Enzymatic Glyco-Modification of Synthetic Membrane Systems
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. LgtC Production
2.3. Enzyme Activity Using UDP-Glo Glycosyltransferase Assay
2.4. Thin Layer Chromatography (TLC)
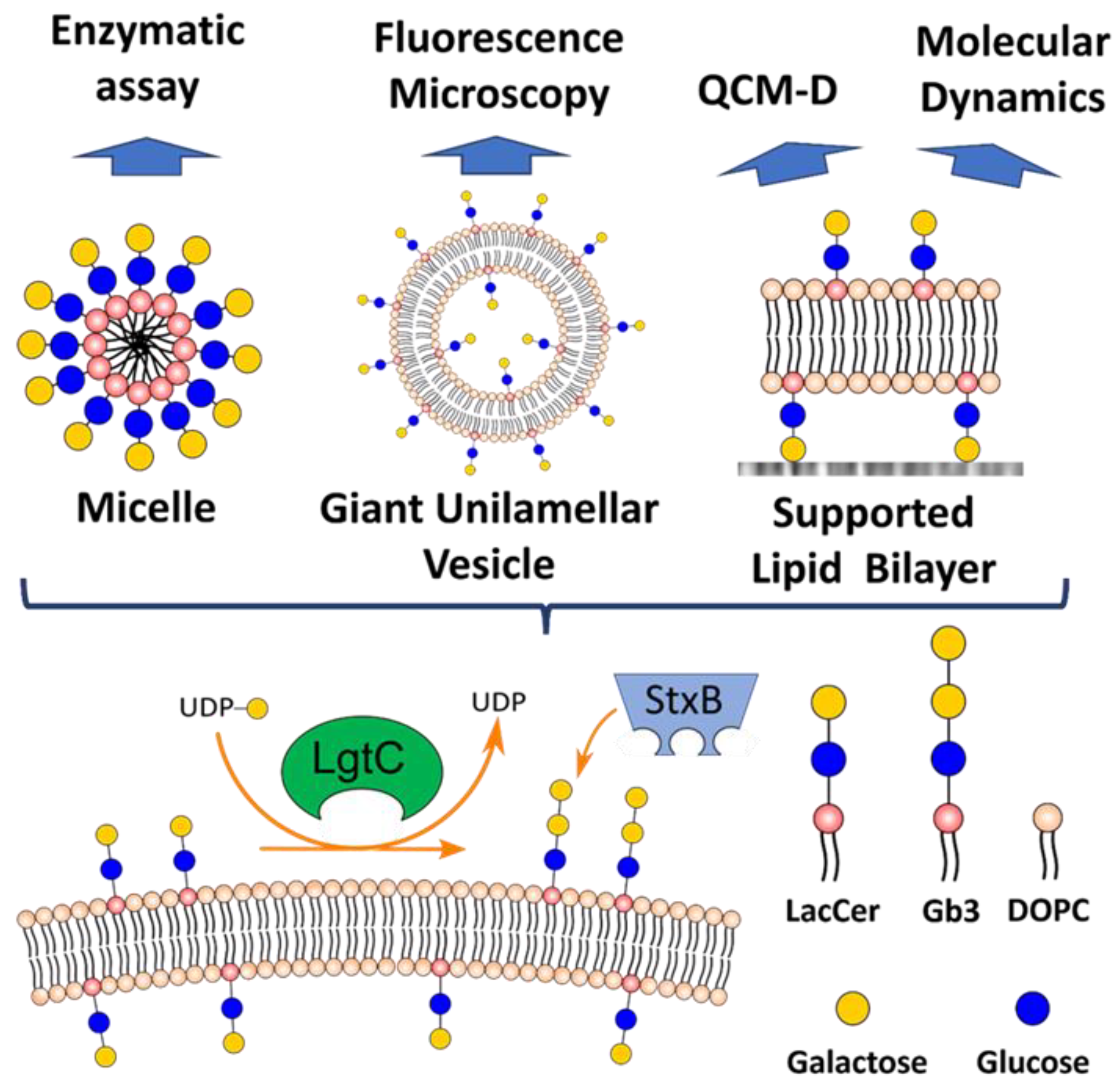
2.5. Giant Unilamellar Vesicles (GUVs)
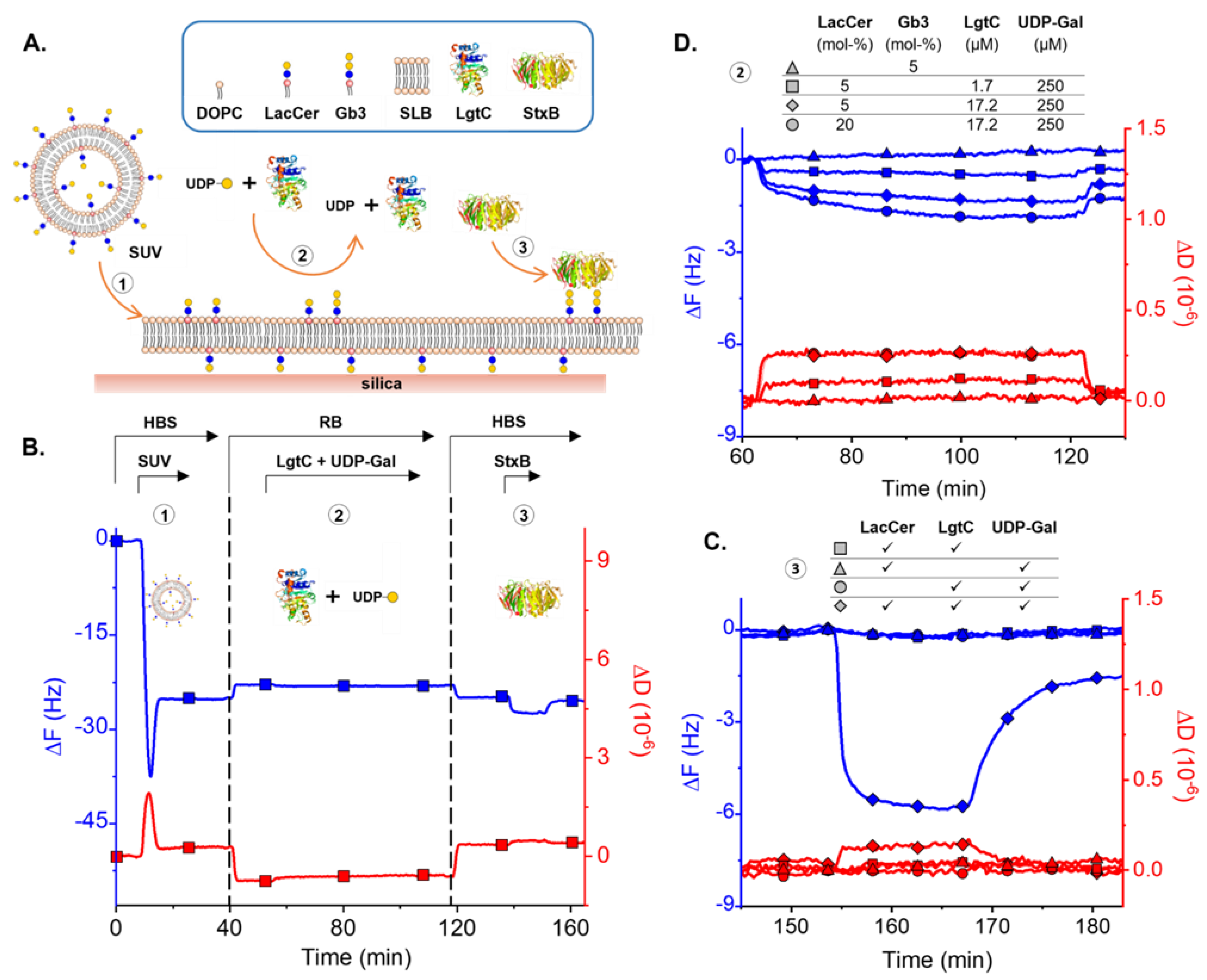
2.6. Quartz Crystal Microbalance with Dissipation Monitoring (QCM-D)
2.7. Small Unilamellar Vesicles (SUVs)
2.8. Computer Simulations
2.9. Molecular Dynamics Simulations
2.10. Molecular Docking
3. Results
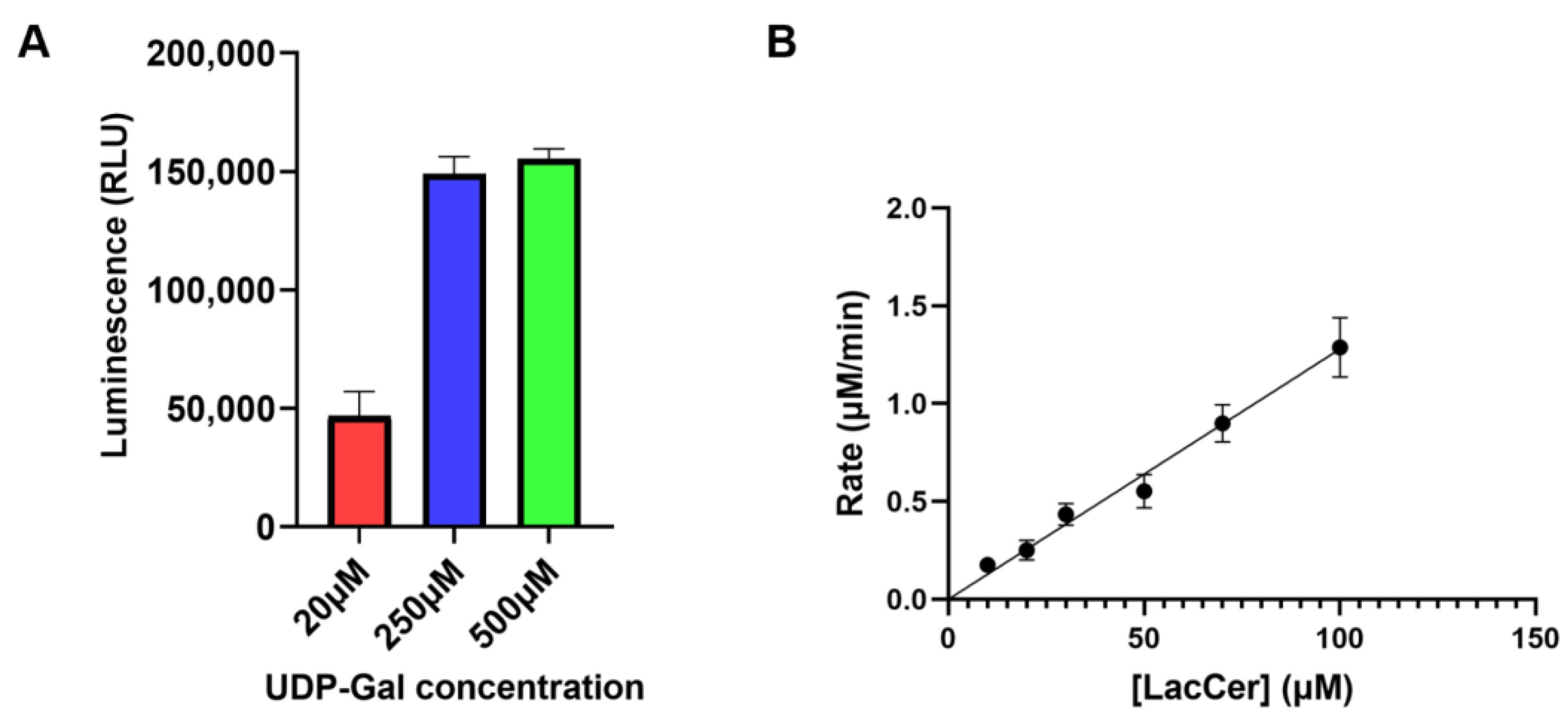
3.1. LgtC Synthesizes Gb3 on LacCer-containing Micelles
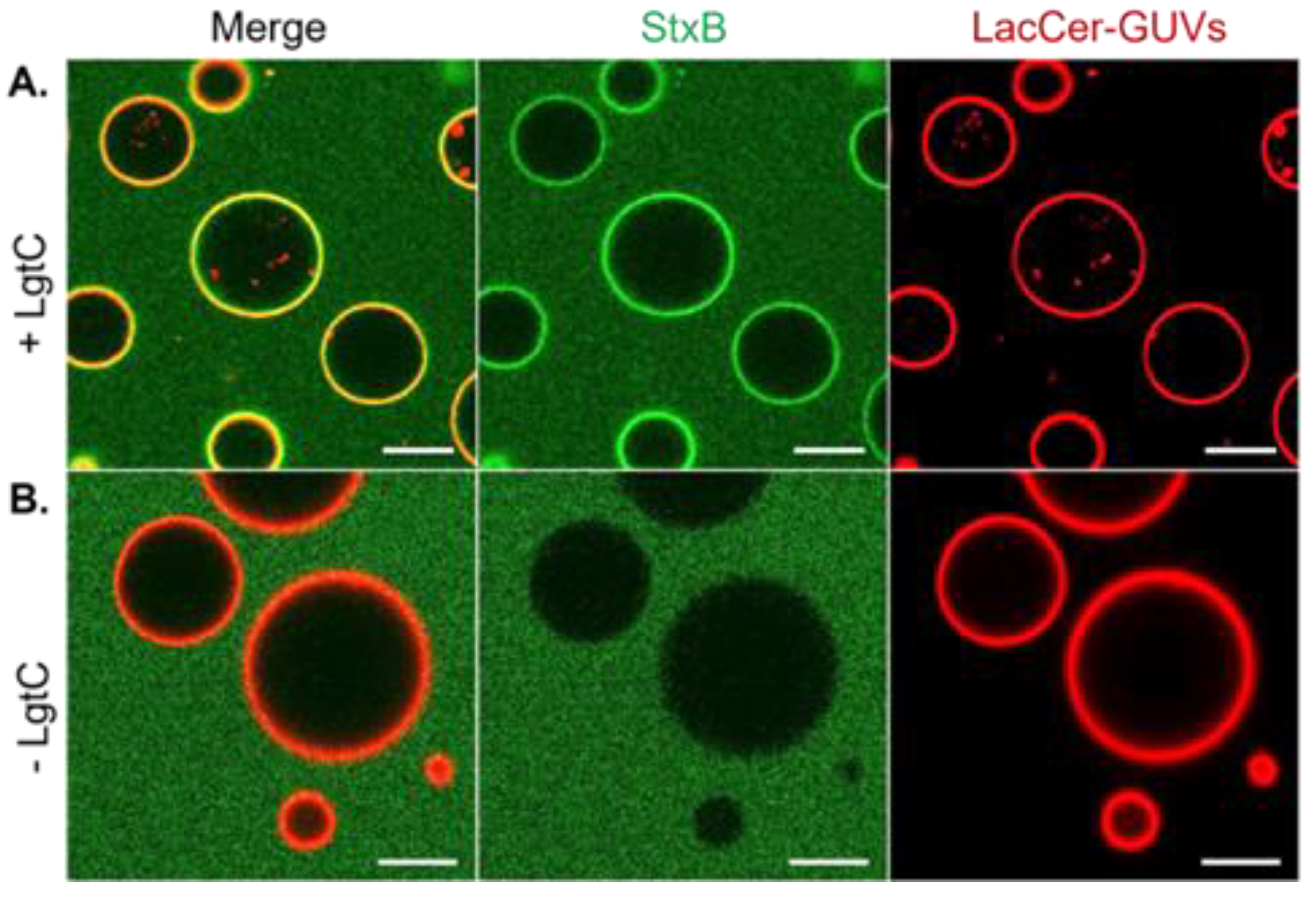
3.2. LgtC Activity on LacCer-Containing GUVs
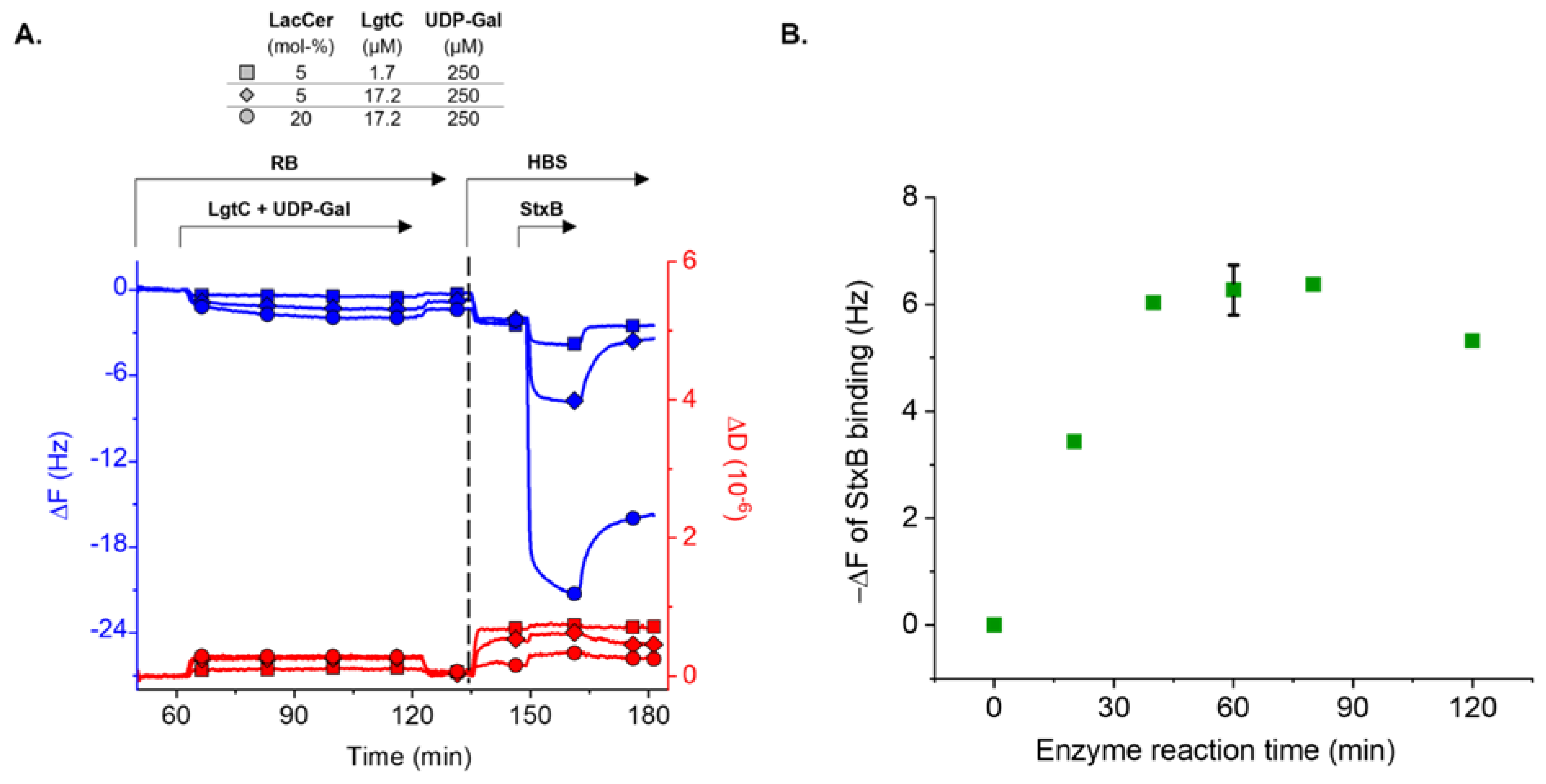
3.3. Enzymatic LacCer Glycosylation Monitored on SLBs
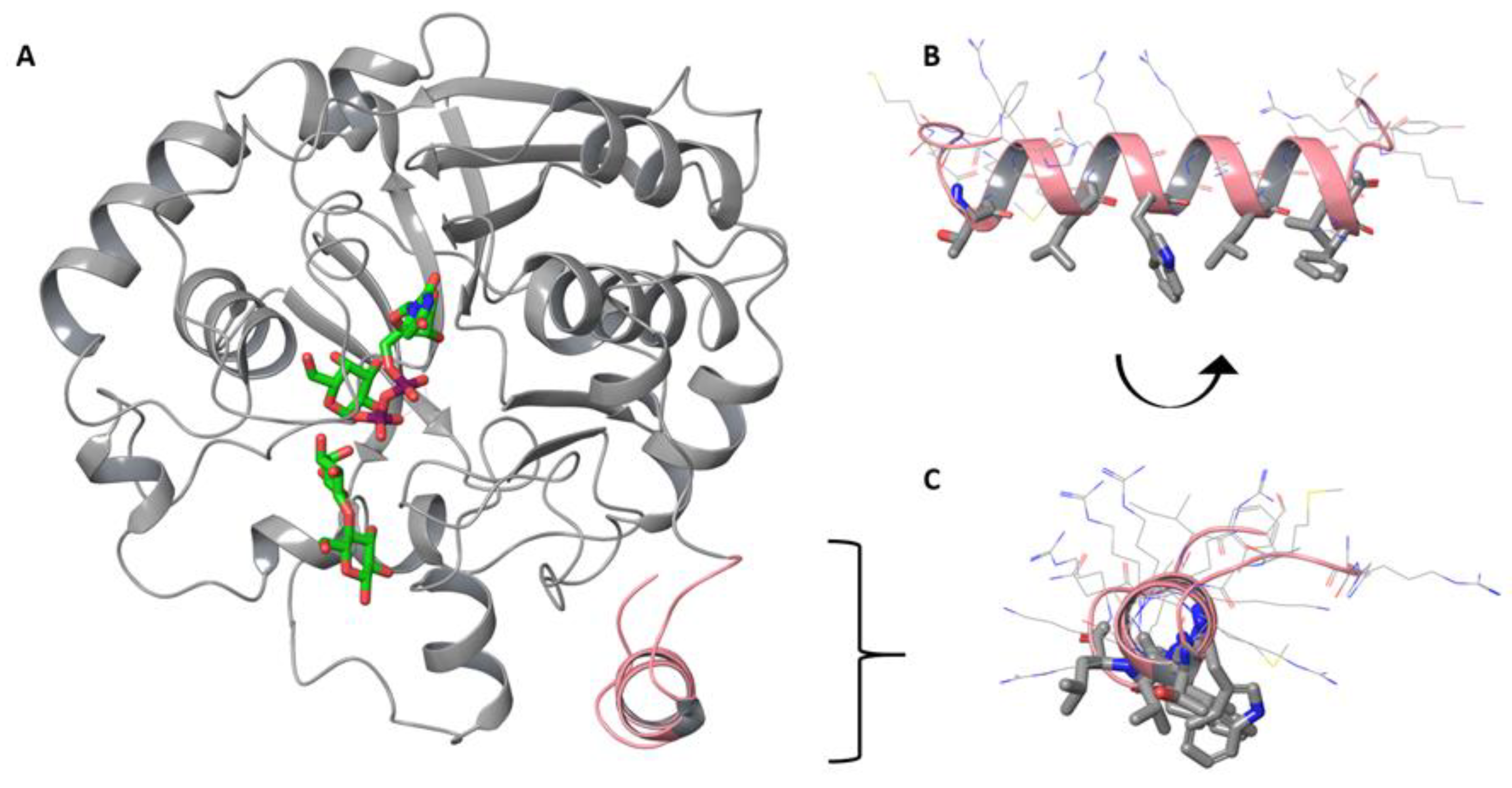
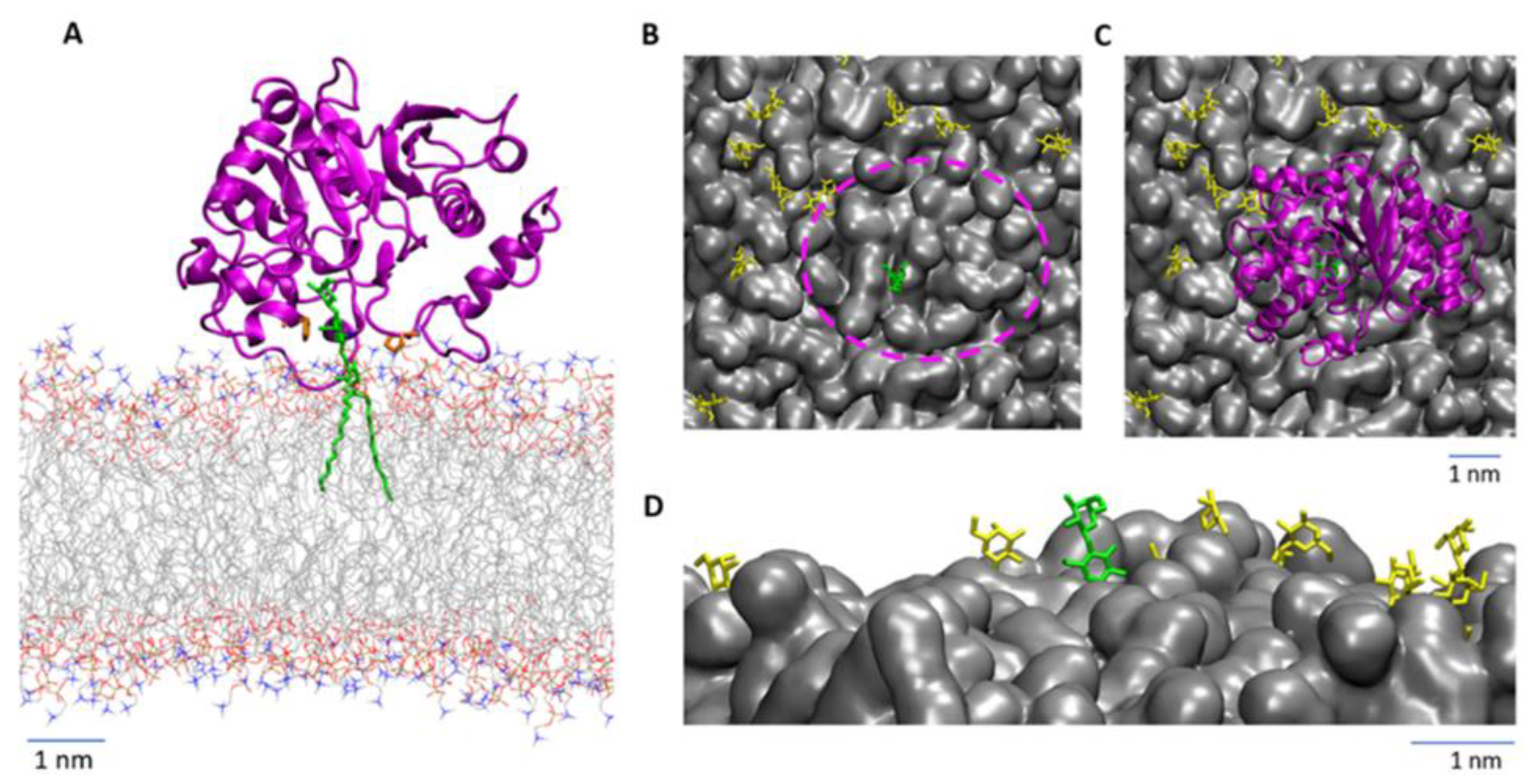
3.4. Computational Modeling Reveals Steric Hindrance between LgtC and the Lipid Bilayer
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix B
Appendix C
Appendix D. Lactose Binding in Solution
| Docking | MD | |||||||
|---|---|---|---|---|---|---|---|---|
| ΔG | ΔH | −TΔS | ΔG | ΔH | −TΔS | |||
| Total | Gas | Desolv | ||||||
| LgtC/βGal(1−4)βGlc | −2.5 | −6.0 | +3.6 | −8.9 ±6.7 | −12.9 ±5.3 | −49.5 ±4.5 | 36.6 ±2.5 | +3.9 ±4.2 |
| LgtC+UDP−Gal/βGal(1−4)βGlc | −2.6 | −6.2 | +3.6 | |||||
| LgtC/αGal(1−4)βGal(1−4)βGlc | −3.0 | −8.4 | +5.4 | |||||
| LgtC+UDP/ αGal(1−4)βGal(1−4)βGlc | −3.2 | −8.5 | +5.4 | |||||
| LgtC/LacCer in membrane (a) | −2.9 ±109 | −528 ±89 | 525 ±64 | NA | ||||
| α3GT+UDP/βGal(1−4)βGlc (b) | −4.0 | −7.6 | +3.6 | |||||
References
- Gagneux, P.; Hennet, T.; Varki, A. Biological functions of glycans. In Essentials of Glycobiology, 4th ed.; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G.W., Aebi, M., Mohnen, D., Kinoshita, T., Packer, N.H., Prestegard, J.H., et al., Eds.; Cold Spring Cold Spring Harbor Laboratory Press: Harbor, NY, USA, 2022. [Google Scholar]
- Möckl, L. The emerging role of the mammalian glycocalyx in functional membrane organization and immune system regulation. Front. Cell Dev. Biol. 2020, 8, 253. [Google Scholar] [PubMed]
- Zeng, Y. Endothelial glycocalyx as a critical signalling platform integrating the extracellular haemodynamic forces and chemical signalling. J. Cell. Mol. Med. 2017, 21, 1457–1462. [Google Scholar] [PubMed]
- Imberty, A.; Varrot, A. Microbial recognition of human cell surface glycoconjugates. Curr. Opin. Struct. Biol. 2008, 18, 567–576. [Google Scholar] [CrossRef]
- Pinho, S.S.; Reis, C.A. Glycosylation in cancer: Mechanisms and clinical implications. Nat. Rev. Cancer 2015, 15, 540–555. [Google Scholar] [CrossRef]
- Diniz, F.; Coelho, P.; Duarte, H.O.; Sarmento, B.; Reis, C.A.; Gomes, J. Glycans as Targets for Drug Delivery in Cancer. Cancers 2022, 14, 911. [Google Scholar] [PubMed]
- Hudak, J.E.; Bertozzi, C.R. Glycotherapy: New advances inspire a reemergence of glycans in medicine. Chem. Biol. 2013, 21, 16–37. [Google Scholar]
- Meléndez, A.V.; Cárdenas, R.M.-H.V.; Lagies, S.; Strietz, J.; Siukstaite, L.; Thomas, O.S.; Tomisch, J.; Weber, W.; Kammerer, B.; Römer, W.; et al. Novel lectin-based chimeric antigen receptors target Gb3-positive tumour cells. Cell. Mol. Life Sci. 2022, 79, 513. [Google Scholar]
- Critcher, M.; O’Leary, T.; Huang, M.L. Glycoengineering: Scratching the surface. Biochem. J. 2021, 478, 703–719. [Google Scholar] [CrossRef]
- Griffin, M.E.; Hsieh-Wilson, L.C. Glycan engineering for cell and developmental biology. Cell Chem. Biol. 2016, 23, 108–121. [Google Scholar] [CrossRef]
- Jöud, M.; Möller, M.; Olsson, M.L. Identification of human glycosyltransferase genes expressed in erythroid cells predicts potential carbohydrate blood group loci. Sci. Rep. 2018, 8, 6040. [Google Scholar] [CrossRef]
- Breton, C.; Fournel-Gigleux, S.; Palcic, M.M. Recent structures, evolution and mechanisms of glycosyltransferases. Curr. Opin. Struct. Biol. 2012, 22, 540–549. [Google Scholar] [PubMed]
- Chaubard, J.-L.; Krishnamurthy, C.; Yi, W.; Smith, D.F.; Hsieh-Wilson, L.C. Chemoenzymatic probes for detecting and imaging fucose-α(1-2)-galactose glycan biomarkers. J. Am. Chem. Soc. 2012, 134, 4489–4492. [Google Scholar] [PubMed]
- Hong, S.; Shi, Y.; Wu, N.C.; Grande, G.; Douthit, L.; Wang, H.; Zhou, W.; Sharpless, K.B.; Wilson, I.A.; Xie, J.; et al. Bacterial glycosyltransferase-mediated cell-surface chemoenzymatic glycan modification. Nat. Commun. 2019, 10, 1799. [Google Scholar] [PubMed]
- Huang, J.; MacKerell, A.D., Jr. CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data. J. Comput. Chem. 2013, 34, 2135–2145. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.C.; Godula, K. Synthetic glycoscapes: Addressing the structural and functional complexity of the glycocalyx. Interface Focus 2019, 9, 20180080. [Google Scholar] [CrossRef]
- Pinnock, F.; Daniel, S. Small tools for sweet challenges: Advances in microfluidic technologies for glycan synthesis. Anal. Bioanal. Chem. 2022, 414, 5139–5163. [Google Scholar] [CrossRef]
- Omidvar, R.; Römer, W. Glycan-decorated protocells: Novel features for rebuilding cellular processes. Interface Focus 2019, 9, 20180084. [Google Scholar]
- Richter, R.P.; Bérat, R.; Brisson, A.R. Formation of solid-supported lipid bilayers: An integrated view. Langmuir 2006, 22, 3497–3505. [Google Scholar]
- Villringer, S.; Madl, J.; Sych, T.; Manner, C.; Imberty, A.; Römer, W. Lectin-mediated protocell crosslinking to mimic cell-cell junctions and adhesion. Sci. Rep. 2018, 8, 1932. [Google Scholar] [CrossRef]
- Schubert, T.; Römer, W. How synthetic membrane systems contribute to the understanding of lipid-driven endocytosis. Biochim. Biophys. Acta 2015, 1853, 2992–3005. [Google Scholar]
- Goronzy, I.N.; Rawle, R.J.; Boxer, S.G.; Kasson, P.M. Cholesterol enhances influenza binding avidity by controlling nanoscale receptor clustering. Chem. Sci. 2018, 9, 2340–2347. [Google Scholar] [CrossRef] [PubMed]
- Noble, G.T.; Craven, F.L.; Voglmeir, J.; Šardzík, R.; Flitsch, S.L.; Webb, S.J. Accelerated enzymatic galactosylation of N-acetylglucosaminolipids in lipid microdomains. J. Am. Chem. Soc. 2012, 134, 13010–13017. [Google Scholar] [CrossRef] [PubMed]
- Alves, C.A.; Pedroso, M.M.; de Moraes, M.C.; Souza, D.H.; Cass, Q.B.; Faria, R.C. Real-time investigation of mannosyltransferase function of a Xylella fastidiosa recombinant GumH protein using QCM-D. Biochem. Biophys. Res. Commun. 2011, 408, 571–575. [Google Scholar] [PubMed]
- BBreton, C.; Mucha, J.; Jeanneau, C. Structural and functional features of glycosyltransferases. Biochimie 2001, 83, 713–718. [Google Scholar]
- Wakarchuk, W.W.; Cunningham, A.; Watson, D.C.; Young, N.M. Role of paired basic residues in the expression of active recombinant galactosyltransferases from the bacterial pathogen Neisseria meningitidis. Protein Eng. 1998, 11, 295–302. [Google Scholar] [CrossRef]
- Siukstaite, L.; Imberty, A.; Römer, W. Structural diversities of lectins binding to the glycosphingolipid Gb3. Front. Mol. Biosci. 2021, 8, 704685. [Google Scholar]
- Kociurzynski, R.; Makshakova, O.N.; Knecht, V.; Römer, W. Multiscale molecular dynamics studies reveal different modes of receptor clustering by Gb3-binding lectins. J. Chem. Theory Comput. 2021, 17, 2488–2501. [Google Scholar] [CrossRef]
- Chan, P.H.W.; Weissbach, S.; Okon, M.; Withers, S.G.; McIntosh, L.P. Nuclear magnetic resonance spectral assignments of α-1,4-galactosyltransferase LgtC from Neisseria meningitidis: Substrate binding and multiple conformational states. Biochemistry 2012, 51, 8278–8292. [Google Scholar] [CrossRef]
- Persson, K.; Ly, H.D.; Dieckelmann, M.; Wakarchuk, W.W.; Withers, S.G.; Strynadka, N.C.J. Crystal structure of the retaining galactosyltransferase LgtC from Neisseria meningitidis in complex with donor and acceptor sugar analogs. Nat. Struct. Mol. Biol. 2001, 8, 166–175. [Google Scholar]
- Madl, J.; Villringer, S.; Römer, W. Delving into lipid-driven endocytic mechanisms using biomimetic membranes. In Chemical and Synthetic Approaches in Membrane Biology; Shukla, A.K., Ed.; Springer Protocols Handbooks; Springer: New York, NY, USA, 2017; pp. 17–36. [Google Scholar]
- Šali, A.; Blundell, T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [PubMed]
- Wu, E.L.; Cheng, X.; Jo, S.; Rui, H.; Song, K.C.; Dávila-Contreras, E.M.; Qi, Y.; Lee, J.; Monje-Galvan, V.; Venable, R.M.; et al. CHARMM-GUI membrane builder toward realistic biological membrane simulations. J. Comput. Chem. 2014, 35, 1997–2004. [Google Scholar] [PubMed]
- Huang, M.L.; Tota, E.M.; Verespy, S., III; Godula, K. Glycocalyx scaffolding to control cell surface glycan displays. Curr. Protoc. Chem. Biol. 2018, 10, e40. [Google Scholar] [CrossRef] [PubMed]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Duan, L.; Liu, X.; Zhang, J.Z.H. Interaction entropy: A new paradigm for highly efficient and reliable computation of protein-ligand binding free energy. J. Am. Chem. Soc. 2016, 138, 5722–5728. [Google Scholar]
- Valdés-Tresanco, M.S.; Valdés-Tresanco, M.E.; Valiente, P.A.; Moreno, E. gmx_MMPBSA: A new tool to perform end-state free energy calculations with GROMACS. J. Chem. Theory Comput. 2021, 17, 6281–6291. [Google Scholar]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Adlercreutz, D.; Weadge, J.T.; Petersen, B.O.; Duus, J.Ø.; Dovichi, N.J.; Palcic, M.M. Enzymatic synthesis of Gb3 and iGb3 ceramides. Carbohydr. Res. 2010, 345, 1384–1388. [Google Scholar] [CrossRef][Green Version]
- Lairson, L.L.; Watts, A.G.; Wakarchuk, W.W.; Withers, S.G. Using substrate engineering to harness enzymatic promiscuity and expand biological catalysis. Nat. Chem. Biol. 2006, 2, 724–728. [Google Scholar] [CrossRef]
- Ling, H.; Boodhoo, A.; Hazes, B.; Cummings, M.D.; Armstrong, G.D.; Brunton, J.L.; Read, R.J. Structure of the Shiga-like toxin I B-pentamer complexed with an analogue of its receptor Gb3. Biochemistry 1998, 37, 1777–1788. [Google Scholar] [CrossRef]
- Gallegos, K.M.; Conrady, D.G.; Karve, S.S.; Gunasekera, T.S.; Herr, A.B.; Weiss, A.A. Shiga toxin binding to glycolipids and glycans. PLoS ONE 2012, 7, e30368. [Google Scholar]
- Perez, S.; Makshakova, O. Multifaceted computational modeling in glycoscience. Chem. Rev. 2022, 122, 15914–15970. [Google Scholar] [PubMed]
- Lete, M.G.; Franconetti, A.; Delgado, S.; Jiménez-Barbero, J.; Ardá, A. Oligosaccharide presentation modulates the molecular recognition of glycolipids by galectins on membrane surfaces. Pharmaceuticals 2022, 15, 145. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Ulrich-Bott, B.; Wiegandt, H. Micellar properties of glycosphingolipids in aqueous media. J. Lipid Res. 1984, 25, 1233–1245. [Google Scholar] [CrossRef]
- Sot, J.; Goñi, F.M.; Alonso, A. Molecular associations and surface-active properties of short- and long-N-acyl chain ceramides. Biochim. Biophys. Acta (BBA)-Biomembr. 2005, 1711, 12–19. [Google Scholar] [CrossRef]
- Reviakine, I.; Johannsmann, D.; Richter, R.P. Hearing What You Cannot See and Visualizing What You Hear: Interpreting Quartz Crystal Microbalance Data from Solvated Interfaces. Anal. Chem. 2011, 83, 8838–8848. [Google Scholar] [CrossRef]
- Boix, E.; Zhang, Y.; Swaminathan, G.J.; Brew, K.; Acharya, K.R. Structural Basis of Ordered Binding of Donor and Acceptor Substrates to the Retaining Glycosyltransferase, α-1,3-Galactosyltransferase. J. Biol. Chem. 2002, 277, 28310–28318. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jabeguero, D.; Siukstaite, L.; Wang, C.; Mitrovic, A.; Pérez, S.; Makshakova, O.; Richter, R.P.; Römer, W.; Breton, C. Enzymatic Glyco-Modification of Synthetic Membrane Systems. Biomolecules 2023, 13, 335. https://doi.org/10.3390/biom13020335
Jabeguero D, Siukstaite L, Wang C, Mitrovic A, Pérez S, Makshakova O, Richter RP, Römer W, Breton C. Enzymatic Glyco-Modification of Synthetic Membrane Systems. Biomolecules. 2023; 13(2):335. https://doi.org/10.3390/biom13020335
Chicago/Turabian StyleJabeguero, Dylan, Lina Siukstaite, Chunyue Wang, Anna Mitrovic, Serge Pérez, Olga Makshakova, Ralf P. Richter, Winfried Römer, and Christelle Breton. 2023. "Enzymatic Glyco-Modification of Synthetic Membrane Systems" Biomolecules 13, no. 2: 335. https://doi.org/10.3390/biom13020335
APA StyleJabeguero, D., Siukstaite, L., Wang, C., Mitrovic, A., Pérez, S., Makshakova, O., Richter, R. P., Römer, W., & Breton, C. (2023). Enzymatic Glyco-Modification of Synthetic Membrane Systems. Biomolecules, 13(2), 335. https://doi.org/10.3390/biom13020335







