitrogen remobilization. Plant Cell. 2015, 27, 1389–1408. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Y.; Guo, M.; Wang, H.; Lu, J.; Liu, J.; Zhang, C.; Gong, Z.; Lu, M. Autophagy, a conserved mechanism for protein degradation, responds to heat, and other abiotic stresses in Capsicum annuum L. Front. Plant Sci. 2016, 7, 131. [Google Scholar] [CrossRef]
- Li, W.; Chen, M.; Wang, E.; Hu, L.; Hawkesford, M.J.; Zhong, L.; Chen, Z.; Xu, Z.; Li, L.; Zhou, Y.; et al. Genome-wide analysis of autophagy-associated genes in foxtail millet (Setaria italica L.) and characterization of the function of SiATG8a in conferring tolerance to nitrogen starvation in rice. BMC Genomics 2016, 17, 797. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Liu, W.; Hu, W.; Liu, G.; Wu, C.; Liu, W.; Zeng, H.; He, C.; Shi, H. Genome-wide analysis of autophagy-related genes in banana highlights MaATG8s in cell death and autophagy in immune response to Fusarium wilt. Plant Cell Rep. 2017, 36, 1237–1250. [Google Scholar] [CrossRef] [PubMed]
- Shangguan, L.; Fang, X.; Chen, L.; Cui, L.; Fang, J. Genome-wide analysis of autophagy-related genes (ARGs) in grapevine and plant tolerance to copper stress. Planta 2018, 247, 1449–1463. [Google Scholar] [CrossRef]
- Fu, X.; Zhou, X.; Xu, Y.; Hui, Q.; Chun, C.; Ling, L.; Peng, L. Comprehensive analysis of autophagy-related genes in sweet orange (Citrus sinensis) highlights their roles in response to abiotic stresses. Int. J. Mol. Sci. 2020, 21, 2699. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Ding, Z.; Gou, M.; Hu, J.; Wang, Y.; Wang, L.; Wang, Y.; Di, T.; Zhang, X.; Hao, X.; et al. Genome-wide identification, characterization, and expression analysis of tea plant autophagy-related genes (CsARGs) demonstrates that they play diverse roles during development and under abiotic stress. BMC Genomics 2021, 22, 121. [Google Scholar] [CrossRef]
- Yang, M.; Wang, L.; Chen, C.; Guo, X.; Lin, C.; Huang, W.; Chen, L. Genome-wide analysis of autophagy-related genes in Medicago truncatula highlights their roles in seed development and response to drought stress. Sci. Rep. 2021, 11, 22933. [Google Scholar] [CrossRef]
- Fan, T.; Yang, W.; Zeng, X.; Xu, X.; Xu, Y.; Fan, X.; Luo, M.; Tian, C.; Xia, K.; Zhang, M. A rice autophagy gene OsATG8b is involved in nitrogen remobilization and control of grain quality. Front. Plant Sci. 2020, 11, 588. [Google Scholar] [CrossRef]
- Yu, J.; Zhen, X.; Li, X.; Li, N.; Xu, F. Increased autophagy of rice can increase yield and nitrogen use efficiency (NUE). Front. Plant Sci. 2019, 10, 584. [Google Scholar] [CrossRef]
- Wang, P.; Sun, X.; Jia, X.; Ma, F. Apple autophagy-related protein MdATG3s afford tolerance to multiple abiotic stresses. Plant Sci. 2017, 256, 53–64. [Google Scholar] [CrossRef]
- Huo, L.; Guo, Z.; Jia, X.; Sun, X.; Wang, P.; Gong, X.; Ma, F. Increased autophagic activity in roots caused by overexpression of the autophagy-related gene MdATG10 in apple enhances salt tolerance. Plant Sci. 2020, 294, 110444. [Google Scholar] [CrossRef]
- Jia, X.; Jia, X.; Li, T.; Wang, Y.; Sun, X.; Huo, L.; Wang, P.; Che, R.; Gong, X.; Ma, F. MdATG5a induces drought tolerance by improving the antioxidant defenses and promoting starch degradation in apple. Plant Sci. 2021, 312, 111052. [Google Scholar] [CrossRef]
- Sun, X.; Pan, B.; Xu, W.; Chen, Q.; Wang, Y.; Ban, Q.; Xing, C.; Zhang, S. Genome-wide identification and expression analysis of the pear autophagy-related gene PbrATG8 and functional verification of PbrATG8c in Pyrus bretschneideri Rehd. Planta 2021, 253, 14. [Google Scholar] [CrossRef]
- Minina, E.A.; Moschou, P.N.; Vetukuri, R.R.; Sanchez-Vera, V.; Cardoso, C.; Liu, Q.S.; Elander, P.H.; Dalman, K.; Beganovic, M.; Yilmaz, J.L.; et al. Transcriptional stimulation of rate-limiting components of the autophagic pathway improves plant fitness. J. Exp. Bot. 2018, 69, 1415–1432. [Google Scholar] [CrossRef]
- Zhang, B.; Shao, L.; Wang, J.; Zhang, Y.; Guo, X.; Peng, Y.; Cao, Y.; Lai, Z. Phosphorylation of ATG18a by BAK1 suppresses autophagy and attenuates plant resistance against necrotrophic pathogens. Autophagy 2020, 17, 2093–2110. [Google Scholar] [CrossRef] [PubMed]
- Rigault, M.; Citerne, S.; Masclaux-Daubresse, C.; Dellagi, A. Salicylic acid is a key player of Arabidopsis autophagy mutant susceptibility to the necrotrophic bacterium Dickeya dadantii. Sci. Rep 2021, 11, 3624. [Google Scholar] [CrossRef]
- Wang, T.; Wang, C.; Gao, Q.; Li, L.; Luan, S. Calcium spikes, waves and oscillations in plant development and biotic interactions. Nat. Plants 2020, 6, 750–759. [Google Scholar] [CrossRef]
- Wang, T.; Hou, C.; Ren, Z.; Wang, C.; Zhao, F.; Dahlbeck, D.; Hu, S.; Zhang, L.; Niu, Q.; Li, L.; et al. A calmodulin-gated calcium channel links pathogen patterns to plant immunity. Nature 2019, 572, 131–135. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Liu, X.; Song, X.; Guo, Q.; Yin, Y.; Zhang, C.; Li, Y. High-vigor maize seeds resist Fusarium graminearum infection through stronger Ca2+ signaling. Agriculture 2022, 12, 992. [Google Scholar] [CrossRef]
- Winkler, A.; Knoche, M. Calcium uptake through skins of sweet cherry fruit: Effects of different calcium salts and surfactants. Sci. Hortic. 2021, 276, 109761. [Google Scholar] [CrossRef]
- Sun, X.; Pan, B.; Wang, Y.; Xu, W.; Zhang, S. Exogenous calcium improved resistance to Botryosphaeria dothidea by increasing autophagy activity and salicylic acid level in pear. Mol. Plant Microbe Interact. 2020, 33, 1150–1160. [Google Scholar] [CrossRef]
- Mostowfizadeh-Ghalamfarsa, R.; Hussaini, K.; Ghasemi-Fasaei, R. Effects of calcium salts in controlling Phytophthora pistaciae, the causal agent of pistachio gummosis. Eur. J. Plant Pathol. 2018, 151, 475–485. [Google Scholar] [CrossRef]
- Kittisak, L.; Saengchai, A.; Paitip, T. Effect of calcium acetate and calcium chloride on grain morphology and antioxidant regulation in rice under ozone stress. J. Plant Growth Regul. 2021, 41, 3138–3152. [Google Scholar] [CrossRef]
- Wang, Y. Studies on the Effects of Calcium on Watercore and Sorbitol Content in ‘Yueguan’ Apple Fruit; Shenyang Agricultural University: Shenyang, China, 2018. [Google Scholar]
- Dean, R.; Van Kan, J.A.L.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Pietro, A.D.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef]
- Kou, X.; Wu, M.; Li, L.; Wang, S.; Xue, Z.; Liu, B. Effects of CaCl2 dipping and pullulan coating on the development of brown spot on ‘Huangguan’ pears during cold storage. Postharvest Biol. Technol. 2015, 99, 63–72. [Google Scholar] [CrossRef]
- Huang, X.; Yan, H.; Zhai, L.; Yang, Z.; Yi, Y. Characterization of the Rosa roxburghii Tratt transcriptome and analysis of MYB genes. PLoS ONE 2019, 14, e0203014. [Google Scholar] [CrossRef]
- Lu, M.; Ma, W.; Liu, Y.; An, H.; Ludlow, R.A. Transcriptome analysis reveals candidate lignin-related genes and transcription factors in Rosa roxburghii during fruit ripening. Plant Mol. Biol. Report. 2020, 38, 331–342. [Google Scholar] [CrossRef]
- Li, J.; Zhou, Q.; Zhou, X.; Wei, B.; Zhao, Y.; Ji, S. Calcium treatment alleviates pericarp browning of ‘Nanguo’ pears by regulating the GABA shunt after cold storage. Front. Plant Sci. 2020, 11, 580986. [Google Scholar] [CrossRef] [PubMed]
- Aldon, D.; Mbengue, M.; Mazars, C.; Galaud, J.P. Calcium signalling in plant biotic interactions. Int. J. Mol. Sci. 2018, 19, 665. [Google Scholar] [CrossRef]
- Xu, Q.; Chen, L.; Ruan, X.; Chen, D.; Zhu, A.; Chen, C.; Bertrand, D.; Jiao, W.; Hao, B.; Lyon, M.P.; et al. The draft genome of sweet orange (Citrus sinensis). Nat. Genet. 2013, 45, 59–66. [Google Scholar] [CrossRef]
- Xiong, Y.; Contento, A.L.; Bassham, D.C. AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J. 2005, 42, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Alkan, N.; Friedlander, G.; Ment, D.; Prusky, D.; Fluhr, R. Simultaneous transcriptome analysis of Colletotrichum gloeosporioides and tomato fruit pathosystem reveals novel fungal pathogenicity and fruit defense strategies. New Phytol. 2015, 205, 801–815. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Huo, L.; Jia, X.; Che, R.; Gong, X.; Wang, P.; Ma, F. Overexpression of MdATG18a in apple improves resistance to Diplocarpon mali infection by enhancing antioxidant activity and salicylic acid levels. Hortic. Res. 2018, 5, 57. [Google Scholar] [CrossRef] [PubMed]
- Kluza, A.; Niedzialkowska, E.; Kurpiewska, K.; Wojdyla, Z.; Quesne, M.; Kot, E.; Porebski, P.J.; Borowski, T. Crystal structure of thebaine 6-O-demethylase from the morphine biosynthesis pathway. J. Struct. Biol. 2018, 202, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Liu, S.; Fan, F.; Yu, Q.; Zhang, P. A moss 2-Oxoglutarate/Fe(II)-dependent dioxygenases (2-ODD) gene of flavonoids biosynthesis positively regulates plants abiotic stress tolerance. Front. Plant Sci. 2022, 13, 850062. [Google Scholar] [CrossRef] [PubMed]


| Gene Name | Arabidopsisa ID | Gene | R. roxburghii ID | Identity to Arabidopsisa(%) | Protein (aa) | Protein Molecular Mass(KDa) | pI | Predicted Localization |
|---|---|---|---|---|---|---|---|---|
| AtATG1a | At3g61960 | RrATG1a | Contig179.812 | 64.78 | 722 | 79.88 | 6.73 | Nuclear |
| AtATG1b | At3g53930 | RrATG1b | Contig110.67 | 61.36 | 138 | 15.41 | 9.88 | Nuclear |
| AtATG1c | At2g37840 | RrATG1c | Contig191.2 | 52.21 | 677 | 74.79 | 6.44 | Nuclear |
| AtATG1d | At1g49180 | RrATG1d | Contig289.274 | 49.16 | 649 | 71.65 | 6.51 | Nuclear |
| AtATG2 | At3g19190 | RrATG2 | Contig161.356 | 49.80 | 1983 | 217.54 | 5.47 | Plasma membrane |
| AtATG3 | At5g61500 | RrATG3 | Contig189.150 | 82.86 | 366 | 41.44 | 4.51 | Cytoskeleton |
| AtATG4a | At2g44140 | RrATG4 | Contig161.437 | 55.85 | 427 | 46.98 | 4.98 | Chloroplast |
| AtATG4b | At3g59950 | NA | NA | NA | NA | NA | NA | NA |
| AtATG5 | At5g17290 | RrATG5a | Contig361.91 | 61.98 | 362 | 40.95 | 4.61 | Cytoplasmic |
| RrATG5b | Contig8.24 | 59.53 | 302 | 33.96 | 5.21 | Cytoplasmic | ||
| RrATG5c | Contig169.100 | 58.92 | 310 | 35.38 | 5.99 | Nuclear | ||
| AtATG6 | At3g61710 | RrATG6 | Contig289.113 | 67.13 | 469 | 52.98 | 5.58 | Cytoplasmic |
| AtATG7 | At5g45900 | RrATG7 | Contig363.72 | 63.96 | 581 | 63.17 | 6.42 | Endoplasmic reticulum |
| AtATG8a | At4g21980 | RrATG8a | Contig179.203 | 63.53 | 110 | 11.91 | 5.02 | Mitochondrial |
| AtATG8b | At4g04620 | RrATG8b | Contig18.48 | 76.07 | 119 | 13.65 | 5.00 | Cytoplasmic |
| AtATG8c | At1g62040 | RrATG8c | Contig360.160 | 91.45 | 119 | 13.72 | 9.29 | Cytoplasmic |
| AtATG8d | At2g05630 | RrATG8d | Contig10.163 | 82.46 | 121 | 13.84 | 9.32 | Cytoplasmic |
| AtATG8e | At2g45170 | NA | NA | NA | NA | NA | NA | NA |
| AtATG8f | At4g16520 | RrATG8f | Contig136.201 | 87.18 | 117 | 13.42 | 9.77 | Cytoplasmic |
| AtATG8g | At3g60640 | NA | NA | NA | NA | NA | NA | NA |
| AtATG8h | At3g06420 | NA | NA | NA | NA | NA | NA | NA |
| AtATG8i | At3g15580 | NA | NA | NA | NA | NA | NA | NA |
| AtATG9 | At2g31260 | RrATG9 | Contig385.359 | 70.63 | 808 | 93.13 | 7.60 | Plasma membrane |
| AtATG10 | At3g07525 | RrATG10 | Contig290.80 | 68.85 | 525 | 58.91 | 8.42 | Chloroplast |
| AtATG11 | At4g30790 | NA | NA | NA | NA | NA | NA | NA |
| AtATG12a | At1g54210 | RrATG12a | Contig428.656 | 81.25 | 110 | 12.70 | 9.12 | Nuclear |
| AtATG12b | At3g13970 | RrATG12b | Contig414.84 | 80.85 | 95 | 10.76 | 10.11 | Chloroplast |
| RrATG12c | Contig401.201 | 80 | 77 | 8.73 | 10.06 | Mitochondrial | ||
| RrATG12d | Contig385.680 | 69.66 | 144 | 16.30 | 10.40 | Chloroplast | ||
| AtATG13a | At3g49590 | RrATG13a | Contig386.98 | 52.04 | 1032 | 115.66 | 9.07 | Nuclear |
| AtATG13b | At3g18770 | RrATG13b | Contig52.3 | 56.70 | 644 | 71.33 | 7.97 | Cytoplasmic |
| RrATG13c | Contig266.16 | 56.24 | 628 | 69.41 | 7.84 | Cytoplasmic | ||
| AtATG16 | At5g50230 | RrATG16 | Contig104.384 | 63.32 | 745 | 82.24 | 8.56 | Nuclear |
| AtATG18a | At3g62770 | RrATG18a | Contig385.717 | 89.50 | 928 | 104.33 | 6.46 | Nuclear |
| AtATG18b | At4g30510 | RrATG18b | Contig405.14 | 74.18 | 371 | 40.31 | 6.61 | Extracellular |
| AtATG18c | At2g40810 | RrATG18c | Contig317.6 | 75.37 | 411 | 45.67 | 7.54 | Nuclear |
| AtATG18d | At3g56440 | RrATG18d | Contig121.44 | 69.37 | 280 | 31.63 | 7.99 | Golgi |
| AtATG18e | At5g05150 | RrATG18e | Contig121.43 | 64 | 783 | 87.27 | 7.88 | Cytoplasmic |
| AtATG18f | At5g54730 | NA | NA | NA | NA | NA | NA | NA |
| AtATG18g | At1g03380 | RrATG18g | Contig385.614 | 61.30 | 1243 | 136.39 | 7.08 | Chloroplast |
| AtATG18h | At1g54710 | RrATG18h | Contig149.53 | 63.74 | 863 | 94.06 | 5.637 | Mitochondrial |
| AtATG20 | At5g06140 | RrATG20 | Contig179.205 | 76.03 | 337 | 38.74 | 9.31 | Chloroplast |
| AtATG101 | At5g66930 | RrATG101 | Contig124.67 | 76.44 | 208 | 24.09 | 6.60 | Cytoplasmic |
| AtTOR | At1g50030 | RrTORa | Contig59.5 | 81.53 | 2459 | 276.22 | 6.83 | Cytoplasmic |
| RrTORb | Contig161.4 | 79.85 | 2449 | 274.88 | 6.82 | Cytoplasmic | ||
| AtVPS15 | At4g29380 | RrVPS15 | Contig354.8 | 70.62 | 1555 | 174.30 | 7.27 | Nuclear |
| AtVPS34 | At1g60490 | RrVPS34 | Contig332.44 | 84.17 | 805 | 91.87 | 7.06 | Cytoplasmic |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, K.; Li, J.; Lu, M.; An, H.; Wu, X. Genome-Wide Identification and Expression Analysis of Rosa roxburghii Autophagy-Related Genes in Response to Top-Rot Disease. Biomolecules 2023, 13, 556. https://doi.org/10.3390/biom13030556
Luo K, Li J, Lu M, An H, Wu X. Genome-Wide Identification and Expression Analysis of Rosa roxburghii Autophagy-Related Genes in Response to Top-Rot Disease. Biomolecules. 2023; 13(3):556. https://doi.org/10.3390/biom13030556
Chicago/Turabian StyleLuo, Kaisha, Jiaohong Li, Min Lu, Huaming An, and Xiaomao Wu. 2023. "Genome-Wide Identification and Expression Analysis of Rosa roxburghii Autophagy-Related Genes in Response to Top-Rot Disease" Biomolecules 13, no. 3: 556. https://doi.org/10.3390/biom13030556
APA StyleLuo, K., Li, J., Lu, M., An, H., & Wu, X. (2023). Genome-Wide Identification and Expression Analysis of Rosa roxburghii Autophagy-Related Genes in Response to Top-Rot Disease. Biomolecules, 13(3), 556. https://doi.org/10.3390/biom13030556





