Construction of Ovarian Cancer Prognostic Model Based on the Investigation of Ferroptosis-Related lncRNA
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Screening for Differentially Expressed Ferroptosis-Related lncRNAs
2.3. Construction and Validation of Prognostic Models
2.4. Construction of a Predictive Nomogram
2.5. Analysis of Immune Cell Infiltration in the Tumor Microenvironment (TME)
2.6. Gene Set Enrichment Analysis (GSEA)
2.7. Cell Culture
2.8. RNA Isolation and Quantitative Reverse Transcriptase Polymerase Chain Reaction (qRT-PCR)
2.9. Data Analysis
3. Results
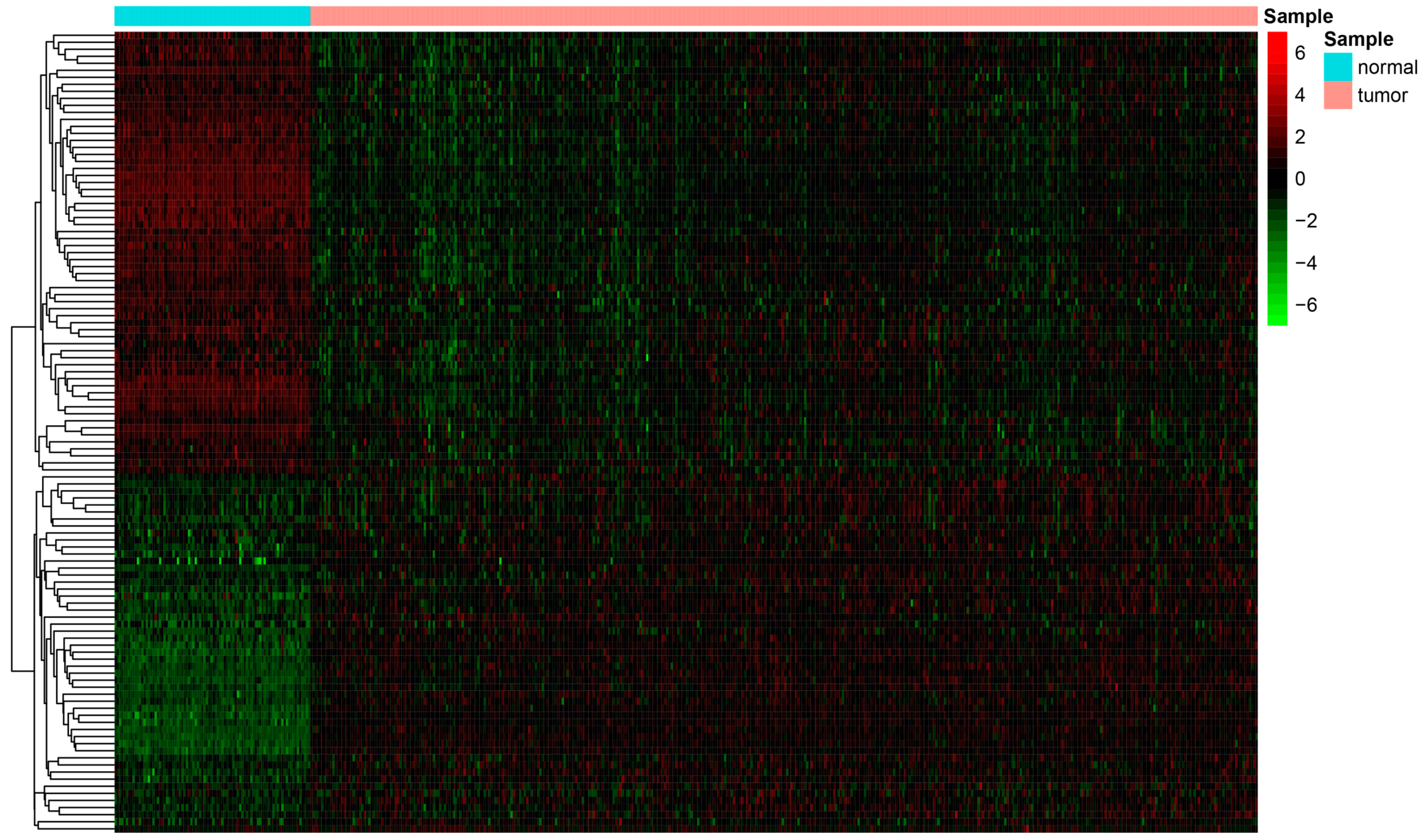
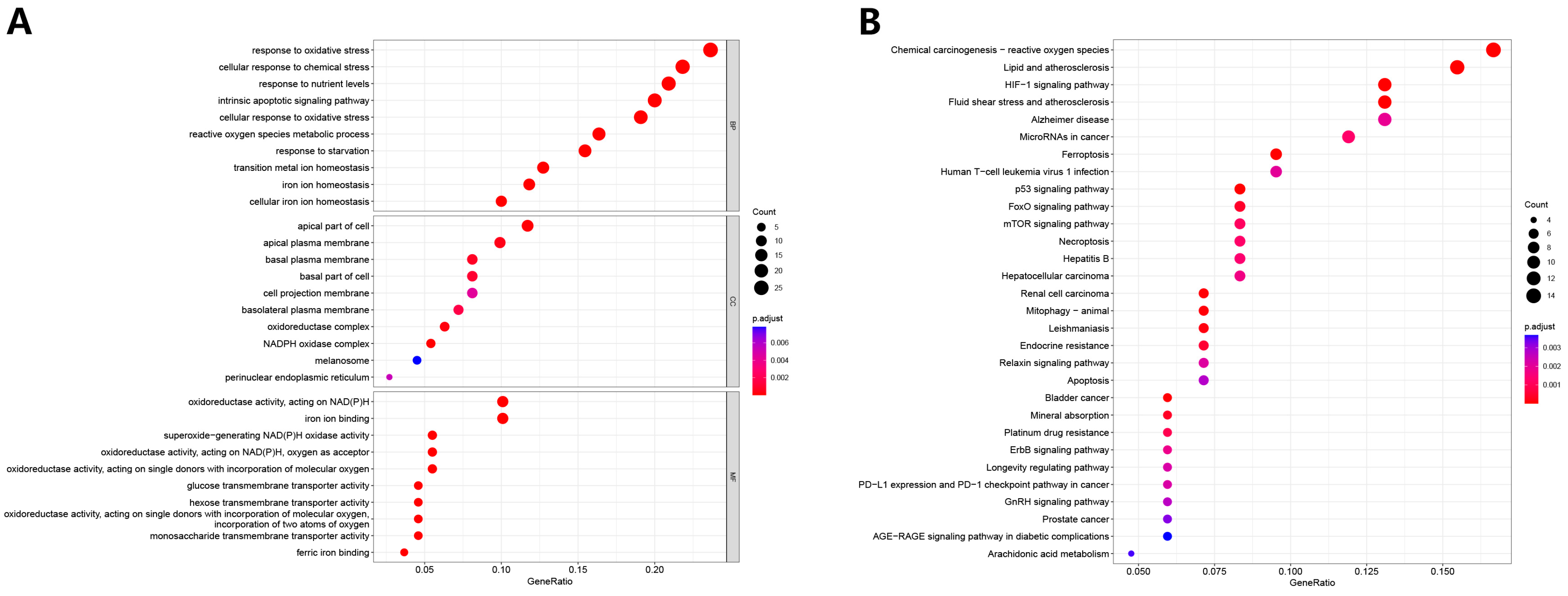
3.1. Differential Analysis and Enrichment Analysis of Ferroptosis-Related Genes
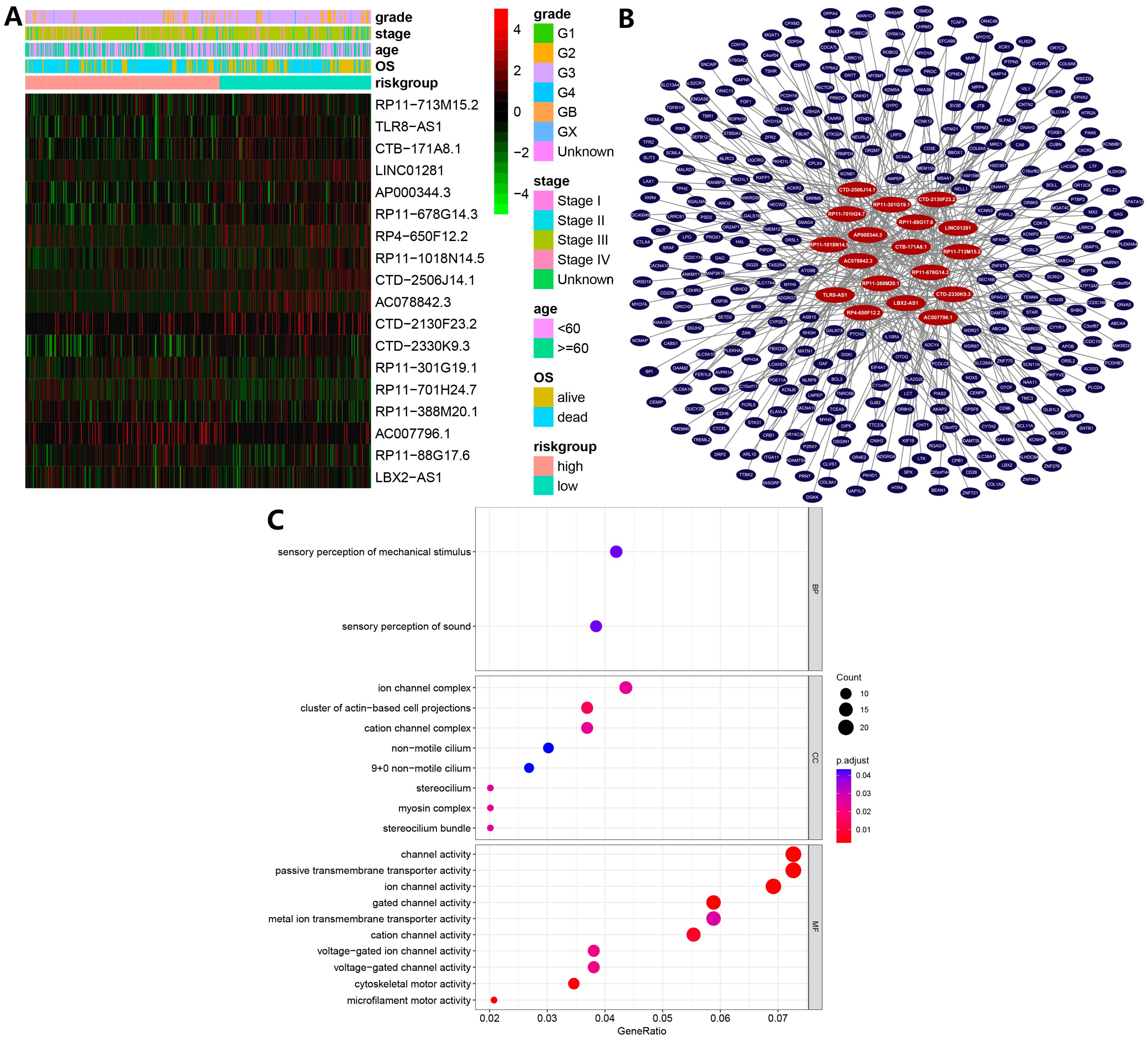
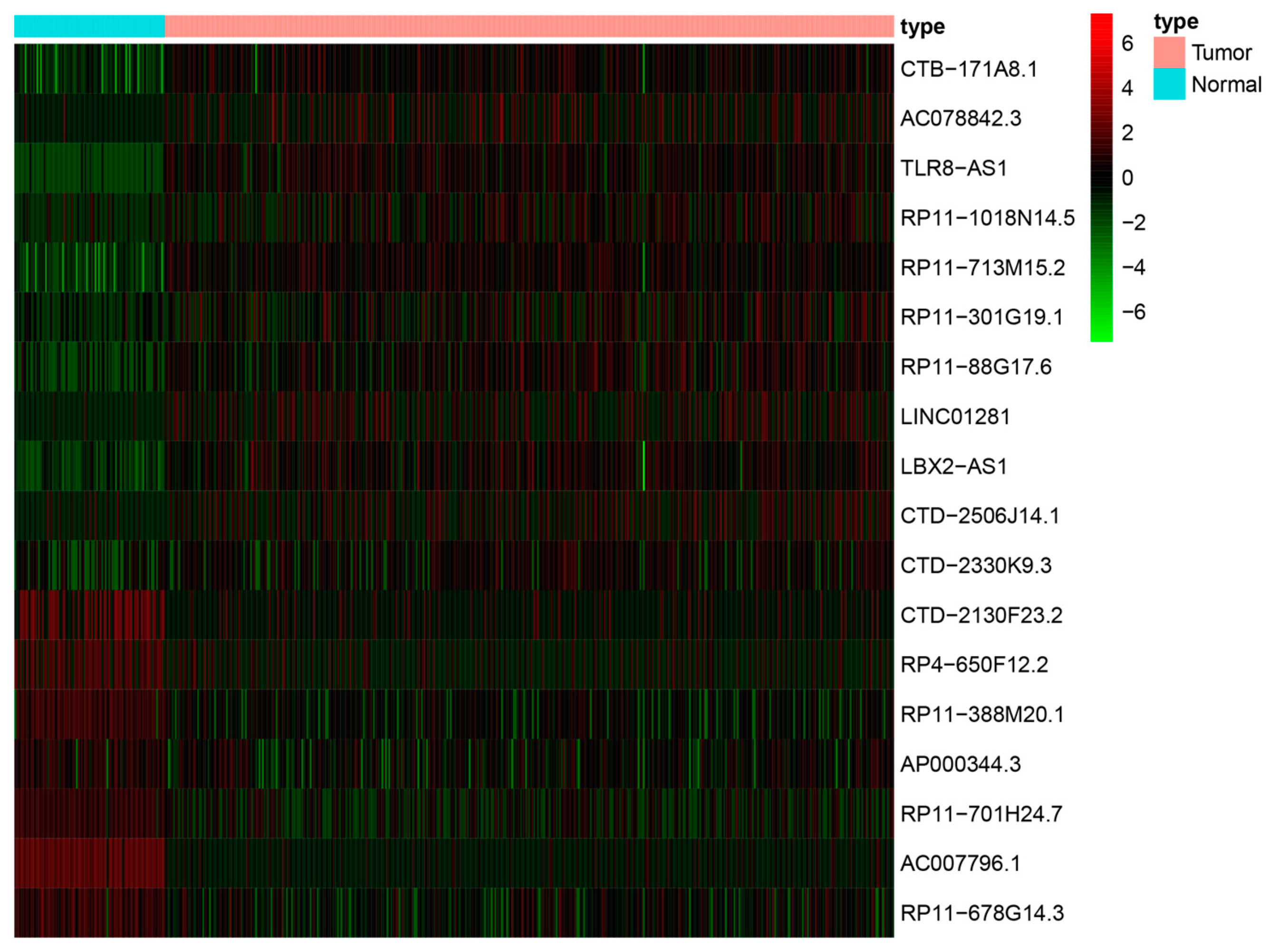
3.2. Ferroptosis-Related lncRNAs-Based Prognostic Signature
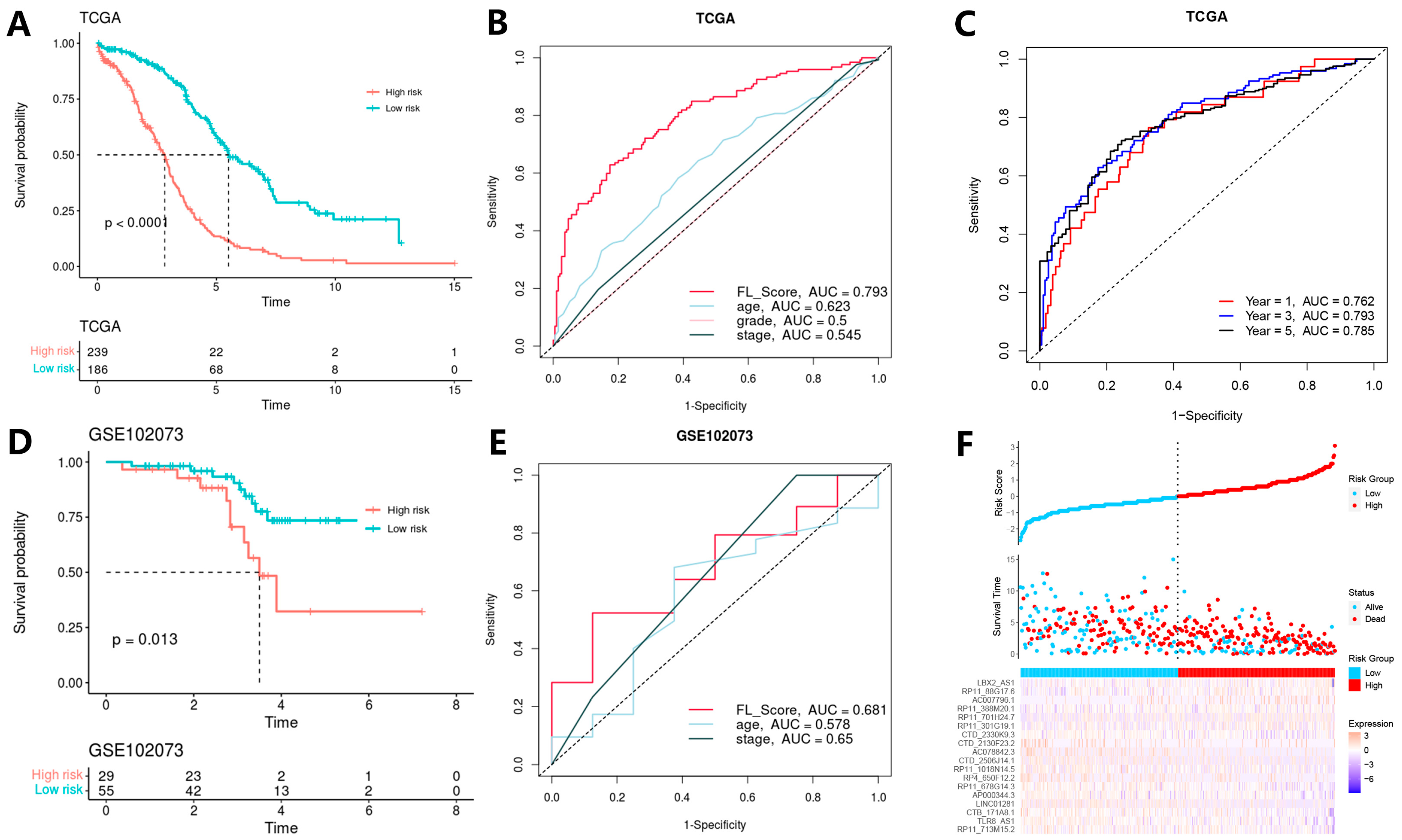
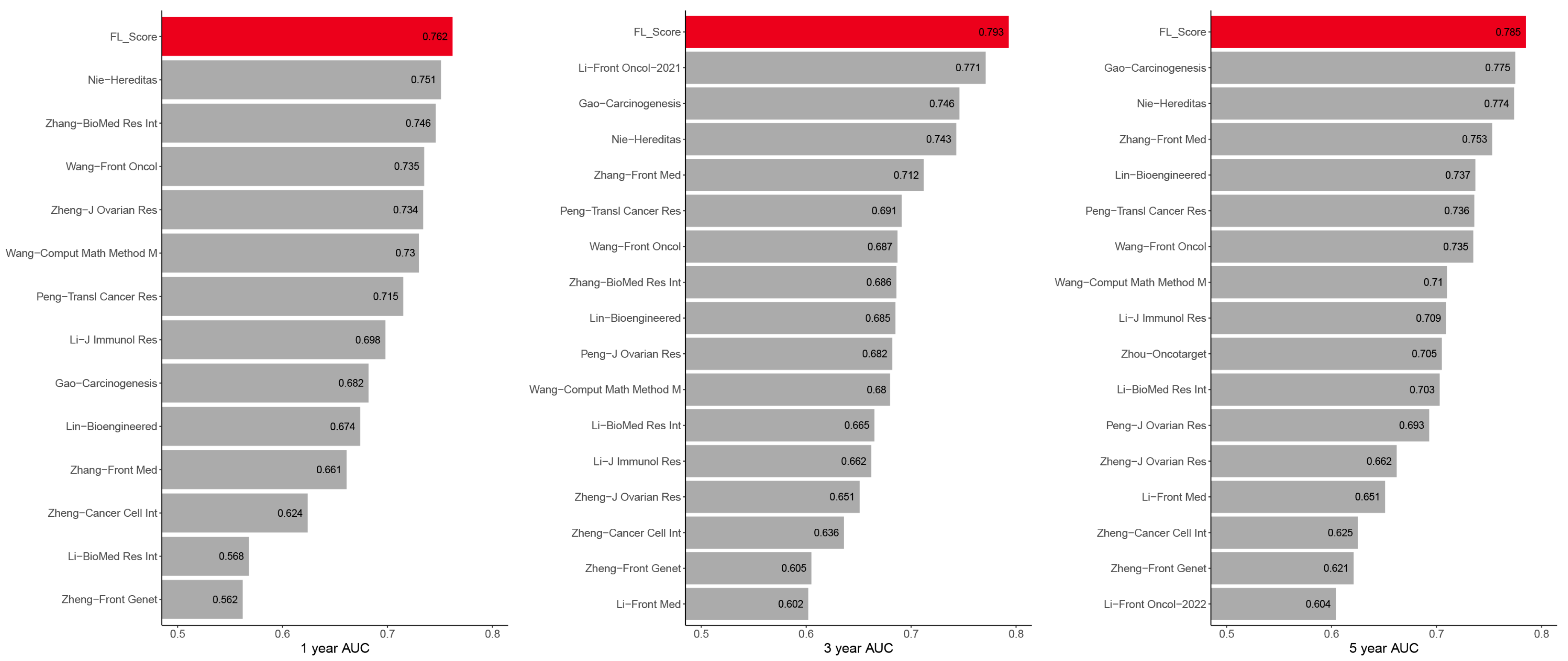
3.3. Model Construction and Validation
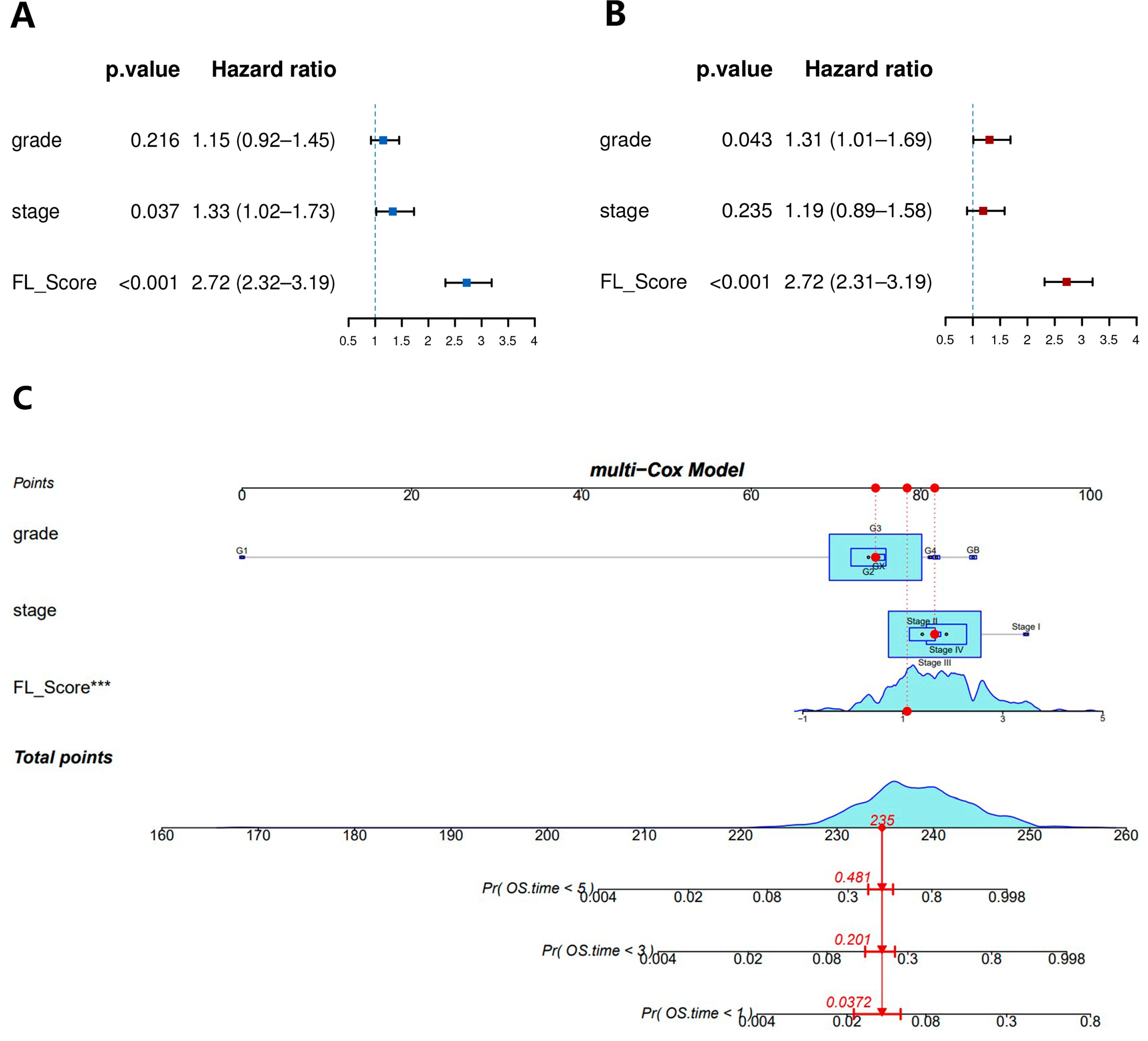
3.4. Construction of a Nomogram for Predicting Prognosis
3.5. Gene Set Enrichment Analysis (GSEA)
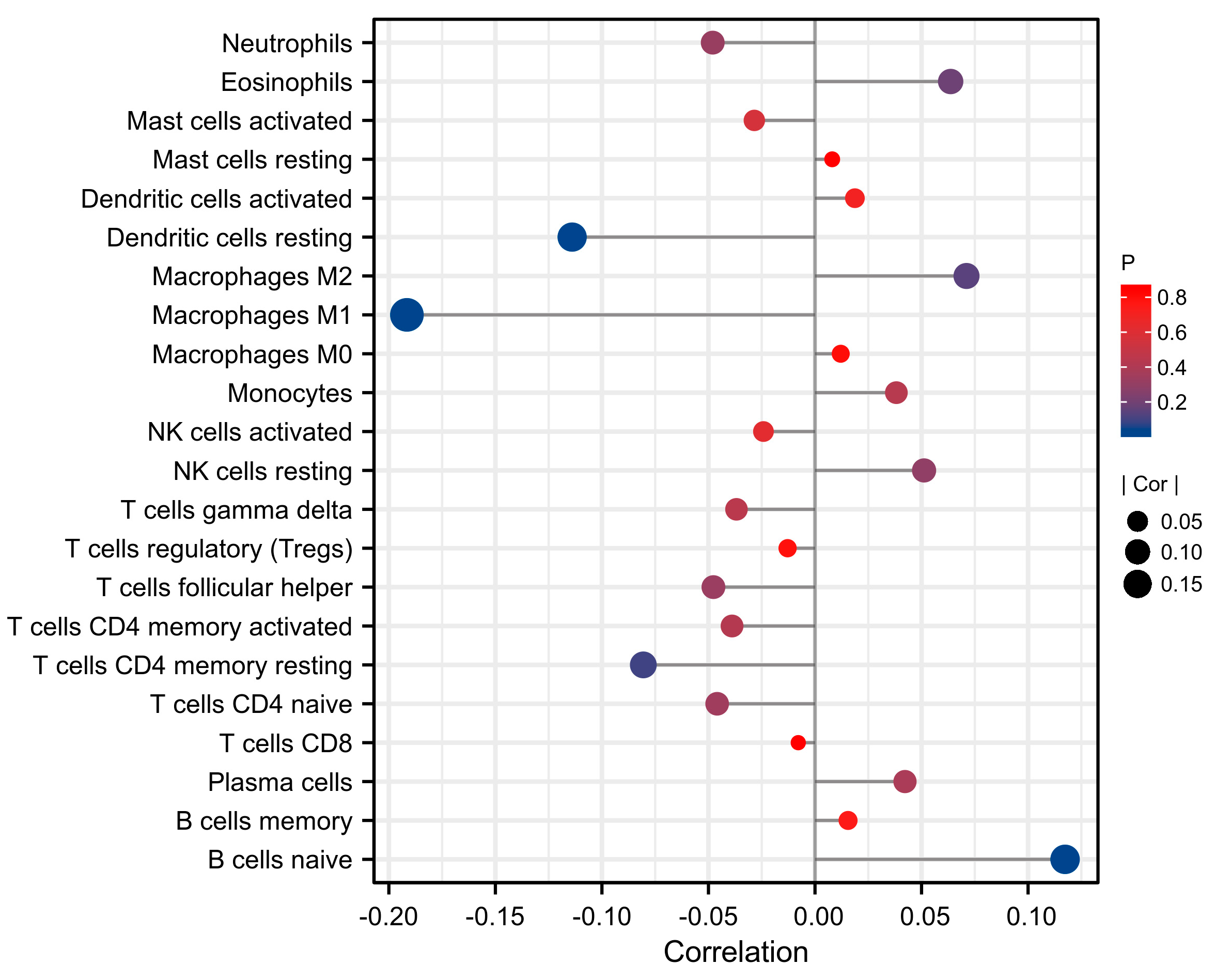
3.6. Correlation between Immune Cell Infiltration and FL Score
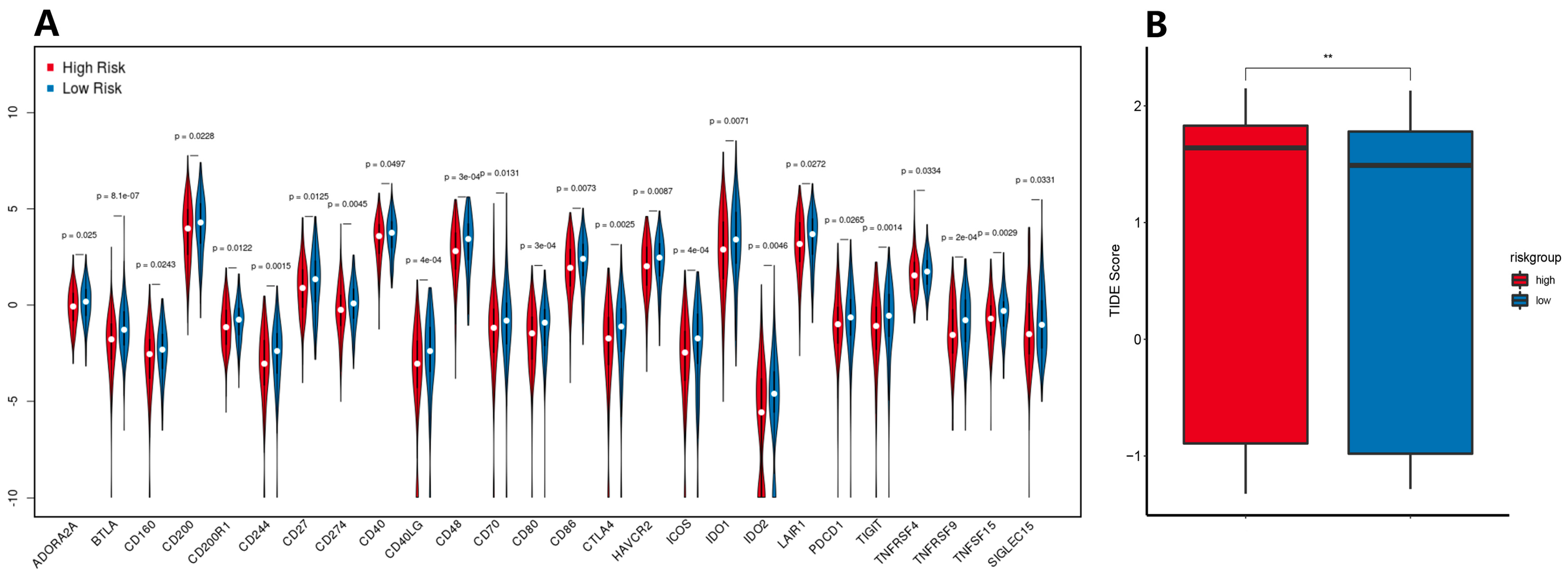
3.7. Immune Checkpoint Expression Levels and ICB Response Prediction
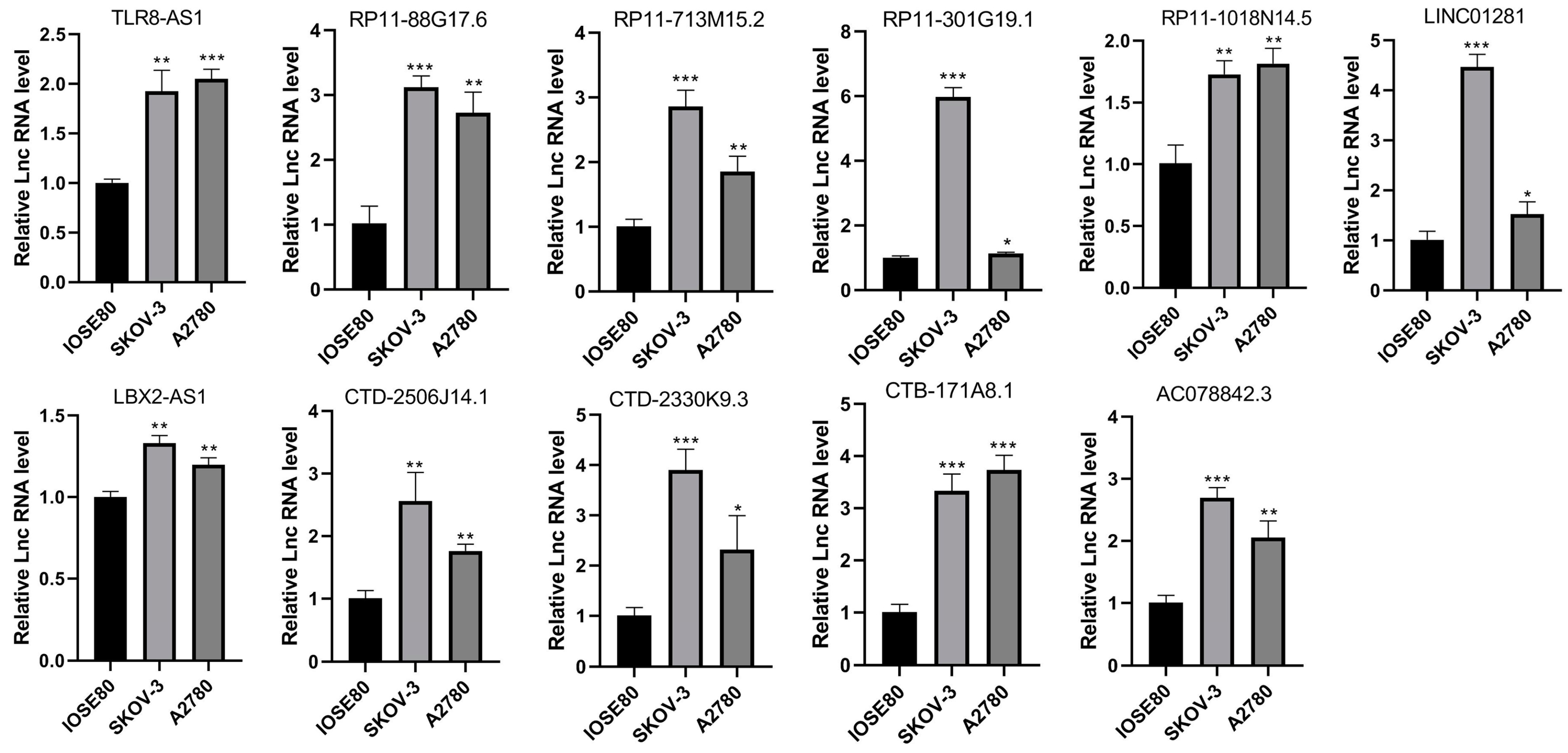
3.8. LncRNA Expression Levels in Cell Lines
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Lheureux, S.; Braunstein, M.; Oza, A.M. Epithelial ovarian cancer: Evolution of management in the era of precision medicine. CA Cancer J. Clin. 2019, 69, 280–304. [Google Scholar] [CrossRef] [PubMed]
- Meyer, L.A.; He, W.; Sun, C.C.; Zhao, H.; Wright, A.A.; Suidan, R.S.; Dottino, J.; Alejandro Rauh-Hain, J.; Lu, K.H.; Giordano, S.H. Neoadjuvant chemotherapy in elderly women with ovarian cancer: Rates of use and effectiveness. Gynecol. Oncol. 2018, 150, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Salminen, L.; Gidwani, K.; Grenman, S.; Carpen, O.; Hietanen, S.; Pettersson, K.; Huhtinen, K.; Hynninen, J. HE4 in the evaluation of tumor load and prognostic stratification of high grade serous ovarian carcinoma. Acta Oncol. 2020, 59, 1461–1468. [Google Scholar] [CrossRef] [PubMed]
- Piatek, S.; Panek, G.; Lewandowski, Z.; Bidzinski, M.; Piatek, D.; Kosinski, P.; Wielgos, M. Rising serum CA-125 levels within the normal range is strongly associated recurrence risk and survival of ovarian cancer. J. Ovarian Res. 2020, 13, 102. [Google Scholar]
- Cescon, D.W.; Bratman, S.V.; Chan, S.M.; Siu, L.L. Circulating tumor DNA and liquid biopsy in oncology. Nat. Cancer 2020, 1, 276–290. [Google Scholar] [CrossRef]
- Hayes, J.; Peruzzi, P.P.; Lawler, S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol. Med. 2014, 20, 460–469. [Google Scholar] [CrossRef]
- Lee, Y.R.; Kim, G.; Tak, W.Y.; Jang, S.Y.; Kweon, Y.O.; Park, J.G.; Lee, H.W.; Han, Y.S.; Chun, J.M.; Park, S.Y.; et al. Circulating exosomal noncoding RNAs as prognostic biomarkers in human hepatocellular carcinoma. Int. J. Cancer 2019, 144, 1444–1452. [Google Scholar] [CrossRef]
- Zhang, X.; Yang, Q. An Immune-Related lncRNA Pairing Model for Predicting the Prognosis and Immune-Infiltrating Cell Condition in Human Ovarian Cancer. Biomed. Res. Int. 2022, 2022, 3168408. [Google Scholar] [CrossRef]
- Li, Y.; Wang, J.; Wang, F.; Gao, C.; Cao, Y.; Wang, J. Development and Verification of an Autophagy-Related lncRNA Signature to Predict Clinical Outcomes and Therapeutic Responses in Ovarian Cancer. Front. Med. 2021, 8, 715250. [Google Scholar] [CrossRef]
- Nie, X.; Tan, J.C. N6-methyladenosine-related lncRNAs is a potential marker for predicting prognosis and immunotherapy in ovarian cancer. Hereditas 2022, 159, 17. [Google Scholar] [CrossRef] [PubMed]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef] [PubMed]
- Doll, S.; Conrad, M. Iron and ferroptosis: A still ill-defined liaison. IUBMB Life 2017, 69, 423–434. [Google Scholar] [CrossRef] [PubMed]
- Chan, D.W.; Yung, M.M.; Chan, Y.S.; Xuan, Y.; Yang, H.; Xu, D.; Zhan, J.B.; Chan, K.K.; Ng, T.B.; Ngan, H.Y. MAP30 protein from Momordica charantia is therapeutic and has synergic activity with cisplatin against ovarian cancer in vivo by altering metabolism and inducing ferroptosis. Pharmacol. Res. 2020, 161, 105157. [Google Scholar] [CrossRef]
- You, Y.; Fan, Q.; Huang, J.Y.; Wu, Y.Q.; Lin, H.Y.; Zhang, Q.X. Ferroptosis-Related Gene Signature Promotes Ovarian Cancer by Influencing Immune Infiltration and Invasion. J. Oncol. 2021, 2021, 9915312. [Google Scholar] [CrossRef]
- Tesfay, L.; Paul, B.T.; Konstorum, A.; Deng, Z.; Cox, A.O.; Lee, J.; Furdui, C.M.; Hegde, P.; Torti, F.M.; Torti, S.V. Stearoyl-CoA Desaturase 1 Protects Ovarian Cancer Cells from Ferroptotic Cell Death. Cancer Res. 2019, 79, 5355–5366. [Google Scholar] [CrossRef]
- Jiang, N.; Zhang, X.; Gu, X.; Li, X.; Shang, L. Progress in understanding the role of lncRNA in programmed cell death. Cell Death Discov. 2021, 7, 30. [Google Scholar] [CrossRef]
- Zheng, Z.; Zhang, Q.; Wu, W.; Xue, Y.; Liu, S.; Chen, Q.; Lin, D. Identification and Validation of a Ferroptosis-Related Long Non-coding RNA Signature for Predicting the Outcome of Lung Adenocarcinoma. Front. Genet. 2021, 12, 690509. [Google Scholar] [CrossRef]
- Tang, Y.; Li, C.; Zhang, Y.J.; Wu, Z.H. Ferroptosis-Related Long Non-Coding RNA signature predicts the prognosis of Head and neck squamous cell carcinoma. Int. J. Biol. Sci. 2021, 17, 702–711. [Google Scholar] [CrossRef]
- Zhou, N.; Bao, J. FerrDb: A manually curated resource for regulators and markers of ferroptosis and ferroptosis-disease associations. Database 2020, 2020, baaa021. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdottir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.G.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453. [Google Scholar] [CrossRef] [PubMed]
- Sturm, G.; Finotello, F.; Petitprez, F.; Zhang, J.D.; Baumbach, J.; Fridman, W.H.; List, M.; Aneichyk, T. Comprehensive evaluation of transcriptome-based cell-type quantification methods for immuno-oncology. Bioinformatics 2019, 35, i436–i445. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Gu, S.; Pan, D.; Fu, J.; Sahu, A.; Hu, X.; Li, Z.; Traugh, N.; Bu, X.; Li, B.; et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018, 24, 1550–1558. [Google Scholar] [CrossRef]
- Fu, J.; Li, K.; Zhang, W.; Wan, C.; Zhang, J.; Jiang, P.; Liu, X.S. Large-scale public data reuse to model immunotherapy response and resistance. Genome Med. 2020, 12, 21. [Google Scholar] [CrossRef] [PubMed]
- Adler, P.; Kolde, R.; Kull, M.; Tkachenko, A.; Peterson, H.; Reimand, J.; Vilo, J. Mining for coexpression across hundreds of datasets using novel rank aggregation and visualization methods. Genome Biol. 2009, 10, R139. [Google Scholar] [CrossRef]
- Peng, Y.; Wang, H.; Huang, Q.; Wu, J.; Zhang, M. A prognostic model based on immune-related long noncoding RNAs for patients with epithelial ovarian cancer. J. Ovarian Res. 2022, 15, 8. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, Z.Y.; Chen, Y.C.; Zhang, X.Y.; Wu, N.; Wang, J. Identification and validation of an immune-related lncRNAs signature to predict the overall survival of ovarian cancer. Front. Oncol. 2022, 12, 999654. [Google Scholar] [CrossRef]
- Zhou, M.; Sun, Y.; Sun, Y.; Xu, W.; Zhang, Z.; Zhao, H.; Zhong, Z.; Sun, J. Comprehensive analysis of lncRNA expression profiles reveals a novel lncRNA signature to discriminate nonequivalent outcomes in patients with ovarian cancer. Oncotarget 2016, 7, 32433–32448. [Google Scholar] [CrossRef]
- Li, H.; Wang, S.; Yao, Q.; Liu, Y.; Yang, J.; Xu, L.; Yang, G. A Combined Long Noncoding RNA Signature as a Candidate Prognostic Biomarker for Ovarian Cancer. Front. Oncol. 2021, 11, 624240. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Hao, Y.; Rao, B.; Zhang, Z. A ferroptosis-related lncRNA signature predicts prognosis in ovarian cancer patients. Transl. Cancer Res. 2021, 10, 4802–4816. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Guo, J.; Zhu, L.; Zhou, Y.; Tong, J. Comprehensive analyses of glycolysis-related lncRNAs for ovarian cancer patients. J. Ovarian Res. 2021, 14, 124. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Huo, F.F.; Wen, Y.Y.; Jiang, M. Screening and Identification of an Immune-Associated lncRNA Prognostic Signature in Ovarian Carcinoma: Evidence from Bioinformatic Analysis. Biomed Res. Int. 2021, 2021, 6680036. [Google Scholar] [CrossRef]
- Zheng, J.; Guo, J.; Zhang, H.; Cao, B.; Xu, G.; Zhang, Z.; Tong, J. Four Prognosis-Associated lncRNAs Serve as Biomarkers in Ovarian Cancer. Front. Genet. 2021, 12, 672674. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Guo, J.; Cao, B.; Zhou, Y.; Tong, J. Identification and validation of lncRNAs involved in m6A regulation for patients with ovarian cancer. Cancer Cell Int. 2021, 21, 363. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Mei, S.; Cai, M.; Zhai, D.; Zhang, D.; Yu, J.; Ni, Z.; Yu, C. Ferroptosis-Related Long Noncoding RNAs as Prognostic Biomarkers for Ovarian Cancer. Front. Oncol. 2022, 12, 888699. [Google Scholar] [CrossRef] [PubMed]
- Lin, N.; Lin, J.Z.; Tanaka, Y.; Sun, P.; Zhou, X. Identification and validation of a five-lncRNA signature for predicting survival with targeted drug candidates in ovarian cancer. Bioengineered 2021, 12, 3263–3274. [Google Scholar] [CrossRef]
- Zhang, Z.; Xu, Z.; Yan, Y. Role of a Pyroptosis-Related lncRNA Signature in Risk Stratification and Immunotherapy of Ovarian Cancer. Front. Med. 2021, 8, 793515. [Google Scholar] [CrossRef]
- Wang, X.; Wang, J.; Yu, M. Immune Subtype Profiling and Establishment of Prognostic Immune-Related lncRNA Pairs in Human Ovarian Cancer. Comput. Math. Methods Med. 2022, 2022, 8338137. [Google Scholar] [CrossRef]
- Li, M.; Zhang, L.; Feng, M.; Huang, X. m6A-Related lncRNA Signature Is Involved in Immunosuppression and Predicts the Patient Prognosis of the Age-Associated Ovarian Cancer. J. Immunol. Res. 2022, 2022, 3258400. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Pang, X.; Ren, F.; Zhu, L. Identification of a ferroptosis-related long non-coding RNA Signature for prognosis prediction of ovarian cancer. Carcinogenesis 2022. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Jia, R.; Li, H.; Yu, H.; Ren, K.; Jia, S.; Li, Y.; Wang, Q. Insight into the Double-Edged Role of Ferroptosis in Disease. Biomolecules 2021, 11, 1790. [Google Scholar] [CrossRef] [PubMed]
- Mou, Y.; Wang, J.; Wu, J.; He, D.; Zhang, C.; Duan, C.; Li, B. Ferroptosis, a new form of cell death: Opportunities and challenges in cancer. J. Hematol. Oncol. 2019, 12, 34. [Google Scholar] [CrossRef]
- Na, K.; Kim, M.; Kim, C.Y.; Lim, J.S.; Cho, J.Y.; Shin, H. Potential Regulatory Role of Human-Carboxylesterase-1 Glycosylation in Liver Cancer Cell Growth. J. Proteome Res. 2020, 19, 4867–4883. [Google Scholar] [CrossRef]
- Carneiro, B.A.; El-Deiry, W.S. Targeting apoptosis in cancer therapy. Nat. Rev. Clin. Oncol. 2020, 17, 395–417. [Google Scholar] [CrossRef] [PubMed]
- Capelletti, M.M.; Manceau, H.; Puy, H.; Peoc’h, K. Ferroptosis in Liver Diseases: An Overview. Int. J. Mol. Sci. 2020, 21, 4908. [Google Scholar] [CrossRef]
- Lachaier, E.; Louandre, C.; Godin, C.; Saidak, Z.; Baert, M.; Diouf, M.; Chauffert, B.; Galmiche, A. Sorafenib induces ferroptosis in human cancer cell lines originating from different solid tumors. Anticancer Res. 2014, 34, 6417–6422. [Google Scholar]
- Chen, G.Q.; Benthani, F.A.; Wu, J.; Liang, D.; Bian, Z.X.; Jiang, X. Artemisinin compounds sensitize cancer cells to ferroptosis by regulating iron homeostasis. Cell Death Differ. 2020, 27, 242–254. [Google Scholar] [CrossRef]
- Eling, N.; Reuter, L.; Hazin, J.; Hamacher-Brady, A.; Brady, N.R. Identification of artesunate as a specific activator of ferroptosis in pancreatic cancer cells. Oncoscience 2015, 2, 517–532. [Google Scholar] [CrossRef]
- Mao, C.; Wang, X.; Liu, Y.; Wang, M.; Yan, B.; Jiang, Y.; Shi, Y.; Shen, Y.; Liu, X.; Lai, W.; et al. A G3BP1-Interacting lncRNA Promotes Ferroptosis and Apoptosis in Cancer via Nuclear Sequestration of p53. Cancer Res. 2018, 78, 3484–3496. [Google Scholar] [CrossRef]
- Wang, M.; Mao, C.; Ouyang, L.; Liu, Y.; Lai, W.; Liu, N.; Shi, Y.; Chen, L.; Xiao, D.; Yu, F.; et al. Long noncoding RNA LINC00336 inhibits ferroptosis in lung cancer by functioning as a competing endogenous RNA. Cell Death Differ. 2019, 26, 2329–2343. [Google Scholar] [CrossRef]
- Macklin, P.S.; McAuliffe, J.; Pugh, C.W.; Yamamoto, A. Hypoxia and HIF pathway in cancer and the placenta. Placenta 2017, 56, 8–13. [Google Scholar] [CrossRef]
- Balamurugan, K. HIF-1 at the crossroads of hypoxia, inflammation, and cancer. Int. J. Cancer 2016, 138, 1058–1066. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Zhao, C.; Zhou, Y.; Zheng, J.; Gao, S.; Lu, Y. HIF-1alpha binding to AEG-1 promoter induced upregulated AEG-1 expression associated with metastasis in ovarian cancer. Cancer Med. 2017, 6, 1072–1081. [Google Scholar] [CrossRef] [PubMed]
- Ai, Z.; Lu, Y.; Qiu, S.; Fan, Z. Overcoming cisplatin resistance of ovarian cancer cells by targeting HIF-1-regulated cancer metabolism. Cancer Lett. 2016, 373, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Huang, J. Current developments of targeting the p53 signaling pathway for cancer treatment. Pharmacol. Ther. 2021, 220, 107720. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Zhu, S.; Song, X.; Sun, X.; Fan, Y.; Liu, J.; Zhong, M.; Yuan, H.; Zhang, L.; Billiar, T.R.; et al. The Tumor Suppressor p53 Limits Ferroptosis by Blocking DPP4 Activity. Cell Rep. 2017, 20, 1692–1704. [Google Scholar] [CrossRef]
- Xu, Q.; Lin, Y.B.; Li, L.; Liu, J. LncRNA TLR8-AS1 promotes metastasis and chemoresistance of ovarian cancer through enhancing TLR8 mRNA stability. Biochem. Biophys. Res. Commun. 2020, 526, 857–864. [Google Scholar] [CrossRef]
- Cao, J.; Wang, H.; Liu, G.Q.; Tang, R.R.; Ding, Y.; Xu, P.F.; Wang, H.Y.; Miao, J.; Gu, X.Y.; Han, S.P. LBX2-AS1 promotes ovarian cancer progression by facilitating E2F2 gene expression via miR-455-5p and miR-491-5p sponging. J. Cell. Mol. Med. 2021, 25, 1178–1189. [Google Scholar] [CrossRef]
- Gu, H.Z.; Lin, R.R.; Zheng, F.Y.; Zhang, Q. ELK1 activated-long noncoding RNA LBX2-AS1 aggravates the progression of ovarian cancer through targeting miR-4784/KDM5C axis. J. Mol. Histol. 2021, 52, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Dong, X.H.; Pu, M.L.; Yang, H.W.; Chang, W.L.; Ji, F.H.; Liu, T.; Wei, C.Q.; Zhang, X.F.; Qiu, X.G. LBX2-AS1/miR-219a-2-3p/FUS/LBX2 positive feedback loop contributes to the proliferation of gastric cancer. Gastric Cancer 2020, 23, 449–463. [Google Scholar] [CrossRef] [PubMed]
- Ma, G.; Dai, W.J.; Zhang, J.; Li, Q.J.; Gu, B.; Song, Y.Q.; Yang, X.Z. ELK1-mediated upregulation of lncRNA LBX2-AS1 facilitates cell proliferation and invasion via regulating miR-491-5p/S100A11 axis in colorectal cancer. Int. J. Mol. Med. 2021, 48, 138. [Google Scholar] [CrossRef] [PubMed]
- Li, W.T.; Soufiany, I.; Lyu, X.; Lu, C.F.; Wei, Y.T.; Shi, Z.M.; You, Y.P. SP1-upregulated LBX2-AS1 promotes the progression of glioma by targeting the miR-491-5p/LIF axis. J. Cancer 2021, 12, 6989–7002. [Google Scholar] [CrossRef]
- Wang, F.M.; Luo, Y.; Zhang, L.; Younis, M.; Yuan, L.D. The LncRNA RP11-301G19.1/miR-582-5p/HMGB2 axis modulates the proliferation and apoptosis of multiple myeloma cancer cells via the PI3K/AKT signalling pathway. Cancer Gene Ther. 2022, 29, 292–303. [Google Scholar] [CrossRef]
- Shen, X.; Zhu, W.; Wang, G.; Li, J.; Deng, Q. Effect of long-chain non-coding RNA AP000344. 3 on the proliferation and invasion of human bladder cancer cells and its mechanism. Chin. J. Clin. Exp. Pathol. 2019, 35, 182–186. [Google Scholar]
- Williams, C.B.; Yeh, E.S.; Soloff, A.C. Tumor-associated macrophages: Unwitting accomplices in breast cancer malignancy. NPJ Breast Cancer 2016, 2, 15025. [Google Scholar] [CrossRef]
- Mantovani, A.; Sica, A. Macrophages, innate immunity and cancer: Balance, tolerance, and diversity. Curr. Opin. Immunol. 2010, 22, 231–237. [Google Scholar] [CrossRef]
- Ou, Z.Y.; Wang, Y.J.; Liu, L.F.; Li, L.; Yeh, S.Y.; Qi, L.; Chang, C.S. Tumor microenvironment B cells increase bladder cancer metastasis via modulation of the IL-8/androgen receptor (AR)/MMPs signals. Oncotarget 2015, 6, 26065–26078. [Google Scholar] [CrossRef]
- Charoentong, P.; Finotello, F.; Angelova, M.; Mayer, C.; Efremova, M.; Rieder, D.; Hackl, H.; Trajanoski, Z. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep. 2017, 18, 248–262. [Google Scholar] [CrossRef]
- Iglesia, M.D.; Parker, J.S.; Hoadley, K.A.; Serody, J.S.; Perou, C.M.; Vincent, B.G. Genomic Analysis of Immune Cell Infiltrates Across 11 Tumor Types. JNCI J. Natl. Cancer Inst. 2016, 108, djw144. [Google Scholar] [CrossRef] [PubMed]
- Woo, J.R.; Liss, M.A.; Muldong, M.T.; Palazzi, K.; Strasner, A.; Ammirante, M.; Varki, N.; Shabaik, A.; Howell, S.; Kane, C.J.; et al. Tumor infiltrating B-cells are increased in prostate cancer tissue. J. Transl. Med. 2014, 12, 30. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, Z.M.A.; Going, J.J.; Edwards, J.; Elsberger, B.; McMillan, D.C. The relationship between lymphocyte subsets and clinico-pathological determinants of survival in patients with primary operable invasive ductal breast cancer. Brit. J. Cancer 2013, 109, 1676–1684. [Google Scholar] [CrossRef]
- Shah, S.; Divekar, A.A.; Hilchey, S.P.; Cho, H.M.; Newman, C.L.; Shin, S.U.; Nechustan, H.; Challita-Eid, P.M.; Segal, B.M.; Yi, K.H.; et al. Increased rejection of primary tumors in mice lacking B cells: Inhibition of anti-tumor CTL and TH1 cytokine responses by B cells. Int. J. Cancer 2005, 117, 574–586. [Google Scholar] [CrossRef] [PubMed]
- Inoue, S.; Leitner, W.W.; Golding, B.; Scott, D. Inhibitory effects of B cells on antitumor immunity. Cancer Res. 2006, 66, 7741–7747. [Google Scholar] [CrossRef]

| TCGA | GSE102073 | ||||
|---|---|---|---|---|---|
| OV Samples | 427 | 85 | |||
| Age, mean/SD | 59.60 | 11.41 | 59 | 8.81 | |
| Stage, n/% | I | 1 | 0.23 | 1 | 1.18 |
| II | 26 | 6.09 | 3 | 3.53 | |
| III | 334 | 78.22 | 56 | 65.88 | |
| IV | 63 | 14.75 | 25 | 29.41 | |
| Grade, n/% | 1 | 1 | 0.23 | NA | NA |
| 2 | 52 | 12.18 | NA | NA | |
| 3 | 363 | 85.01 | NA | NA | |
| 4 | 1 | 0.23 | NA | NA | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, S.; Ji, J.; Wang, M.; Nie, J.; Wang, S. Construction of Ovarian Cancer Prognostic Model Based on the Investigation of Ferroptosis-Related lncRNA. Biomolecules 2023, 13, 306. https://doi.org/10.3390/biom13020306
Yang S, Ji J, Wang M, Nie J, Wang S. Construction of Ovarian Cancer Prognostic Model Based on the Investigation of Ferroptosis-Related lncRNA. Biomolecules. 2023; 13(2):306. https://doi.org/10.3390/biom13020306
Chicago/Turabian StyleYang, Shaoyi, Jie Ji, Meng Wang, Jinfu Nie, and Shujie Wang. 2023. "Construction of Ovarian Cancer Prognostic Model Based on the Investigation of Ferroptosis-Related lncRNA" Biomolecules 13, no. 2: 306. https://doi.org/10.3390/biom13020306
APA StyleYang, S., Ji, J., Wang, M., Nie, J., & Wang, S. (2023). Construction of Ovarian Cancer Prognostic Model Based on the Investigation of Ferroptosis-Related lncRNA. Biomolecules, 13(2), 306. https://doi.org/10.3390/biom13020306






