Prognostic Biomarker SPOCD1 and Its Correlation with Immune Infiltrates in Colorectal Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Acquisition and Preprocessing
2.2. Survival Analysis
2.3. Enrichment Analysis
2.4. Analyses Immune Infiltration
2.5. Patients and Clinical Specimens
2.6. Immunohistochemistry (IHC) Staining
2.7. Immunofluorescence Staining
2.8. Prediction of Drug Sensitivity
2.9. Statistical Analysis
3. Results
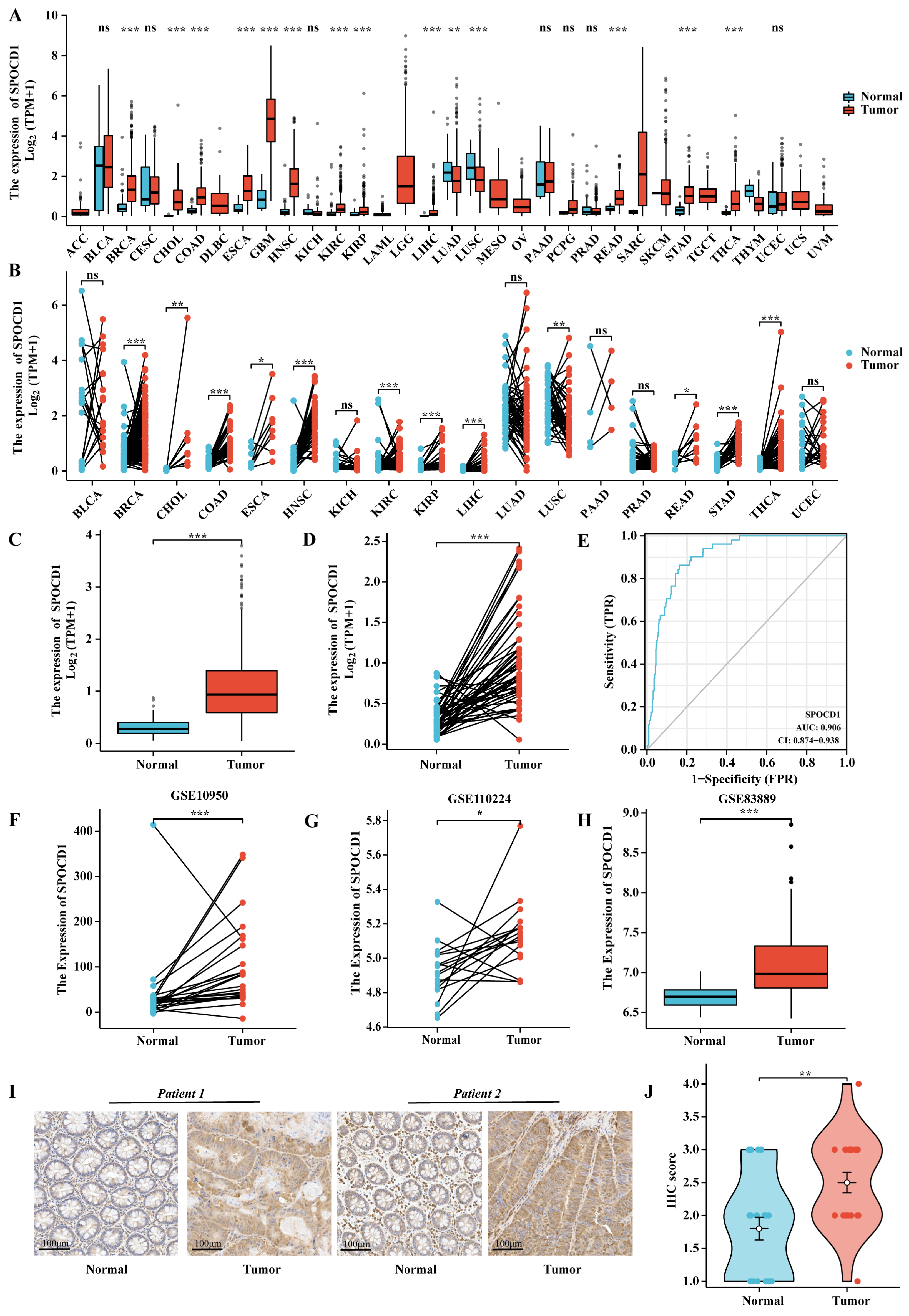
3.1. The mRNA and Protein Expression Levels of SPOCD1 Are Upregulated in CRC
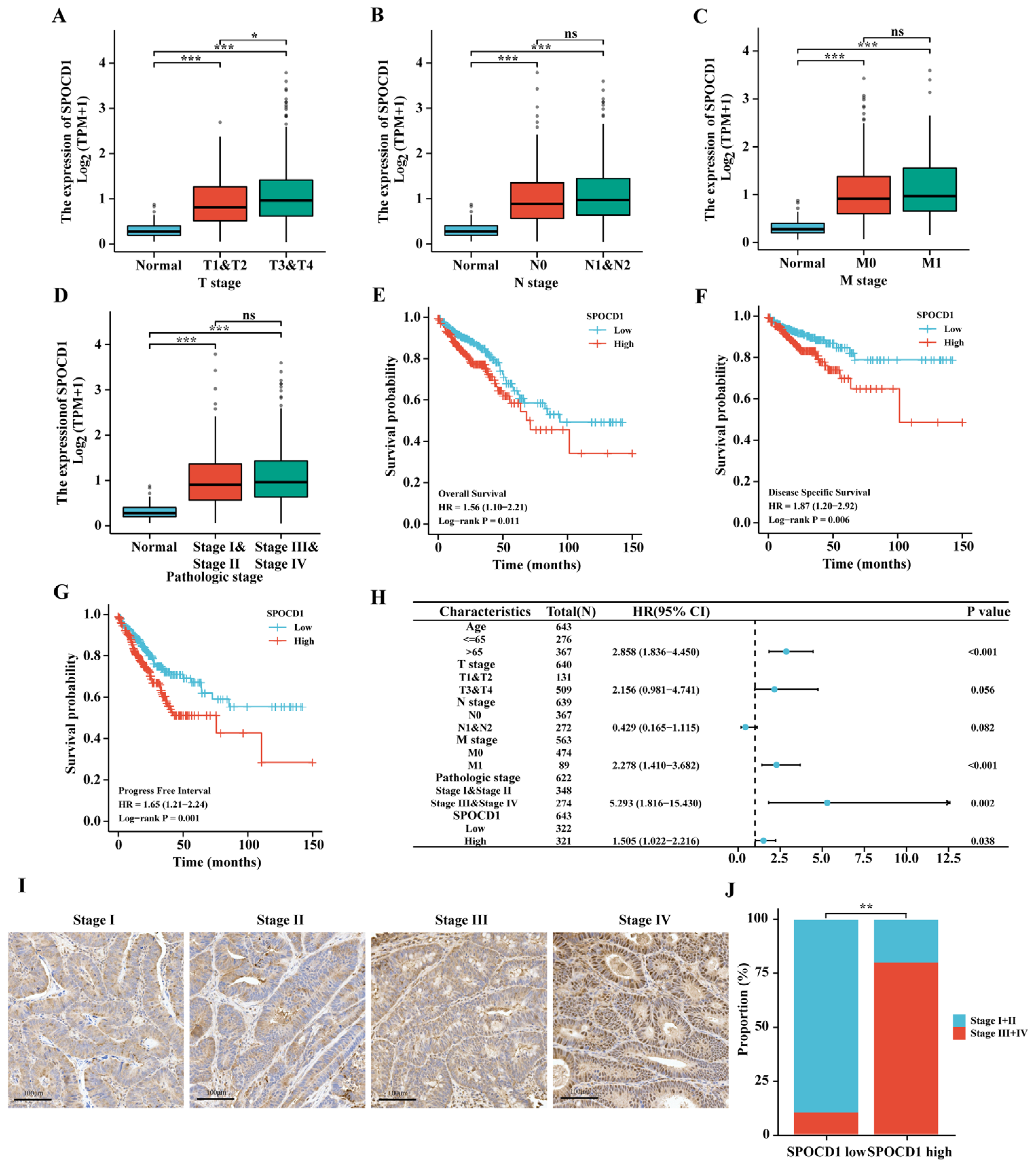
3.2. SPOCD1 Expression Correlates with the Tumor Pathological Stage of CRC
3.3. High SPOCD1 Expression Predicts Poor Prognosis in CRC Patients
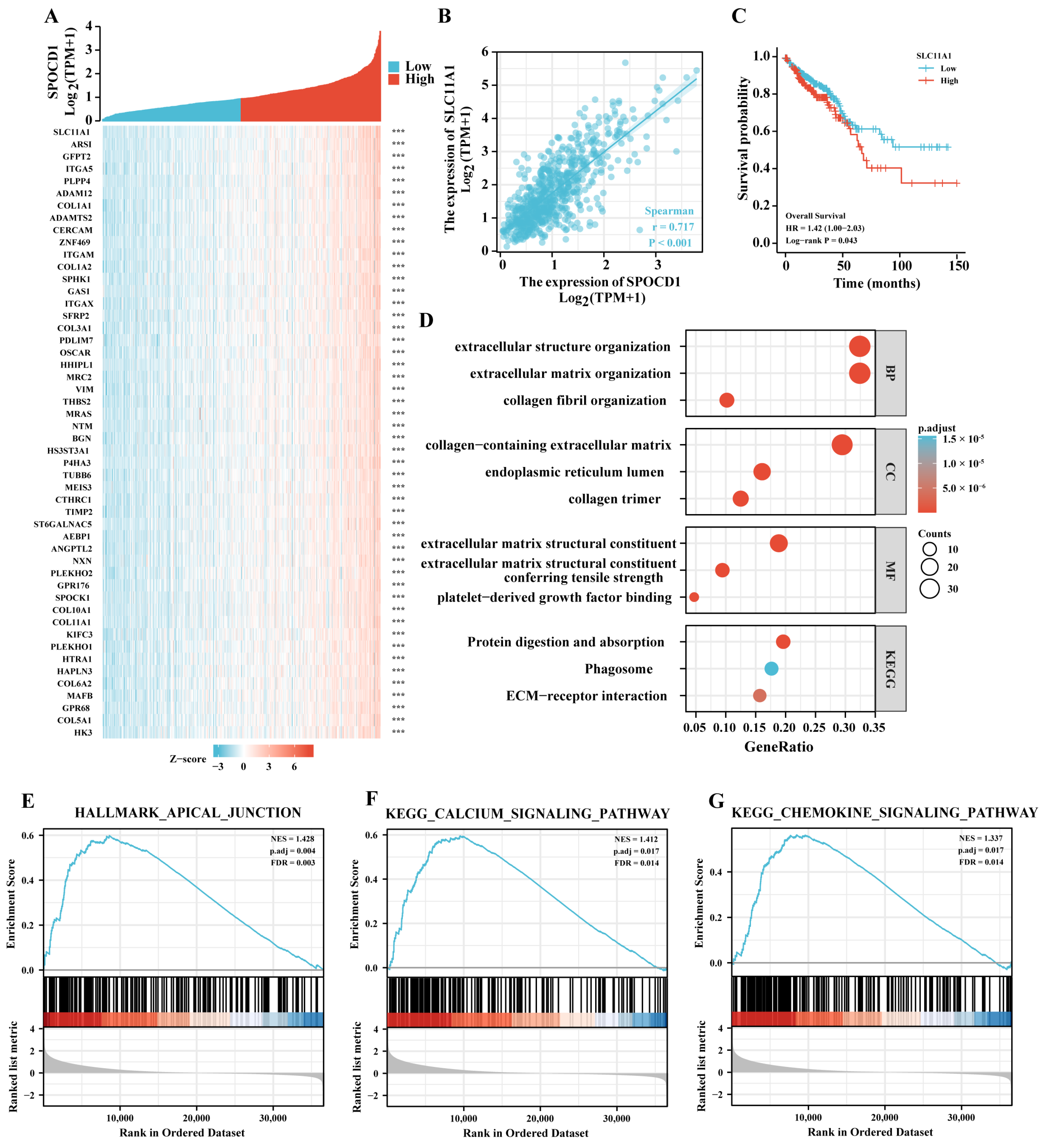
3.4. SPOCD1 May Be Involved in Malignant Progression of CRC
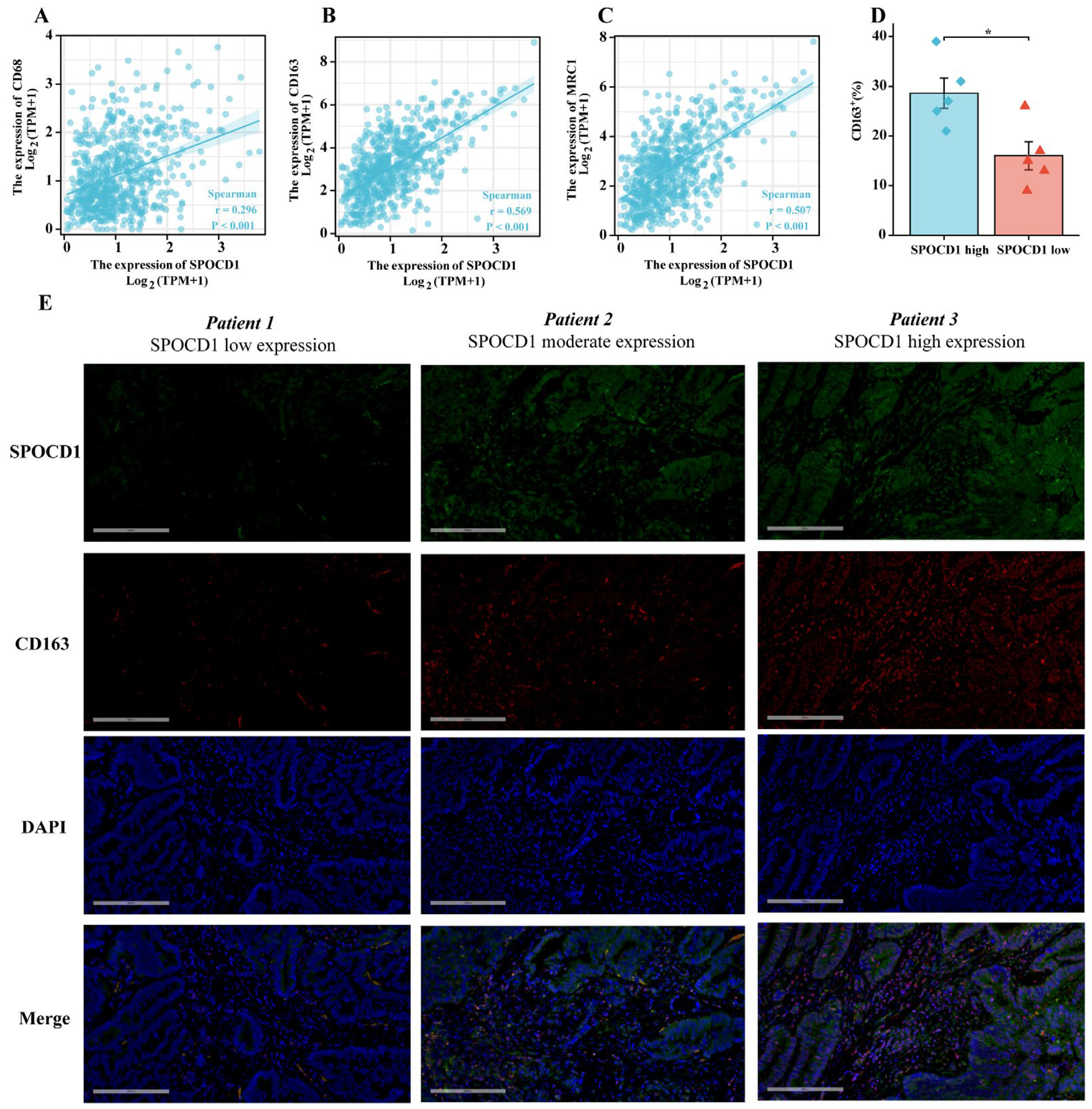
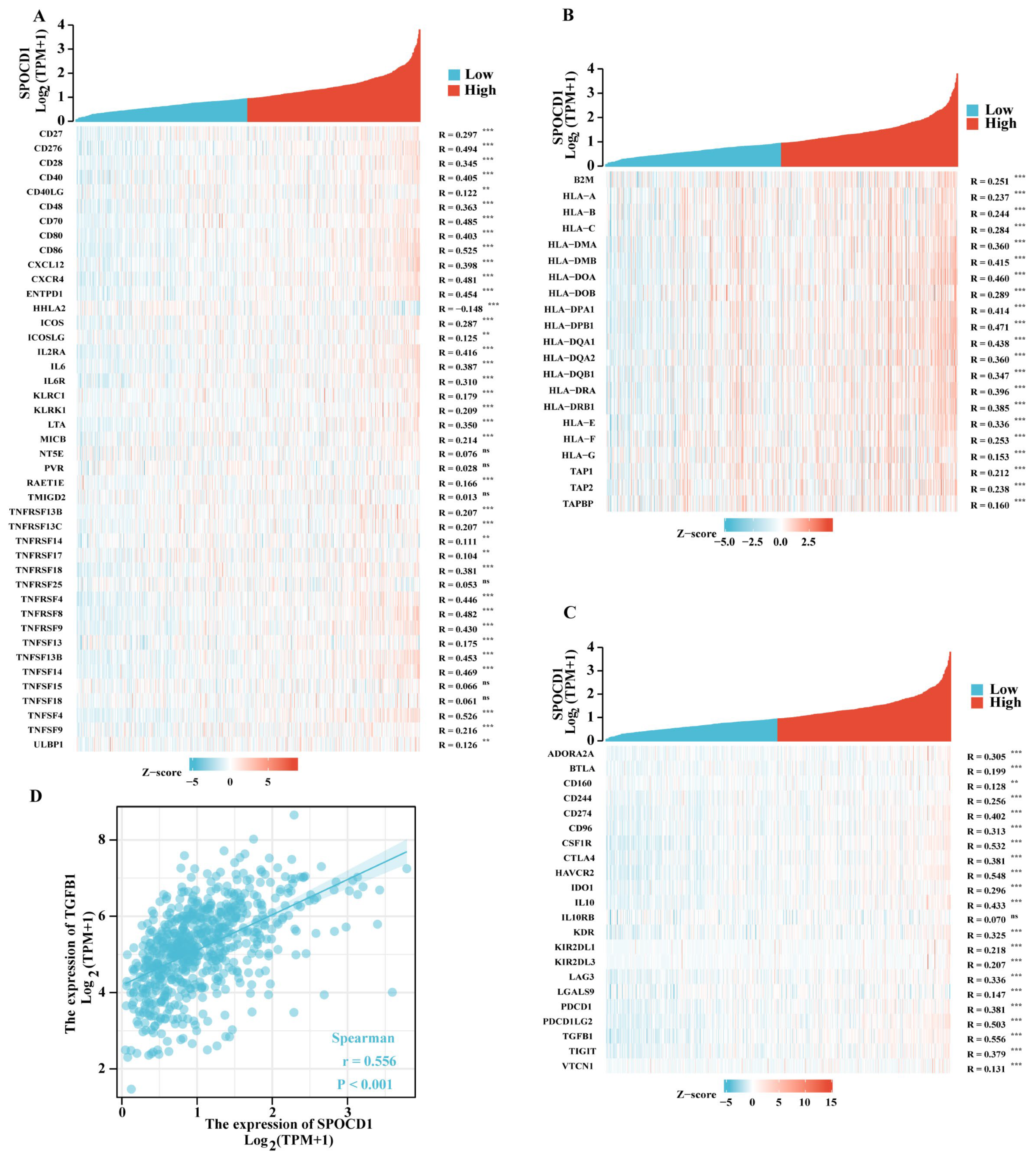
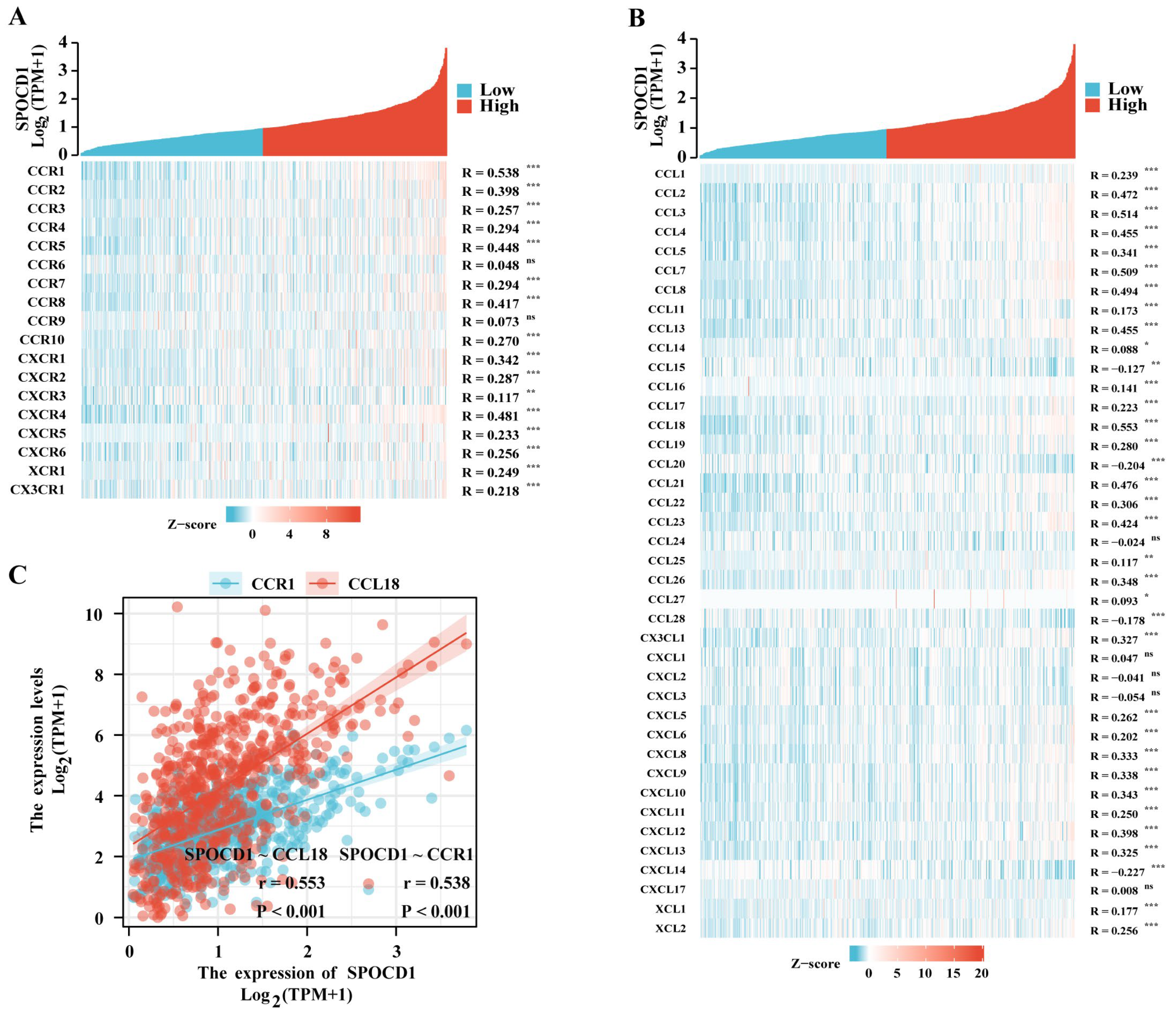
3.5. SPOCD1 Is Associated with CRC Immune Infiltration
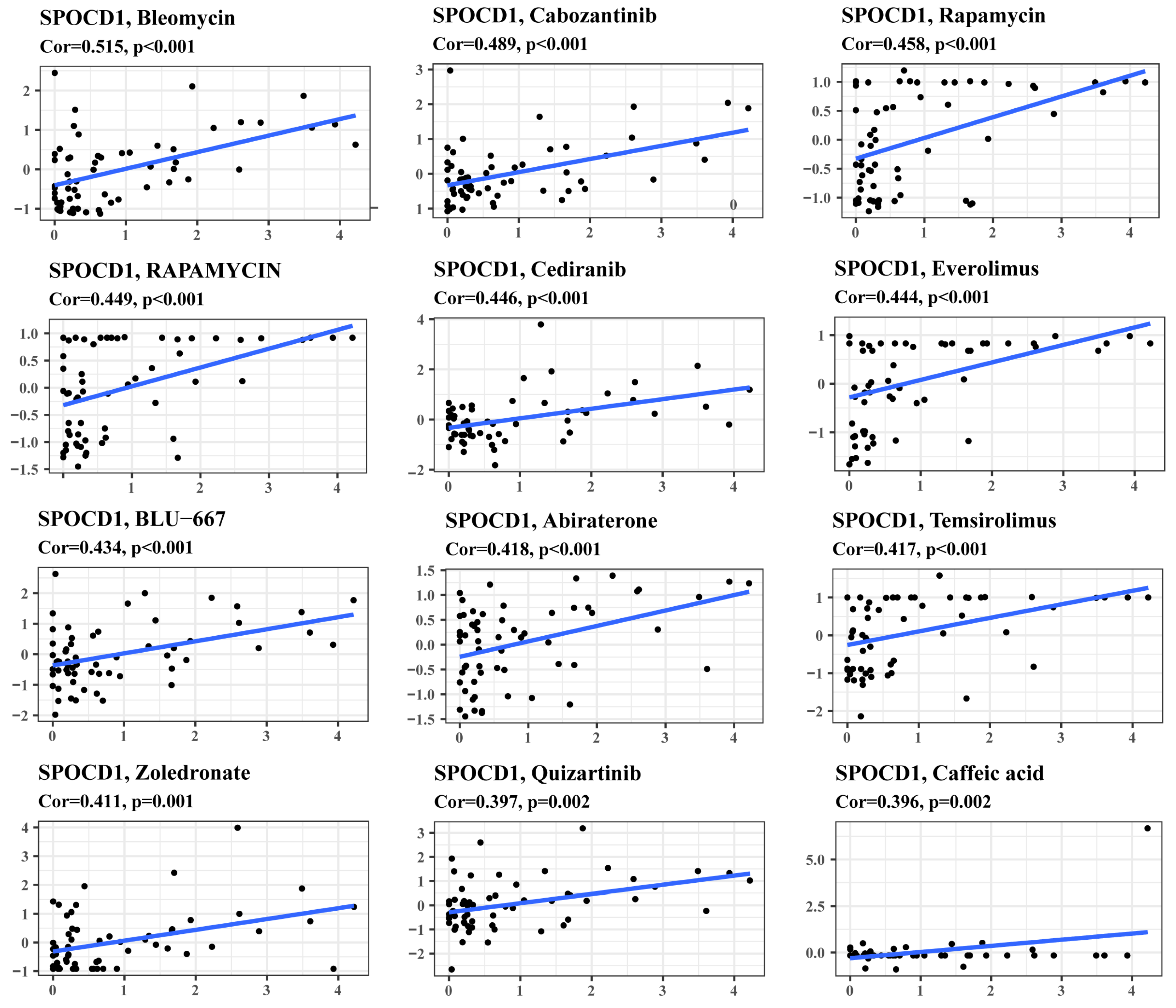
3.6. SPOCD1 Expression Is a Potential Indicator of Drug Sensitivity
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer. J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Bresalier, R.S. Colorectal Cancer Screening in a Changing World. Gastroenterol. Clin. N. Am. 2022, 51, 577–591. [Google Scholar] [CrossRef] [PubMed]
- Zoch, A.; Auchynnikava, T.; Berrens, R.V.; Kabayama, Y.; Schöpp, T.; Heep, M.; Vasiliauskaitė, L.; Pérez-Rico, Y.A.; Cook, A.G.; Shkumatava, A.; et al. SPOCD1 is an essential executor of piRNA-directed de novo DNA methylation. Nature 2020, 584, 635–639. [Google Scholar] [CrossRef]
- Zhu, M.; Yan, C.; Ren, C.; Huang, X.; Zhu, X.; Gu, H.; Wang, M.; Wang, S.; Gao, Y.; Ji, Y.; et al. Exome Array Analysis Identifies Variants in SPOCD1 and BTN3A2 That Affect Risk for Gastric Cancer. Gastroenterology 2017, 152, 2011–2021. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Yang, Y.; Yan, A.; Yang, Y. SPOCD1 accelerates ovarian cancer progression and inhibits cell apoptosis via the PI3K/AKT pathway. Onco. Targets Ther. 2020, 13, 351–359. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, X.Y.; Qin, Y.Y.; Yan, X.L.; Chen, H.M.; Huang, Q.D.; Chen, J.K.; Zheng, J.M. SPOCD1 promotes the proliferation and metastasis of glioma cells by up-regulating PTX3. Am. J. Cancer Res. 2018, 8, 624–635. [Google Scholar]
- Liang, J.; Zhao, H.; Hu, J.; Liu, Y.; Li, Z. SPOCD1 promotes cell proliferation and inhibits cell apoptosis in human osteosarcoma. Mol. Med. Rep. 2018, 17, 3218–3225. [Google Scholar] [CrossRef]
- van der Heijden, A.G.; Mengual, L.; Lozano, J.J.; Ingelmo-Torres, M.; Ribal, M.J.; Fernández, P.L.; Oosterwijk, E.; Schalken, J.A.; Alcaraz, A.; Witjes, J.A. A five-gene expression signature to predict progression in T1G3 bladder cancer. Eur. J. Cancer 2016, 64, 127–136. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Yoshihara, K.; Shahmoradgoli, M.; Martínez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Treviño, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Tosolini, M.; Kirilovsky, A.; Waldner, M.; Obenauf, A.C.; Angell, H.; Fredriksen, T.; Lafontaine, L.; Berger, A.; et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013, 39, 782–795. [Google Scholar] [CrossRef]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef]
- Shankavaram, U.T.; Varma, S.; Kane, D.; Sunshine, M.; Chary, K.K.; Reinhold, W.C.; Pommier, Y.; Weinstein, J.N. CellMiner: A relational database and query tool for the NCI-60 cancer cell lines. BMC Genom. 2009, 10, 277. [Google Scholar] [CrossRef]
- Reinhold, W.C.; Sunshine, M.; Liu, H.; Varma, S.; Kohn, K.W.; Morris, J.; Doroshow, J.; Pommier, Y. CellMiner: A web-based suite of genomic and pharmacologic tools to explore transcript and drug patterns in the NCI-60 cell line set. Cancer Res. 2012, 72, 3499–3511. [Google Scholar] [CrossRef]
- Mantovani, A.; Allavena, P.; Marchesi, F.; Garlanda, C. Macrophages as tools and targets in cancer therapy. Nat. Rev. Drug Discov. 2022, 21, 1–22. [Google Scholar] [CrossRef]
- Akhtar, R.; Chandel, S.; Sarotra, P.; Medhi, B. Current status of pharmacological treatment of colorectal cancer. World J. Gastrointest. Oncol. 2014, 6, 177–183. [Google Scholar] [CrossRef]
- Archer, N.S.; Nassif, N.T.; O’Brien, B.A. Genetic variants of SLC11A1 are associated with both autoimmune and infectious diseases: Systematic review and meta-analysis. Genes Immun. 2015, 16, 275–283. [Google Scholar] [CrossRef]
- De Rosa, M.; Pace, U.; Rega, D.; Costabile, V.; Duraturo, F.; Izzo, P.; Delrio, P. Genetics, diagnosis and management of colorectal cancer (Review). Oncol. Rep. 2015, 34, 1087–1096. [Google Scholar] [CrossRef]
- Weng, J.; Li, S.; Zhu, Z.; Liu, Q.; Zhang, R.; Yang, Y.; Li, X. Exploring immunotherapy in colorectal cancer. J. Hematol. Oncol. 2022, 15, 95. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zheng, X.; Wu, C. The Role of the Tumor Microenvironment and Treatment Strategies in Colorectal Cancer. Front. Immunol. 2021, 12, 792691. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Cao, X. The origin and function of tumor-associated macrophages. Cell Mol. Immunol. 2015, 12, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Italiani, P.; Boraschi, D. From Monocytes to M1/M2 Macrophages: Phenotypical vs. Functional Differentiation. Front. Immunol. 2014, 5, 514. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Tian, T.; Zhang, J. Tumor-Associated Macrophages (TAMs) in Colorectal Cancer (CRC): From Mechanism to Therapy and Prognosis. Int. J. Mol. Sci. 2021, 22, 8470. [Google Scholar] [CrossRef]
- Coates, R.F.; Gardner, J.A.; Gao, Y.; Cortright, V.M.; Mitchell, J.M.; Ashikaga, T.; Skelly, J.; Yang, M.X. Significance of positive and inhibitory regulators in the TGF-β signaling pathway in colorectal cancers. Hum. Pathol. 2017, 66, 34–39. [Google Scholar] [CrossRef]
- Asiri, A.; Raposo, T.P.; Alfahed, A.; Ilyas, M. TGFβ1-induced cell motility but not cell proliferation is mediated through Cten in colorectal cancer. Int. J. Exp. Pathol. 2018, 99, 323–330. [Google Scholar] [CrossRef]
- Chen, Y.; Di, C.; Zhang, X.; Wang, J.; Wang, F.; Yan, J.F.; Xu, C.; Zhang, J.; Zhang, Q.; Li, H.; et al. Transforming growth factor β signaling pathway: A promising therapeutic target for cancer. J. Cell Physiol. 2020, 235, 1903–1914. [Google Scholar] [CrossRef]
- Liu, L.; Cheng, M.; Zhang, T.; Chen, Y.; Wu, Y.; Wang, Q. Mesenchymal stem cell-derived extracellular vesicles prevent glioma by blocking M2 polarization of macrophages through a miR-744-5p/TGFB1-dependent mechanism. Cell. Biol. Toxicol. 2022, 38, 649–665. [Google Scholar] [CrossRef]
- Nguyen, T.N.; Nguyen-Tran, H.H.; Chen, C.Y.; Hsu, T. IL6 and CCL18 Mediate Cross-talk between VHL-Deficient Kidney Cells and Macrophages during Development of Renal Cell Carcinoma. Cancer Res. 2022, 82, 2716–2733. [Google Scholar] [CrossRef]
- Eum, H.H.; Kwon, M.; Ryu, D.; Jo, A.; Chung, W.; Kim, N.; Hong, Y.; Son, D.S.; Kim, S.T.; Lee, J.; et al. Tumor-promoting macrophages prevail in malignant ascites of advanced gastric cancer. Exp. Mol. Med 2020, 52, 1976–1988. [Google Scholar] [CrossRef]
- Bastrup, F.A.; Vissing, M.; Gehl, J. Electrochemotherapy with intravenous bleomycin for patients with cutaneous malignancies, across tumour histology: A systematic review. Acta Oncol. 2022, 61, 1093–1104. [Google Scholar] [CrossRef]
- Maroto, P.; Porta, C.; Capdevila, J.; Apolo, A.B.; Viteri, S.; Rodriguez-Antona, C.; Martin, L.; Castellano, D. Cabozantinib for the treatment of solid tumors: A systematic review. Ther. Adv. Med. Oncol. 2022, 14, 17588359221107112. [Google Scholar] [CrossRef]
- Orbegoso, C.; Marquina, G.; George, A.; Banerjee, S. The role of Cediranib in ovarian cancer. Expert Opin. Pharmacother. 2017, 18, 1637–1648. [Google Scholar] [CrossRef]

| Characteristic | Levels | SPOCD1 | p | Statistic | |
|---|---|---|---|---|---|
| Low Expression | High Expression | ||||
| n | 322 | 322 | |||
| Age, n (%) | <=65 | 128 (19.9%) | 148 (23%) | 0.130 | 2.29 |
| >65 | 194 (30.1%) | 174 (27%) | |||
| Gender, n (%) | Female | 150 (23.3%) | 151 (23.4%) | 1.000 | 0 |
| Male | 172 (26.7%) | 171 (26.6%) | |||
| T stage, n (%) | T1 | 14 (2.2%) | 6 (0.9%) | 0.014 | 10.55 |
| T2 | 66 (10.3%) | 45 (7%) | |||
| T3 | 209 (32.6%) | 227 (35.4%) | |||
| T4 | 30 (4.7%) | 44 (6.9%) | |||
| n stage, n (%) | N0 | 193 (30.2%) | 175 (27.3%) | 0.043 | 6.29 |
| N1 | 79 (12.3%) | 74 (11.6%) | |||
| N2 | 47 (7.3%) | 72 (11.2%) | |||
| M stage, n (%) | M0 | 243 (43.1%) | 232 (41.1%) | 0.248 | 1.33 |
| M1 | 39 (6.9%) | 50 (8.9%) | |||
| Pathologic stage, n (%) | Stage I | 68 (10.9%) | 43 (6.9%) | 0.037 | 8.46 |
| Stage II | 113 (18.1%) | 125 (20.1%) | |||
| Stage III | 90 (14.4%) | 94 (15.1%) | |||
| Stage IV | 38 (6.1%) | 52 (8.3%) | |||
| Characteristics | Total (n) | Univariate Analysis | Multivariate Analysis | ||
|---|---|---|---|---|---|
| HR (95% CI) | p | HR (95% CI) | p | ||
| Gender | 643 | ||||
| Female | 301 | Reference | |||
| Male | 342 | 1.054 (0.744–1.491) | 0.769 | ||
| Age | 643 | ||||
| <=65 | 276 | Reference | |||
| >65 | 367 | 1.939 (1.320–2.849) | <0.001 | 2.858 (1.836–4.450) | <0.001 |
| T stage | 640 | ||||
| T1 & T2 | 131 | Reference | |||
| T3 & T4 | 509 | 2.468 (1.327–4.589) | 0.004 | 2.156 (0.981–4.741) | 0.056 |
| N stage | 639 | ||||
| N0 | 367 | Reference | |||
| N1 & N2 | 272 | 2.627 (1.831–3.769) | <0.001 | 0.429 (0.165–1.115) | 0.082 |
| M stage | 563 | ||||
| M0 | 474 | Reference | |||
| M1 | 89 | 3.989 (2.684–5.929) | <0.001 | 2.278 (1.410–3.682) | <0.001 |
| Pathologic stage | 622 | ||||
| Stage I & II | 348 | Reference | |||
| Stage III & IV | 274 | 2.988 (2.042–4.372) | <0.001 | 5.293 (1.816–15.430) | 0.002 |
| SPOCD1 | 643 | ||||
| Low | 322 | Reference | |||
| High | 321 | 1.575 (1.109–2.238) | 0.011 | 1.505 (1.022–2.216) | 0.038 |
| Characteristics | Total (n) | Univariate Analysis | Multivariate Analysis | ||
|---|---|---|---|---|---|
| HR (95% CI) | p | HR (95% CI) | p | ||
| Gender | 621 | ||||
| Female | 290 | Reference | |||
| Male | 331 | 1.207 (0.769–1.895) | 0.412 | ||
| Age | 621 | ||||
| <=65 | 273 | Reference | |||
| >65 | 348 | 1.421 (0.894–2.257) | 0.137 | ||
| T stage | 618 | ||||
| T1 & T2 | 129 | Reference | |||
| T3 & T4 | 489 | 6.440 (2.029–20.441) | 0.002 | 2.675 (0.812–8.808) | 0.106 |
| N stage | 617 | ||||
| N0 | 358 | Reference | |||
| N1 & N2 | 259 | 4.119 (2.496–6.797) | <0.001 | 0.570 (0.219–1.483) | 0.250 |
| M stage | 542 | ||||
| M0 | 455 | Reference | |||
| M1 | 87 | 7.471 (4.647–12.012) | <0.001 | 3.834 (2.136–6.881) | <0.001 |
| Pathologic stage | 601 | ||||
| Stage I & II | 339 | Reference | |||
| Stage III & IV | 262 | 5.716 (3.240–10.083) | <0.001 | 4.339 (1.324–14.226) | 0.015 |
| SPOCD1 | 621 | ||||
| Low | 307 | Reference | |||
| High | 314 | 1.888 (1.190–2.996) | 0.007 | 1.370 (0.843–2.226) | 0.203 |
| Characteristics | Total (n) | Univariate Analysis | Multivariate Analysis | ||
|---|---|---|---|---|---|
| HR (95% CI) | p | HR (95% CI) | p | ||
| Gender | 643 | ||||
| Female | 301 | Reference | |||
| Male | 342 | 1.217 (0.892–1.660) | 0.216 | ||
| Age | 643 | ||||
| <=65 | 276 | Reference | |||
| >65 | 367 | 1.006 (0.737–1.371) | 0.972 | ||
| T stage | 640 | ||||
| T1 & T2 | 131 | Reference | |||
| T3 & T4 | 509 | 3.198 (1.814–5.636) | <0.001 | 1.796 (0.991–3.255) | 0.053 |
| N stage | 639 | ||||
| N0 | 367 | Reference | |||
| N1 & N2 | 272 | 2.624 (1.916–3.592) | <0.001 | 0.911 (0.389–2.138) | 0.831 |
| M stage | 563 | ||||
| M0 | 474 | Reference | |||
| M1 | 89 | 5.577 (3.945–7.884) | <0.001 | 4.375 (2.803–6.827) | <0.001 |
| Pathologic stage | 622 | ||||
| Stage I & II | 348 | Reference | |||
| Stage III & IV | 274 | 2.924 (2.115–4.044) | <0.001 | 1.415 (0.547–3.662) | 0.474 |
| SPOCD1 | 643 | ||||
| Low | 322 | Reference | |||
| High | 321 | 1.659 (1.215–2.266) | 0.001 | 1.390 (0.998–1.935) | 0.051 |
| Immune Cell Type | Coefficient of Correlation | p Value |
|---|---|---|
| DC | 0.417 | <0.001 |
| Activated DC (aDC) | 0.302 | <0.001 |
| Immature DC (iDC) | 0.481 | <0.001 |
| Plasmacytoid DC (pDC) | 0.312 | <0.001 |
| NK cells | 0.500 | <0.001 |
| NK CD56 bright cells | 0.083 | 0.035 |
| NK CD56 dim cells | 0.187 | <0.001 |
| B cells | 0.136 | <0.001 |
| Eosinophils | 0.289 | <0.001 |
| Macrophages | 0.636 | <0.001 |
| Mast cells | 0.420 | <0.001 |
| Neutrophils | 0.459 | <0.001 |
| Cytotoxic cells | 0.330 | <0.001 |
| T cells | 0.216 | <0.001 |
| CD8 T cells | 0.187 | <0.001 |
| T helper cells | −0.002 | 0.967 |
| T central memory (Tcm) | 0.045 | 0.251 |
| T effector memory (Tem) | 0.387 | <0.001 |
| T follicular helper (TFH) | 0.300 | <0.001 |
| T gamma delta (Tgd) | 0.270 | <0.001 |
| Th1 cells | 0.455 | <0.001 |
| Th17 cells | −0.233 | <0.001 |
| Th2 cells | −0.007 | 0.866 |
| TReg | 0.296 | <0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gan, L.; Yang, C.; Zhao, L.; Wang, S.; Gao, Z.; Ye, Y. Prognostic Biomarker SPOCD1 and Its Correlation with Immune Infiltrates in Colorectal Cancer. Biomolecules 2023, 13, 209. https://doi.org/10.3390/biom13020209
Gan L, Yang C, Zhao L, Wang S, Gao Z, Ye Y. Prognostic Biomarker SPOCD1 and Its Correlation with Immune Infiltrates in Colorectal Cancer. Biomolecules. 2023; 13(2):209. https://doi.org/10.3390/biom13020209
Chicago/Turabian StyleGan, Lin, Changjiang Yang, Long Zhao, Shan Wang, Zhidong Gao, and Yingjiang Ye. 2023. "Prognostic Biomarker SPOCD1 and Its Correlation with Immune Infiltrates in Colorectal Cancer" Biomolecules 13, no. 2: 209. https://doi.org/10.3390/biom13020209
APA StyleGan, L., Yang, C., Zhao, L., Wang, S., Gao, Z., & Ye, Y. (2023). Prognostic Biomarker SPOCD1 and Its Correlation with Immune Infiltrates in Colorectal Cancer. Biomolecules, 13(2), 209. https://doi.org/10.3390/biom13020209








