Abstract
The formation and function of membrane-less organelles (MLOs) is one of the main driving forces in the molecular life of the cell. These processes are based on the separation of biopolymers into phases regulated by multiple specific and nonspecific inter- and intramolecular interactions. Among the realm of MLOs, a special place is taken by the promyelocytic leukemia nuclear bodies (PML-NBs or PML bodies), which are the intranuclear compartments involved in the regulation of cellular metabolism, transcription, the maintenance of genome stability, responses to viral infection, apoptosis, and tumor suppression. According to the accepted models, specific interactions, such as SUMO/SIM, the formation of disulfide bonds, etc., play a decisive role in the biogenesis of PML bodies. In this work, a number of bioinformatics approaches were used to study proteins found in the proteome of PML bodies for their tendency for spontaneous liquid–liquid phase separation (LLPS), which is usually caused by weak nonspecific interactions. A total of 205 proteins found in PML bodies have been identified. It has been suggested that UBC9, P53, HIPK2, and SUMO1 can be considered as the scaffold proteins of PML bodies. It was shown that more than half of the proteins in the analyzed proteome are capable of spontaneous LLPS, with 85% of the analyzed proteins being intrinsically disordered proteins (IDPs) and the remaining 15% being proteins with intrinsically disordered protein regions (IDPRs). About 44% of all proteins analyzed in this study contain SUMO binding sites and can potentially be SUMOylated. These data suggest that weak nonspecific interactions play a significantly larger role in the formation and biogenesis of PML bodies than previously expected.
1. Introduction
Fundamental changes in ideas about the spatiotemporal organization of intracellular space and the organization and control of biochemical processes in the cell occurred in the 2010s [1,2,3]. It has become clear that the formation and function of biomolecular condensates—membrane-less organelles (MLOs)—is one of the main driving forces in the molecular life of a cell. These processes are based on the separation of biopolymers, such as primarily intrinsically disordered proteins (IDPs) and RNA, into phases regulated by multiple specific/nonspecific inter- and intramolecular interactions [4,5]. IDPs and proteins containing intrinsically disordered protein regions (IDPRs) are known to play a decisive role in liquid–liquid phase separation (LLPS), leading to the formation of MLOs [6]. This is because, as a rule, IDPs/IDPRs contain blocks of the same type of amino acid residues, multiple weak nonspecific interactions (electrostatic, π–π, cation–π) between which, under conditions of macromolecular crowding, can help to overcome the potential energy barrier required for the transition of IDPs to the condensed liquid-droplet phase [7,8].
MLOs include nuclear PML bodies (also known as ND10)—compartments involved in the regulation of cellular metabolism, transcription, the maintenance of genome stability, responses to viral infection, apoptosis, and tumor suppression [9,10]. PML bodies are subnuclear spherical and toroidal structures with a diameter of 0.1–2 μm [11]. They are present in most types of mammalian cells and tissues. Depending on the cell type, cell cycle phase, and differentiation stage, there may be about 5–30 PML bodies per cell [12]. Based on an immunofluorescence analysis, PML bodies exist as nuclear dot-shaped spherical structures residing in the interchromatin nuclear space [13]. As a rule, large PML bodies have a toroidal structure formed of a ≈100 nm thick shell at the periphery and a more mobile internal core [14]. According to the accepted models, canonical PML bodies are protein assemblages that do not contain nucleic acids [13]. However, specific types of PML bodies are known that are capable of incorporating DNA/RNA into their composition [9]:
- -
- Viral DNA-containing PML bodies;
- -
- Giant PML bodies containing satellite DNA;
- -
- Alternative lengthening of telomeres (ALT)-associated PML bodies.
The major scaffolding protein of PML bodies is promyelocytic leukemia protein (PML) [15]. Due to alternative splicing, the PML protein is represented by seven main isoforms transcribed from one gene: six with nuclear localization (PML-I to PML-VI) due to the presence of the nuclear localization signal (NLS) in exon 6, and one with cytoplasmic localization (PML-VII). The sequences encoded by the first four exons of the PML gene (418 amino acids with the conserved RBCC motif consisting of a RING finger domain (R), followed by two cysteine/histidine-rich B-box domains (B) and an α-helical coiled-coil domain (CC)) are common to all isoforms and contain structurally conserved domains: proline-rich (1–45 a.a.), RING (45–105 a.a.), B1-box (124–166 a.a.), B2-box (184–229 a.a.), and coiled-coil (229–323 a.a.) [16] (Figure 1).
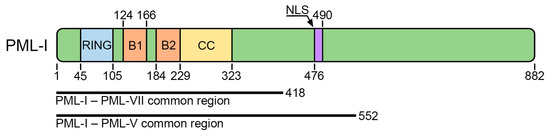
Figure 1.
PML protein domain organization scheme. All PML isoforms have the same N-terminal part that contains RING (R), B-Box1 (B1), B-Box2 (B2), and coiled-coil (CC) domains. The majority of PML isoforms contain a nuclear localization signal (NLS).
According to modern understanding, specific interactions play a decisive role in the formation and biogenesis of PML bodies [17]. It is assumed that the oxidative formation of disulfide bonds between PML molecules and non-covalent interactions in the region of the ordered RBCC motif of PML trigger the oligomerization of non-SUMOylated PML proteins, which subsequently undergo UBC9-mediated (poly-)SUMOylation and recruit PML-body client proteins via SUMO–SIM interactions [17,18].
We have previously shown that a certain role in the formation of PML bodies is played by nonspecific intra- and intermolecular interactions that can lead to the spontaneous phase separation of at least the disordered C-terminal domains of the PML-II and PML-V isoforms [19]. In this regard, we decided to analyze the proteins included in the proteome of PML bodies for their tendency to spontaneously separate into phases, and, consequently, for their ability to be nonspecifically included in PML bodies.
2. Materials and Methods
2.1. Study Design
The identification of the proteins included in the PML-body proteome was carried out using two approaches:
- (i).
- An analysis of the PML-body proteome and the PML protein interactome in the BIOGRID (v.4.4) databases https://thebiogrid.org/ (accessed on 10 October 2023), and resources https://amigo.geneontology.org/amigo/term/GO:0016605 (accessed on 10 October 2023) (GO:0016605) (PML NB/ND10) and SL-0465 (https://www.uniprot.org/locations/SL-0465 (accessed on 10 October 2023);
- (ii).
- An analysis of data from the literature.
For further analysis, 205 proteins were selected that could be localized in PML bodies according to fluorescent microscopy or electron microscopy data. Figure 2 schematically represents the design of our study.
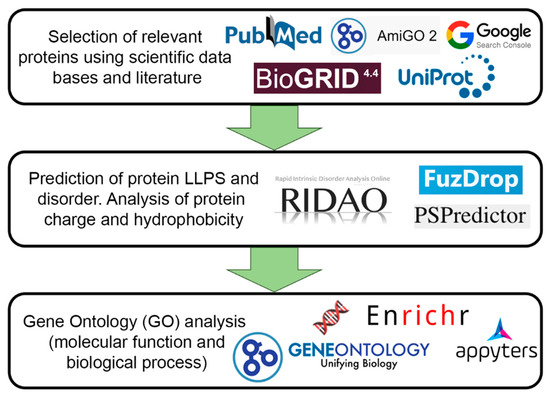
Figure 2.
Scheme illustrating the design of our study.
2.2. Intrinsic Disorder Prediction
An analysis of the intrinsic disorder predispositions of the studied proteins was performed using the RIDAO online platform [20]. This platform predicts potential per-residue disorder in proteins based on their amino acid sequences, using predictors based on different algorithms such as PONDR® FIT, PONDR® VLXT, PONDR® VLS2, PONDR® VL3, IUPred-Long, and IUPred-Short [21,22,23,24,25,26]. The PONDR® VSL2 algorithm showed the most correct prediction of disorder for a large data set. Consequently, the PONDR® VSL2 (%), the percentage of predicted intrinsically disordered residues (PPIDR), was employed as an indicator of protein structural disorder. This indicator reflects the percentage of residues in a protein’s amino acid sequence for which the disorder prediction (disorder score) is >0.5 (“probably disordered” according to the algorithm).
Typically, a PPIDR value below 10% indicates a protein with high order, while a PPIDR ranging between 10% and 30% is assigned to proteins characterized as moderately disordered. Proteins with a PPIDR exceeding 30% are classified as highly disordered [27,28]. Also, the mean disorder score (MDS) was computed for each queried protein as a length-normalized sum of the per-residue disorder scores. Proteins were grouped based on their respective MDS values, with those having an MDS of less than 0.15 considered highly ordered; an MDS between 0.15 and 0.5 categorized them as moderately disordered or flexible, and those with a MDS equal to or greater than 0.5 were designated as highly disordered.
The intrinsic disorder predisposition of the analyzed data set was further evaluated via the CH-CDF analysis, the data for which were generated using RIDAO [21]. The analysis integrates outcomes derived from two binary predictors of disorder, namely the cumulative distribution function (CDF) and charge–hydropathy (CH) plot analysis [29,30]. The combination of data generated using these approaches results in the ΔCH-ΔCDF plot, allowing discrimination between flavors of disorder based on the positions of proteins within the quadrants of the ΔCH-ΔCDF phase space [31,32]. In this context, proteins positioned in the top-left quadrant are anticipated to exhibit disorder according to both the CH and CDF predictions, while those in the bottom-left quadrant are projected to be compact or ordered based on CH but disordered according to CDF. Proteins situated in the top-right quadrant are expected to be disordered according to CH but ordered based on the CDF, whereas those in the bottom-right quadrant are predicted to be ordered based on both the CH and CDF predictors [31,32].
2.3. LLPS Predisposition Analysis
The FuzDrop [33] and PSPredictor [34] predictors were used for the LLPS analysis of the studied proteins. The proteins with a PSPredcitor score > 0.5 and FuzDrop score > 0.6 were marked as prone to LLPS. If the results of the FuzDrop and PSPredictor analyses disagreed, the analyzed proteins were classified as the “controversial LLPS” group.
The FuzDrop predictor also allows one to predict the ability of the analyzed protein to undergo spontaneous and interaction-induced LLPS. Proteins that spontaneously undergo liquid–liquid phase separation are termed as droplet-driving proteins. Proteins that require additional interactions to form droplets are termed as clients. Proteins with a pLPS ≥ 0.60 likely drive liquid–liquid phase separation. Proteins with droplet-promoting regions, defined as consecutive residues with a pDP ≥ 0.60, will likely serve as droplet clients.
2.4. Evaluation of Protein Charge and Hydrophobicity
The charge of the examined proteins at pH 7.0 was determined as the average charge across the protein sequence, considering the charge values of amino acids: D = −1, E = −1, H = 0.5, R = 1, K = 1 [29]. The average hydrophobicity of the investigated proteins was computed using the normalized Kite and Doolittle scale [35].
2.5. Determination of Proteins Targeted for SUMOylation
This was performed by visiting the UniProt website (https://www.uniprot.org/, accessed on 10 September 2023) [36] and then searching for mentions of SUMO in the PTM/Processing section of the corresponding UniProt entry.
2.6. Aggregation Propensity Prediction
The assessment of the proteins’ propensity for aggregation and the formation of amyloid fibrils was conducted through the AggreScan package [37], enabling the identification of aggregation hot spots—regions of the protein that facilitate its aggregation.
2.7. Determination of the Biological Processes and Molecular Functions of Proteins
Gene Ontology (GO) was performed using the Enrichr resource (https://maayanlab.cloud/Enrichr/, accessed on 10 September 2023), and 10 terms with the lowest p-values were selected [38]. The p-values were computed using the Fisher exact test, a statistical method employed to determine the presence of significant non-random associations between two categorical variables.
3. Results and Discussion
3.1. Finding Proteins Included in the PML-Body Proteome
PML bodies take part in a large number of intracellular processes [9,10,39,40,41,42,43]. In this regard, the composition of the client proteins of PML bodies varies significantly. In addition, proteins that interact with PML are often included in the PML-body proteome [15,44]. However, PML protein is localized in the cell, not just in PML bodies. It is known that several isoforms of the PML protein can be localized in the cytoplasm [16,45], and the PML bodies themselves pass into the cytoplasm during mitosis [40]. Taken together, this complicates the identification of proteins that are uniquely present in PML bodies.
In 2010, mainly based on an analysis of data from the literature, the so-called PML-body interactome was proposed, combining 166 proteins, most of which are part of the SUMO conjugation pathway [44]. A 2015 paper [15] discussed a network of 120 proteins that are involved in direct physical interactions with PML, most of which are part of PML bodies. This set included proteins whose interaction with PML was confirmed via affinity capture followed by Western blotting (BIOGRID data). Proteins associated with PML, identified using high-throughput methods, were not discussed in this work [15].
An analysis of more than 150 works [44,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136,137,138,139,140,141,142,143,144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160,161,162,163,164,165,166,167,168,169,170,171,172,173,174,175,176,177,178,179,180,181,182,183,184,185,186,187,188,189,190,191,192,193,194,195,196,197,198,199,200,201,202,203,204,205,206,207,208,209,210,211,212,213,214,215,216,217,218] allowed us to identify 205 proteins that can be localized in PML bodies according to fluorescent microscopy or electron microscopy data. These proteins were selected for further analysis.
Also, we estimated the ratio of proteins capable of directly interacting with PML protein in a selected array. For this purpose, we analyzed the PML interactome from the BIOGRID database (v.4.4) [219], selecting proteins involved in the biogenesis of these membrane-less organelles. From the 707 proteins included in the network of molecular interactions of the PML protein, we selected 267 proteins for which an interaction with PML was shown at least twice. Using information obtained from the resources https://amigo.geneontology.org/amigo/term/GO:0016605 (accessed on 10 October 2023) (GO:0016605) (PML NB/ND10) and SL-0465 (https://www.uniprot.org/locations/SL-0465 (accessed on 10 October 2023)), for further analysis, we selected 104 proteins that are potentially included in human PML bodies.
A comparison of the obtained data showed that about of quarter of the proteins selected via the literature analysis (52 of 205) were not identified in analyzed bioinformatics databases as PML-binding proteins (Figure 3).
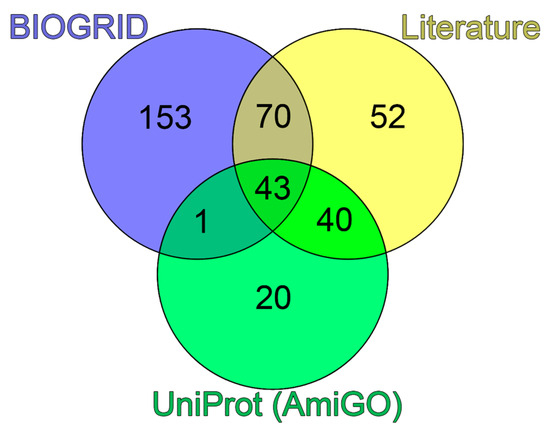
Figure 3.
Venn diagram of three sets of proteins potentially included in PML bodies. Blue circle represents the set based on the analysis of the PML interactome in the BIOGRID database. Green circle represents the set according to the analysis of the GO:0016605 and SL-0465 databases. Yellow circle represents the set collected during the analysis of literary sources.
3.2. PML Bodies’ Scaffold Proteins
PML bodies have complex a structure, formed of scaffold proteins that are constantly present in PML bodies and client proteins that are temporarily localized in PML bodies. It is suggested that the scaffold proteins drive the PML bodies’ formation.
It is known that the scaffold proteins of PML bodies include PML, death domain-associated protein (DAXX), cyclic AMP-responsive element-binding protein (CREB)-binding protein (CREBBP), and speckled 100 kDa (SP100, also known as nuclear dot-associated Sp100 protein or nuclear autoantigen Sp-100) [220]. These proteins were also included in the analyzed array, selected based on data from the literature.
We analyzed the interactomes of these four proteins and found that, according to the BIOGRID database, the proteins ubiquitin carrier protein 9 (UBC9, also known as SUMO-conjugating enzyme UBC9), small ubiquitin-related modifier 1 (SUMO1), p53, and homeodomain-interacting protein kinase 2 (HIPK2) are included in all four interactomes analyzed (Figure 4).
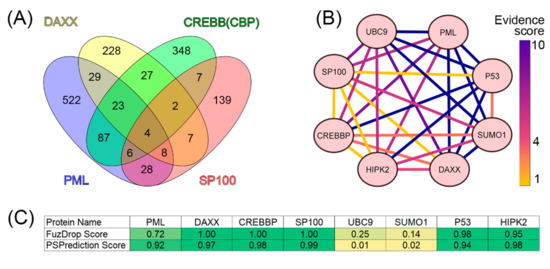
Figure 4.
Analysis of the interactome of PML bodies’ scaffold proteins. (A) Venn diagram represents the intersection of the interactomes of four PML bodies’ scaffold proteins (PML, DAXX, SP100, and CREBB(CBP)). Data were obtained using BioGRID v4.4. (B) Interaction map for eight potential PML body envelope proteins based on the statistical representation in the BIOGRID (evidence score) database. Yellow shows results with weak statistics (data from one experiment), blue shows data with strong statistics (>10 experiments). The heat map was constructed using CytoScape [221]. (C) Table of results for predicting the propensity of detected proteins to undergo spontaneous phase separation using FuzDrop and PSPrediction scores.
The analysis of the literature data indicates that these proteins are one way or another included in the biogenesis of PML bodies (UBC9 [222], SUMO1 [223], HIPK2 [147], and p53 [224,225,226]). As is known, PML bodies are a platform for the SUMOylation and ubiquitination of various proteins [227]. The SUMO-conjugating enzyme, UBC9, accepts ubiquitin-like proteins SUMO1, SUMO2, SUMO3, SUMO4, and SUMO1 pseudogene 1 (SUMO1P1/SUMO5) from the ubiquitin-like 1-activating enzyme E1A–ubiquitin-like 1-activating enzyme E1B (UBLE1A-UBLE1B) complex and catalyzes their covalent attachment to other proteins using E3 ligases, such as Ran-binding protein 2 (RANBP2, also known as E3 SUMO-protein ligase RanBP2 or nucleoporin Nup358), chromobox protein homolog 4 (CBX4, also known as E3 SUMO-protein ligase CBX4 or Polycomb 2 homolog, Pc2), and zinc finger protein 451 (ZNF451, also known as E3 SUMO-protein ligase ZNF451) [227,228]. UBC9 can catalyze the formation of poly-SUMO chains. It is known that the SUMOylation of proteins included in PML bodies occurs in a UBC9-dependent manner [229]. The conjugation of SUMO substrates often requires an E3 ligase, which provides substrate specificity by simultaneously binding UBC9 and the substrate. E3 SUMO ligases typically use the RING domain to interact with UBC9. The PML protein is considered a possible SUMO ligase [220]. The antiparallel PML dimer model has been proposed to potentially support the accessibility of the PML B-box1 domain for UBC9 binding [229].
PML bodies are also a platform that provides post-translational modifications (PTMs) of the tumor suppressor p53, allowing this protein to take part in the regulation of a number of intracellular processes, in particular, senescence [230]. It is worth noting that one of the p53 regulatory proteins that serves as a part of PML bodies is the ubiquitin carboxyl-terminal hydrolase 7 (USP7) protein [231]. Homeodomain-interacting protein kinase 2 (HIPK2) promotes the p300-mediated acetylation of p53 at K382 and influences the selective transactivation of proapoptotic p53 target genes. To cause the HIPK2-mediated induction of pro-apoptotic p53 genes, p53 must be modified by both S46 phosphorylation and K382 acetylation [232]. It is suggested that SP100 acts as a coactivator of the HIPK2-mediated activation of p53, and thus has an essential role in p53-dependent gene expression [233]. p300 (CREBBP homolog) and CBP (cyclic AMP response element-binding protein-binding protein) directly bind to p53 and acetylate several lysine residues located in the C-terminal domain of this protein: K370, K372, K373, K381, K382, and K386. Furthermore, four additional lysine residues, K101, K139, K164, and K305, can be acetylated by p300/CBP. It has been suggested that the acetylation of these sites enhances the DNA-binding ability of p53 and promotes its transcription, leading to growth arrest or apoptosis [232].
Taken together, these data suggest that UBC9, SUMO1, p53, and HIPK2 can be included in the set of PML scaffolding proteins.
3.3. Proteome Analysis of PML Bodies
We analyzed 205 proteins that were identified as proteins of the PML-body proteome for their predisposition for intrinsic disorder and tendency. The results of this analysis are summarized in Figure 5.
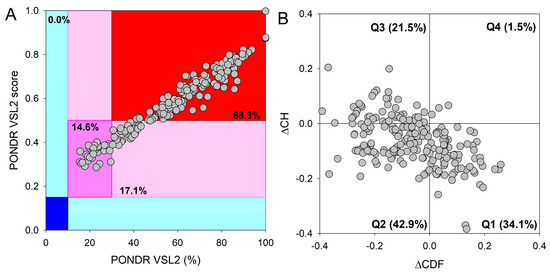
Figure 5.
Analysis of global protein disorder within the PML-body proteome. (A) The output of PONDR® VSL2, where the PONDR® VSL2 score denotes the mean disorder score (MDS) for a given protein. This score is derived by summing up the per-residue disorder scores and dividing this sum by the number of residues in a query sequence. The PONDR® VSL2 (%) corresponds to the percentage of predicted intrinsically disordered residues (PPIDR), indicating the proportion of residues with disorder scores above 0.5. Color-coded blocks depict distinct regions within the MDS-PPIDR phase space based on the degree of protein (dis)order: mostly disordered proteins are found in the red colored block, moderately disordered proteins are within the pink and light-pink blocks, and predominantly ordered proteins are positioned in the blue and light-blue blocks. Dark-colored background areas (blue, pink, and red) indicate concordance between the two parameters; i.e., MDS and PPIDR, while light-blue and light-pink colors signify areas where only one criterion aligns. The delineation of colored regions is contingent upon the arbitrary yet accepted cutoffs for PPDR (x-axis) and MDS (y-axis). (B) The cumulative distribution function and charge–hydropathy plot (ΔCDF-ΔCH) for the nuclear PML-body proteome comprising 205 proteins. The Y-coordinate is determined by the distance of each protein from the boundary in the CH plot, while the X-coordinate is calculated as the average distance of the protein’s CDF curve from the CDF boundary. The protein classification is based on the quadrant in which it is positioned. In Quadrant 1 (Q1), 70 proteins (34.1%) are projected to be ordered according to CDF and exhibit compact characteristics in the CH plot. In Quadrant 2 (Q2), 88 proteins (42.9%) are anticipated to be ordered/compact based on the CH plot but display disorder as per the CDF plot. Quadrant 3 (Q3) encompasses 44 proteins (21.5%) predicted to be disordered based on the CH plot and CDF. Quadrant 4 (Q4) includes 3 proteins (1.5%) anticipated to be disordered according to the CH plot but ordered according to the CDF analysis.
The integrative disorder analysis of the proteins that can be localized in PML bodies revealed an exceptionally high prevalence of intrinsic disorder. Approximately 14.6% of these proteins (Figure 5A) were predicted to be moderately disordered (dark-pink segment). An additional 17.1% of proteins were anticipated to be moderately disordered based on their MDS values (segment with light-pink color), while the majority (68.3%) of proteins in the PML-body proteome were expected to be mostly disordered (located in the red-colored block). Importantly, a significant proportion of these mostly disordered proteins (131 out of 140 or 93.6%) exhibited PPIDR values ≥50% and MDS values ≥0.5, indicating their highly disordered nature. This global disorder content in the PML-body proteome dramatically exceeds that of the human proteome, where the moderately and highly disordered proteins account for 51.7% and 39.8%, respectively [234].
Additional evidence supporting the remarkably high disorder status of human proteins within the PML-body proteome was obtained by analyzing the combined outcomes of two binary disorder predictors: cumulative distribution function (CDF) and charge–hydropathy (CH) plot analysis, as depicted in Figure 5B. Figure 5B highlights that only 34.3% of these proteins (in contrast to 59.1% in the entire human proteome [234]) refer to the Q1 quadrant (bottom-right corner) that includes proteins predicted to be ordered. Meanwhile, 65.7% of these proteins are located outside the Q1 quadrant, suggesting their elevated disorder levels. A total of 42.9% of the studied proteins are located within the Q2 quadrant (bottom-left corner), corresponding to molten globular or hybrid proteins (in comparison to 21.5% in the human proteome [234]). Additionally, 21.5% of the proteins analyzed in this study are found in the Q3 quadrant (top-left corner), indicating a prediction of high disorder levels with both predictors. In quadrant Q4 (top-right corner), only 1.5% of proteins are located (compared to 3.1% in the human proteome [234]).
This hypothesis was supported by the analysis of the predisposition of human proteins from the PML-body proteome to undergo spontaneous LLPS.
The conducted analysis revealed (Figure 6, Table S1) that there are practically no globular proteins in the analyzed data set, which differs significantly from the human proteome (the proportion of IDPs in which is about 40% [234]). This is also consistent with the data that 52% of the analyzed proteins in the PML-body proteome are highly likely to be capable of spontaneous phase separation (Figure 6E). Furthermore, according to the FuzDrop analysis, the number of proteins that are “LLPS drivers”, i.e., proteins capable of spontaneous phase separation in the proteome of PML bodies, is almost twice as large as the number of client proteins, i.e., proteins capable of LLPS upon interaction with some partners (Figure 6C, Table S1). Scaffold proteins of PML bodies (including the main PML-body isoforms) also are presented as predominantly LLPS-drivers and IDPs (Table 1 and Table 2).
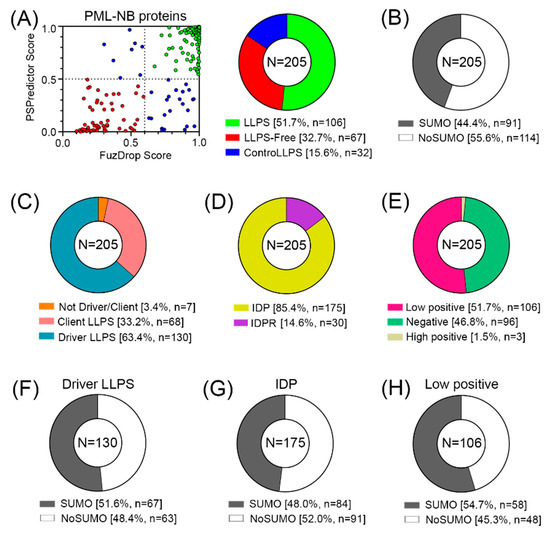
Figure 6.
Proteome analysis of PML bodies. (A) Correlation of the results of two predictors (FuzDrop and PSPredictor) and the results in the form of a pie diagram. (B) Proportion of proteins potentially capable (SUMO) and incapable (NoSUMO) of SUMOylation in the analyzed set of proteins. (C) Share of LLPS drivers/clients. (D) Proportion of disordered proteins (IDPs) and proteins with intrinsically disordered protein regions (IDPRs). (E) Proportion of proteins potentially carrying a negative charge, a weak positive charge, and a positive charge. Proportion of proteins potentially capable (SUMO) and incapable (NoSUMO) of SUMOylation for LLPS (F), IDP (G) and weakly positive (H) drivers.

Table 1.
Comparative characteristics of the scaffold proteins of PML bodies (except PML protein). DPRs are droplet-promoting regions, AHSs are aggregation hot spots.

Table 2.
Comparative characteristics of the main nuclear PML isoforms. DPRs are droplet-promoting regions, AHSs are aggregation hot spots.
However, only 44% of all proteins studied contain SUMO binding sites and can potentially be SUMOylated (Figure 6B).
The data obtained indicate that the proportion of proteins in the proteome of PML bodies that are capable of nonspecific inclusion in these structures is at least no less than the proportion of proteins included in these MLOs due to SUMO/SIM interactions.
Proteins prone to spontaneous LLPS are mainly represented by transcription factors, regulatory elements, DNA-damage response (DDR) proteins, and proteins involved in the alternative lengthening of telomeres (Figure 7). About 10% of LLPS-prone proteins are DNA-binding proteins.
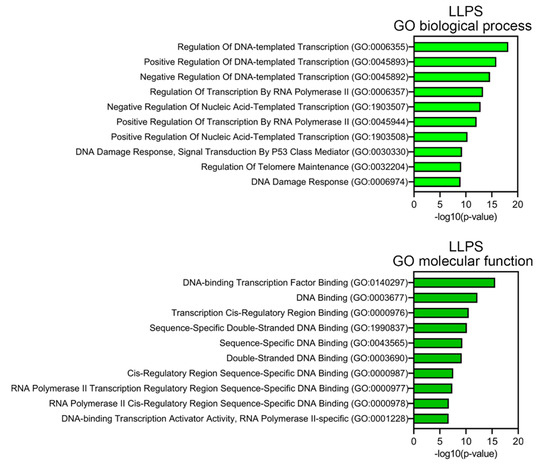
Figure 7.
Result of GO analysis for proteins potentially prone to LLPS. Biological process and molecular functions are shown.
This abundance of DNA-binding proteins in the proteome of PML bodies suggests that DNA is somehow involved in the biogenesis of these MLOs. This assumption is also supported by the fact that more than half of the proteins studied are positively charged, which may indicate their nonspecific interaction with negatively charged nucleic acids. Furthermore, the proportion of such proteins among proteins that do not directly interact with PML is 8% higher than in the full data set.
There are a number of works that discuss the influence of DNA on the biogenesis of PML bodies. For example, in 2004, it was shown [235] that the structural integrity of nuclear PML bodies depends not only on PTMs of PML, but also on the integrity and state of chromatin condensation. In cells not treated with transcription inhibitors, PML bodies are surrounded by chromatin, which explains the stability of their position; direct physical contact between the protein core and chromatin fibers predominate. The inhibition of transcription causes a reduction in these contacts due to chromatin condensation.
PML bodies have been shown to degrade in response to changes in chromatin integrity (demonstrated by introducing exogenous nuclease DNase I into cells) [235]. A 2009 study showed that PML bodies are surrounded by decondensed and condensed chromatin [236]. It has been suggested that PML regulates the incorporation of H3.3 between loose and compact chromatin compartments [237]. In doing so, PML regulates the routing of H3.3 to chromatin by diverting the H3.3 pool and limiting its deposition in chromatin. In addition, there is evidence that PML bodies are involved not only in gene silencing, but also as a potential regulator of epigenetic conditions [237].
4. Conclusions
In some of our previous works, we have already raised the question of the need to change existing ideas about the formation and biogenesis of PML bodies [16,19]. In particular, our analysis of the amino acid sequences of PML isoforms showed that the variable C-terminal domains of almost all isoforms of this protein have properties that are characteristic of protein sequences that are potentially capable of phase separation due to electrostatic interactions.
The results obtained indicate a high content of disordered proteins in the composition of PML bodies that are potentially capable of spontaneous separation into phases, and, therefore, allow us to raise the question of changing ideas about the role of nonspecific protein–protein interactions and, possibly, protein–DNA interactions in the formation and biogenesis of PML bodies.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/biom13121805/s1: Table S1: Comparative characteristics of the proteins that are localized in PML bodies.
Author Contributions
Conceptualization, A.V.F.; methodology, A.V.F. and V.N.U.; validation, I.M.K., K.K.T., V.N.U. and A.V.F.; formal analysis, S.A.S., Y.I.M., E.M.N., E.Y.S., V.N.U. and A.V.F.; investigation, S.A.S., Y.I.M., E.M.N., E.Y.S., V.N.U. and A.V.F.; writing—original draft preparation, S.A.S., Y.I.M., E.M.N., E.Y.S., I.M.K., K.K.T., V.N.U. and A.V.F.; writing—review and editing, S.A.S., Y.I.M., E.M.N., E.Y.S., I.M.K., K.K.T., V.N.U. and A.V.F.; visualization, S.A.S., Y.I.M., E.M.N., E.Y.S., V.N.U. and A.V.F.; supervision, A.V.F. and V.N.U.; funding acquisition, K.K.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Russian Science Foundation, grant number 22-15-00429 (K.K.T.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article or Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Wang, B.; Zhang, L.; Dai, T.; Qin, Z.; Lu, H.; Zhang, L.; Zhou, F. Liquid-liquid phase separation in human health and diseases. Signal Transduct. Target. Ther. 2021, 6, 290. [Google Scholar] [CrossRef] [PubMed]
- Shin, Y.; Brangwynne, C.P. Liquid phase condensation in cell physiology and disease. Science 2017, 357, eaaf4382. [Google Scholar] [CrossRef] [PubMed]
- Antifeeva, I.A.; Fonin, A.V.; Fefilova, A.S.; Stepanenko, O.V.; Povarova, O.I.; Silonov, S.A.; Kuznetsova, I.M.; Uversky, V.N.; Turoverov, K.K. Liquid-liquid phase separation as an organizing principle of intracellular space: Overview of the evolution of the cell compartmentalization concept. Cell. Mol. Life Sci. 2022, 79, 251. [Google Scholar] [CrossRef] [PubMed]
- Berry, J.; Weber, S.C.; Vaidya, N.; Haataja, M.; Brangwynne, C.P. RNA transcription modulates phase transition-driven nuclear body assembly. Proc. Natl. Acad. Sci. USA 2015, 112, E5237–E5245. [Google Scholar] [CrossRef] [PubMed]
- Berchtold, D.; Battich, N.; Pelkmans, L. A Systems-Level Study Reveals Regulators of Membrane-less Organelles in Human Cells. Mol. Cell 2018, 72, 1035–1049.e5. [Google Scholar] [CrossRef]
- Brangwynne, C.P.; Tompa, P.; Pappu, R.V. Polymer physics of intracellular phase transitions. Nat. Phys. 2015, 11, 899–904. [Google Scholar] [CrossRef]
- Dignon, G.L.; Best, R.B.; Mittal, J. Biomolecular Phase Separation: From Molecular Driving Forces to Macroscopic Properties. Annu. Rev. Phys. Chem. 2020, 71, 53–75. [Google Scholar] [CrossRef]
- Adame-Arana, O.; Weber, C.A.; Zaburdaev, V.; Prost, J.; Jülicher, F. Liquid Phase Separation Controlled by pH. Biophys. J. 2020, 119, 1590–1605. [Google Scholar] [CrossRef]
- Corpet, A.; Kleijwegt, C.; Roubille, S.; Juillard, F.; Jacquet, K.; Texier, P.; Lomonte, P. PML nuclear bodies and chromatin dynamics: Catch me if you can! Nucleic Acids Res. 2020, 48, 11890–11912. [Google Scholar] [CrossRef]
- Niwa-Kawakita, M.; Wu, H.C.; Thé, H.; Lallemand-Breitenbach, V. PML nuclear bodies, membrane-less domains acting as ROS sensors? Semin. Cell Dev. Biol. 2018, 80, 29–34. [Google Scholar] [CrossRef]
- Lallemand-Breitenbach, V. PML nuclear bodies. Cold Spring Harb. Perspect. Biol. 2010, 2, a000661. [Google Scholar] [CrossRef] [PubMed]
- Prikrylova, T.; Pachernik, J.; Kozubek, S.; Bartova, E. Epigenetics and chromatin plasticity in embryonic stem cells. World J. Stem Cells 2013, 5, 73–85. [Google Scholar] [CrossRef] [PubMed]
- Boisvert, F.M.; Hendzel, M.J.; Bazett-Jones, D.P. Promyelocytic leukemia (PML) nuclear bodies are protein structures that do not accumulate RNA. J. Cell Biol. 2000, 148, 283–292. [Google Scholar] [CrossRef] [PubMed]
- Hands, K.J.; Cuchet-Lourenco, D.; Everett, R.D.; Hay, R.T. PML isoforms in response to arsenic: High-resolution analysis of PML body structure and degradation characteristics. J. Cell Sci. 2014, 127, 365–375. [Google Scholar] [CrossRef] [PubMed]
- Guan, D.; Kao, H.Y. The function, regulation and therapeutic implications of the tumor suppressor protein, PML. Cell Biosci. 2015, 5, 60. [Google Scholar] [CrossRef] [PubMed]
- Fonin, A.V.; Silonov, S.A.; Shpironok, O.G.; Antifeeva, I.A.; Petukhov, A.V.; Romanovich, A.E.; Kuznetsova, I.M.; Uversky, V.N.; Turoverov, K.K. The Role of Non-Specific Interactions in Canonical and ALT-Associated PML-Bodies Formation and Dynamics. Int. J. Mol. Sci. 2021, 22, 5821. [Google Scholar] [CrossRef]
- Li, Y.; Ma, X.; Wu, W.; Chen, Z.; Meng, G. PML Nuclear Body Biogenesis, Carcinogenesis, and Targeted Therapy. Trends Cancer 2020, 6, 889–906. [Google Scholar] [CrossRef]
- Sahin, U.; De Thé, H.; Lallemand-Breitenbach, V. PML nuclear bodies: Assembly and oxidative stress-sensitive sumoylation. Nucleus 2014, 5, 499–507. [Google Scholar] [CrossRef]
- Fonin, A.V.; Silonov, S.A.; Fefilova, A.S.; Stepanenko, O.V.; Gavrilova, A.A.; Petukhov, A.V.; Romanovich, A.E.; Modina, A.L.; Zueva, T.S.; Nedelyaev, E.M.; et al. New Evidence of the Importance of Weak Interactions in the Formation of PML-Bodies. Int. J. Mol. Sci. 2022, 23, 1613. [Google Scholar] [CrossRef]
- Dayhoff, G.W., 2nd; Uversky, V.N. Rapid prediction and analysis of protein intrinsic disorder. Protein Sci. 2022, 31, e4496. [Google Scholar] [CrossRef]
- Meszaros, B.; Erdos, G.; Dosztanyi, Z. IUPred2A: Context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 2018, 46, W329–W337. [Google Scholar] [CrossRef] [PubMed]
- Obradovic, Z.; Peng, K.; Vucetic, S.; Radivojac, P.; Dunker, A.K. Exploiting heterogeneous sequence properties improves prediction of protein disorder. Proteins Struct. Funct. Bioinform. 2005, 61, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Peng, K.; Radivojac, P.; Vucetic, S.; Dunker, A.K.; Obradovic, Z. Length-dependent prediction of protein intrinsic disorder. BMC Bioinform. 2006, 7, 208. [Google Scholar] [CrossRef] [PubMed]
- Peng, K.; Vucetic, S.; Radivojac, P.; Brown, C.J.; Dunker, A.K.; Obradovic, Z. Optimizing long intrinsic disorder predictors with protein evolutionary information. J. Bioinform. Comput. Biol. 2005, 3, 35–60. [Google Scholar] [CrossRef] [PubMed]
- Romero, P.; Obradovic, Z.; Li, X.; Garner, E.C.; Brown, C.J.; Dunker, A.K. Sequence complexity of disordered protein. Proteins 2001, 42, 38–48. [Google Scholar] [CrossRef]
- Xue, B.; Dunbrack, R.L.; Williams, R.W.; Dunker, A.K.; Uversky, V.N. PONDR-FIT: A meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta 2010, 1804, 996–1010. [Google Scholar] [CrossRef]
- Rajagopalan, K.; Mooney, S.M.; Parekh, N.; Getzenberg, R.H.; Kulkarni, P. A majority of the cancer/testis antigens are intrinsically disordered proteins. J. Cell. Biochem. 2011, 112, 3256–3267. [Google Scholar] [CrossRef]
- Uversky, V.N. Analyzing IDPs in interactomes. In Intrinsically Disordered Proteins; Kragelund, B.B., Skriver, K., Eds.; Volume Methods in Molecular Biology; Humana: New York, NY, USA, 2020; pp. 895–945. [Google Scholar]
- Uversky, V.N.; Gillespie, J.R.; Fink, A.L. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins 2000, 41, 415–427. [Google Scholar] [CrossRef]
- Oldfield, C.J.; Cheng, Y.; Cortese, M.S.; Brown, C.J.; Uversky, V.N.; Dunker, A.K. Comparing and combining predictors of mostly disordered proteins. Biochemistry 2005, 44, 1989–2000. [Google Scholar] [CrossRef]
- Mohan, A.; Sullivan, W.J., Jr.; Radivojac, P.; Dunker, A.K.; Uversky, V.N. Intrinsic disorder in pathogenic and non-pathogenic microbes: Discovering and analyzing the unfoldomes of early-branching eukaryotes. Mol. Biosyst. 2008, 4, 328–340. [Google Scholar] [CrossRef]
- Huang, F.; Oldfield, C.; Meng, J.; Hsu, W.L.; Xue, B.; Uversky, V.N.; Romero, P.; Dunker, A.K. Subclassifying disordered proteins by the CH-CDF plot method. In Proceedings of the Pacific Symposium on Biocomputing, Kohala Coast, HI, USA, 3–7 January 2012; pp. 128–139. [Google Scholar]
- Hardenberg, M.; Horvath, A.; Ambrus, V.; Fuxreiter, M.; Vendruscolo, M. Widespread occurrence of the droplet state of proteins in the human proteome. Proc. Natl. Acad. Sci. USA 2020, 117, 33254–33262. [Google Scholar] [CrossRef] [PubMed]
- Chu, X.; Sun, T.; Li, Q.; Xu, Y.; Zhang, Z.; Lai, L.; Pei, J. Prediction of liquid-liquid phase separating proteins using machine learning. BMC Bioinform. 2022, 23, 72. [Google Scholar] [CrossRef] [PubMed]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef] [PubMed]
- UniProt, C. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [Google Scholar] [CrossRef]
- Conchillo-Solé, O.; de Groot, N.S.; Avilés, F.X.; Vendrell, J.; Daura, X.; Ventura, S. AGGRESCAN: A server for the prediction and evaluation of “hot spots” of aggregation in polypeptides. BMC Bioinform. 2007, 8, 65. [Google Scholar] [CrossRef] [PubMed]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed]
- Guan, D.; Lim, J.H.; Peng, L.; Liu, Y.; Lam, M.; Seto, E.; Kao, H.Y. Deacetylation of the tumor suppressor protein PML regulates hydrogen peroxide-induced cell death. Cell Death Dis. 2014, 5, e1340. [Google Scholar] [CrossRef]
- Lång, A.; Lång, E.; Bøe, S.O. PML Bodies in Mitosis. Cells 2019, 8, 893. [Google Scholar] [CrossRef]
- Missiroli, S.; Bonora, M.; Patergnani, S.; Poletti, F.; Perrone, M.; Gafà, R.; Magri, E.; Raimondi, A.; Lanza, G.; Tacchetti, C.; et al. PML at Mitochondria-Associated Membranes Is Critical for the Repression of Autophagy and Cancer Development. Cell Rep. 2016, 16, 2415–2427. [Google Scholar] [CrossRef]
- Giorgi, C.; Ito, K.; Lin, H.K.; Santangelo, C.; Wieckowski, M.R.; Lebiedzinska, M.; Bononi, A.; Bonora, M.; Duszynski, J.; Bernardi, R.; et al. PML regulates apoptosis at endoplasmic reticulum by modulating calcium release. Science 2010, 330, 1247–1251. [Google Scholar] [CrossRef]
- Cheng, X.; Kao, H.Y. Post-translational modifications of PML: Consequences and implications. Front. Oncol. 2012, 2, 210. [Google Scholar] [CrossRef] [PubMed]
- Van Damme, E.; Laukens, K.; Dang, T.H.; Van Ostade, X. A manually curated network of the PML nuclear body interactome reveals an important role for PML-NBs in SUMOylation dynamics. Int. J. Biol. Sci. 2010, 6, 51–67. [Google Scholar] [CrossRef] [PubMed]
- Jin, G.; Gao, Y.; Lin, H.K. Cytoplasmic PML: From molecular regulation to biological functions. J. Cell. Biochem. 2014, 115, 812–818. [Google Scholar] [CrossRef] [PubMed]
- Clausen, T.H.; Lamark, T.; Isakson, P.; Finley, K.; Larsen, K.B.; Brech, A.; Øvervatn, A.; Stenmark, H.; Bjørkøy, G.; Simonsen, A.; et al. p62/SQSTM1 and ALFY interact to facilitate the formation of p62 bodies/ALIS and their degradation by autophagy. Autophagy 2010, 6, 330–344. [Google Scholar] [CrossRef] [PubMed]
- Korioth, F.; Gieffers, C.; Maul, G.G.; Frey, J. Molecular characterization of NDP52, a novel protein of the nuclear domain 10, which is redistributed upon virus infection and interferon treatment. J. Cell Biol. 1995, 130, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ahmady, E.; Deeke, S.A.; Rabaa, S.; Kouri, L.; Kenney, L.; Stewart, A.F.; Burgon, P.G. Identification of a novel muscle A-type lamin-interacting protein (MLIP). J. Biol. Chem. 2011, 286, 19702–19713. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Luo, Z.; Jiang, S.; Li, F.; Han, X.; Hu, Y.; Wang, D.; Zhao, Y.; Ma, W.; Liu, D.; et al. The telomere-associated homeobox-containing protein TAH1/HMBOX1 participates in telomere maintenance in ALT cells. J. Cell Sci. 2013, 126, 3982–3989. [Google Scholar] [CrossRef]
- Lopez, P.; Vidal, F.; Martin, L.; Lopez-Fernandez, L.A.; Rual, J.F.; Rosen, B.S.; Cuzin, F.; Rassoulzadegan, M. Gene control in germinal differentiation: RNF6, a transcription regulatory protein in the mouse sertoli cell. Mol. Cell. Biol. 2002, 22, 3488–3496. [Google Scholar] [CrossRef]
- Cheung, B.B.; Bell, J.; Raif, A.; Bohlken, A.; Yan, J.; Roediger, B.; Poljak, A.; Smith, S.; Lee, M.; Thomas, W.D.; et al. The estrogen-responsive B box protein is a novel regulator of the retinoid signal. J. Biol. Chem. 2006, 281, 18246–18256. [Google Scholar] [CrossRef]
- Gao, C.; Cheng, X.; Lam, M.; Liu, Y.; Liu, Q.; Chang, K.S.; Kao, H.Y. Signal-dependent regulation of transcription by histone deacetylase 7 involves recruitment to promyelocytic leukemia protein nuclear bodies. Mol. Biol. Cell 2008, 19, 3020–3027. [Google Scholar] [CrossRef][Green Version]
- Green, L.M.; Wagner, K.J.; Campbell, H.A.; Addison, K.; Roberts, S.G. Dynamic interaction between WT1 and BASP1 in transcriptional regulation during differentiation. Nucleic Acids Res. 2009, 37, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.C.; Lee, C.C.; Yao, Y.L.; Lai, C.C.; Schmitz, M.L.; Yang, W.M. SUMO5, a Novel Poly-SUMO Isoform, Regulates PML Nuclear Bodies. Sci. Rep. 2016, 6, 26509. [Google Scholar] [CrossRef] [PubMed]
- Tse, W.T.; Tang, J.; Jin, O.; Korsgren, C.; John, K.M.; Kung, A.L.; Gwynn, B.; Peters, L.L.; Lux, S.E. A new spectrin, beta IV, has a major truncated isoform that associates with promyelocytic leukemia protein nuclear bodies and the nuclear matrix. J. Biol. Chem. 2001, 276, 23974–23985. [Google Scholar] [CrossRef] [PubMed]
- Roussigne, M.; Cayrol, C.; Clouaire, T.; Amalric, F.; Girard, J.P. THAP1 is a nuclear proapoptotic factor that links prostate-apoptosis-response-4 (Par-4) to PML nuclear bodies. Oncogene 2003, 22, 2432–2442. [Google Scholar] [CrossRef]
- Boisvert, F.M.; Hendzel, M.J.; Masson, J.Y.; Richard, S. Methylation of MRE11 regulates its nuclear compartmentalization. Cell Cycle 2005, 4, 981–989. [Google Scholar] [CrossRef]
- Lee, J.; Lee, Y.; Lee, M.J.; Park, E.; Kang, S.H.; Chung, C.H.; Lee, K.H.; Kim, K. Dual modification of BMAL1 by SUMO2/3 and ubiquitin promotes circadian activation of the CLOCK/BMAL1 complex. Mol. Cell. Biol. 2008, 28, 6056–6065. [Google Scholar] [CrossRef]
- Karvonen, U.; Jääskeläinen, T.; Rytinki, M.; Kaikkonen, S.; Palvimo, J.J. ZNF451 is a novel PML body- and SUMO-associated transcriptional coregulator. J. Mol. Biol. 2008, 382, 585–600. [Google Scholar] [CrossRef]
- Yang, S.; Kuo, C.; Bisi, J.E.; Kim, M.K. PML-dependent apoptosis after DNA damage is regulated by the checkpoint kinase hCds1/Chk2. Nat. Cell Biol. 2002, 4, 865–870. [Google Scholar] [CrossRef]
- Sano, M.; Tokudome, S.; Shimizu, N.; Yoshikawa, N.; Ogawa, C.; Shirakawa, K.; Endo, J.; Katayama, T.; Yuasa, S.; Ieda, M.; et al. Intramolecular control of protein stability, subnuclear compartmentalization, and coactivator function of peroxisome proliferator-activated receptor gamma coactivator 1alpha. J. Biol. Chem. 2007, 282, 25970–25980. [Google Scholar] [CrossRef]
- Miki, T.; Xu, Z.; Chen-Goodspeed, M.; Liu, M.; Van Oort-Jansen, A.; Rea, M.A.; Zhao, Z.; Lee, C.C.; Chang, K.S. PML regulates PER2 nuclear localization and circadian function. EMBO J. 2012, 31, 1427–1439. [Google Scholar] [CrossRef]
- Li, Q.; He, Y.; Wei, L.; Wu, X.; Wu, D.; Lin, S.; Wang, Z.; Ye, Z.; Lin, S.C. AXIN is an essential co-activator for the promyelocytic leukemia protein in p53 activation. Oncogene 2011, 30, 1194–1204. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kaytor, M.D.; Duvick, L.A.; Skinner, P.J.; Koob, M.D.; Ranum, L.P.; Orr, H.T. Nuclear localization of the spinocerebellar ataxia type 7 protein, ataxin-7. Hum. Mol. Genet. 1999, 8, 1657–1664. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jang, S.M.; Nathans, J.F.; Fu, H.; Redon, C.E.; Jenkins, L.M.; Thakur, B.L.; Pongor, L.S.; Baris, A.M.; Gross, J.M.; O’Neill, M.J.; et al. The RepID-CRL4 ubiquitin ligase complex regulates metaphase to anaphase transition via BUB3 degradation. Nat. Commun. 2020, 11, 24. [Google Scholar] [CrossRef] [PubMed]
- Marcello, A.; Ferrari, A.; Pellegrini, V.; Pegoraro, G.; Lusic, M.; Beltram, F.; Giacca, M. Recruitment of human cyclin T1 to nuclear bodies through direct interaction with the PML protein. EMBO J. 2003, 22, 2156–2166. [Google Scholar] [CrossRef]
- Wu, G.; Lee, W.H.; Chen, P.L. NBS1 and TRF1 colocalize at promyelocytic leukemia bodies during late S/G2 phases in immortalized telomerase-negative cells. Implication of NBS1 in alternative lengthening of telomeres. J. Biol. Chem. 2000, 275, 30618–30622. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.M.; Nomura, T.; Kim, H.; Kaul, S.C.; Wadhwa, R.; Shinagawa, T.; Ichikawa-Iwata, E.; Zhong, S.; Pandolfi, P.P.; Ishii, S. Role of PML and PML-RARalpha in Mad-mediated transcriptional repression. Mol. Cell 2001, 7, 1233–1243. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.R.; Conn, K.L.; Wasson, P.; Charman, M.; Tong, L.; Grant, K.; McFarlane, S.; Boutell, C. SUMO Ligase Protein Inhibitor of Activated STAT1 (PIAS1) Is a Constituent Promyelocytic Leukemia Nuclear Body Protein That Contributes to the Intrinsic Antiviral Immune Response to Herpes Simplex Virus 1. J. Virol. 2016, 90, 5939–5952. [Google Scholar] [CrossRef]
- Li, C.; McManus, F.P.; Plutoni, C.; Pascariu, C.M.; Nelson, T.; Alberici Delsin, L.E.; Emery, G.; Thibault, P. Quantitative SUMO proteomics identifies PIAS1 substrates involved in cell migration and motility. Nat. Commun. 2020, 11, 834. [Google Scholar] [CrossRef]
- Xu, G.L.; Pan, Y.K.; Wang, B.Y.; Huang, L.; Tian, L.; Xue, J.L.; Chen, J.Z.; Jia, W. TTRAP is a novel PML nuclear bodies-associated protein. Biochem. Biophys. Res. Commun. 2008, 375, 395–398. [Google Scholar] [CrossRef]
- Vilotti, S.; Biagioli, M.; Foti, R.; Dal Ferro, M.; Lavina, Z.S.; Collavin, L.; Del Sal, G.; Zucchelli, S.; Gustincich, S. The PML nuclear bodies-associated protein TTRAP regulates ribosome biogenesis in nucleolar cavities upon proteasome inhibition. Cell Death Differ. 2012, 19, 488–500. [Google Scholar] [CrossRef]
- Nayak, A.; Glöckner-Pagel, J.; Vaeth, M.; Schumann, J.E.; Buttmann, M.; Bopp, T.; Schmitt, E.; Serfling, E.; Berberich-Siebelt, F. Sumoylation of the transcription factor NFATc1 leads to its subnuclear relocalization and interleukin-2 repression by histone deacetylase. J. Biol. Chem. 2009, 284, 10935–10946. [Google Scholar] [CrossRef]
- Cairo, S.; De Falco, F.; Pizzo, M.; Salomoni, P.; Pandolfi, P.P.; Meroni, G. PML interacts with Myc, and Myc target gene expression is altered in PML-null fibroblasts. Oncogene 2005, 24, 2195–2203. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cobb, A.M.; De Silva, S.A.; Hayward, R.; Sek, K.; Ulferts, S.; Grosse, R.; Shanahan, C.M. Filamentous nuclear actin regulation of PML NBs during the DNA damage response is deregulated by prelamin A. Cell Death Dis. 2022, 13, 1042. [Google Scholar] [CrossRef] [PubMed]
- Fogal, V.; Gostissa, M.; Sandy, P.; Zacchi, P.; Sternsdorf, T.; Jensen, K.; Pandolfi, P.P.; Will, H.; Schneider, C.; Del Sal, G. Regulation of p53 activity in nuclear bodies by a specific PML isoform. EMBO J. 2000, 19, 6185–6195. [Google Scholar] [CrossRef] [PubMed]
- Marks, D.; Heinen, N.; Bachmann, L.; Meermeyer, S.; Werner, M.; Gallego, L.; Hemmerich, P.; Bader, V.; Winklhofer, K.F.; Schröder, E.; et al. Amyloid precursor protein elevates fusion of promyelocytic leukemia nuclear bodies in human hippocampal areas with high plaque load. Acta Neuropathol. Commun. 2021, 9, 66. [Google Scholar] [CrossRef] [PubMed]
- Alcalay, M.; Tomassoni, L.; Colombo, E.; Stoldt, S.; Grignani, F.; Fagioli, M.; Szekely, L.; Helin, K.; Pelicci, P.G. The promyelocytic leukemia gene product (PML) forms stable complexes with the retinoblastoma protein. Mol. Cell. Biol. 1998, 18, 1084–1093. [Google Scholar] [CrossRef]
- Cohen, N.; Sharma, M.; Kentsis, A.; Perez, J.M.; Strudwick, S.; Borden, K.L. PML RING suppresses oncogenic transformation by reducing the affinity of eIF4E for mRNA. EMBO J. 2001, 20, 4547–4559. [Google Scholar] [CrossRef]
- Kentsis, A.; Dwyer, E.C.; Perez, J.M.; Sharma, M.; Chen, A.; Pan, Z.Q.; Borden, K.L. The RING domains of the promyelocytic leukemia protein PML and the arenaviral protein Z repress translation by directly inhibiting translation initiation factor eIF4E. J. Mol. Biol. 2001, 312, 609–623. [Google Scholar] [CrossRef]
- Dahle, Ø.; Bakke, O.; Gabrielsen, O.S. c-Myb associates with PML in nuclear bodies in hematopoietic cells. Exp. Cell Res. 2004, 297, 118–126. [Google Scholar] [CrossRef]
- Matsuzaki, K.; Minami, T.; Tojo, M.; Honda, Y.; Uchimura, Y.; Saitoh, H.; Yasuda, H.; Nagahiro, S.; Saya, H.; Nakao, M. Serum response factor is modulated by the SUMO-1 conjugation system. Biochem. Biophys. Res. Commun. 2003, 306, 32–38. [Google Scholar] [CrossRef]
- Gupta, P.; Ho, P.C.; Ha, S.G.; Lin, Y.W.; Wei, L.N. HDAC3 as a molecular chaperone for shuttling phosphorylated TR2 to PML: A novel deacetylase activity-independent function of HDAC3. PLoS ONE 2009, 4, e4363. [Google Scholar] [CrossRef] [PubMed]
- Cao, T.; Duprez, E.; Borden, K.L.; Freemont, P.S.; Etkin, L.D. Ret finger protein is a normal component of PML nuclear bodies and interacts directly with PML. J. Cell Sci. 1998, 111 Pt 10, 1319–1329. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.S.; Xu, Z.X.; Ran, R.; Meng, F.; Chang, K.S. Promyelocytic leukemia protein PML inhibits Nur77-mediated transcription through specific functional interactions. Oncogene 2002, 21, 3925–3933. [Google Scholar] [CrossRef] [PubMed]
- Dent, A.L.; Yewdell, J.; Puvion-Dutilleul, F.; Koken, M.H.; de The, H.; Staudt, L.M. LYSP100-associated nuclear domains (LANDs): Description of a new class of subnuclear structures and their relationship to PML nuclear bodies. Blood 1996, 88, 1423–1426. [Google Scholar] [CrossRef] [PubMed]
- Warren, D.T.; Tajsic, T.; Mellad, J.A.; Searles, R.; Zhang, Q.; Shanahan, C.M. Novel nuclear nesprin-2 variants tether active extracellular signal-regulated MAPK1 and MAPK2 at promyelocytic leukemia protein nuclear bodies and act to regulate smooth muscle cell proliferation. J. Biol. Chem. 2010, 285, 1311–1320. [Google Scholar] [CrossRef]
- Ulbricht, T.; Alzrigat, M.; Horch, A.; Reuter, N.; von Mikecz, A.; Steimle, V.; Schmitt, E.; Krämer, O.H.; Stamminger, T.; Hemmerich, P. PML promotes MHC class II gene expression by stabilizing the class II transactivator. J. Cell Biol. 2012, 199, 49–63. [Google Scholar] [CrossRef]
- Dhordain, P.; Albagli, O.; Honore, N.; Guidez, F.; Lantoine, D.; Schmid, M.; The, H.D.; Zelent, A.; Koken, M.H. Colocalization and heteromerization between the two human oncogene POZ/zinc finger proteins, LAZ3 (BCL6) and PLZF. Oncogene 2000, 19, 6240–6250. [Google Scholar] [CrossRef]
- Buchberger, E.; El Harchi, M.; Payrhuber, D.; Zommer, A.; Schauer, D.; Simonitsch-Klupp, I.; Bilban, M.; Brostjan, C. Overexpression of the transcriptional repressor complex BCL-6/BCoR leads to nuclear aggregates distinct from classical aggresomes. PLoS ONE 2013, 8, e76845. [Google Scholar] [CrossRef]
- Peche, L.Y.; Scolz, M.; Ladelfa, M.F.; Monte, M.; Schneider, C. MageA2 restrains cellular senescence by targeting the function of PMLIV/p53 axis at the PML-NBs. Cell Death Differ. 2012, 19, 926–936. [Google Scholar] [CrossRef]
- Johnson, F.B.; Lombard, D.B.; Neff, N.F.; Mastrangelo, M.A.; Dewolf, W.; Ellis, N.A.; Marciniak, R.A.; Yin, Y.; Jaenisch, R.; Guarente, L. Association of the Bloom syndrome protein with topoisomerase IIIalpha in somatic and meiotic cells. Cancer Res. 2000, 60, 1162–1167. [Google Scholar]
- Kruse, M.L.; Arlt, A.; Sieke, A.; Grohmann, F.; Grossmann, M.; Minkenberg, J.; Fölsch, U.R.; Schäfer, H. Immediate early gene X1 (IEX-1) is organized in subnuclear structures and partially co-localizes with promyelocytic leukemia protein in HeLa cells. J. Biol. Chem. 2005, 280, 24849–24856. [Google Scholar] [CrossRef] [PubMed]
- Levy, D.; Adamovich, Y.; Reuven, N.; Shaul, Y. The Yes-associated protein 1 stabilizes p73 by preventing Itch-mediated ubiquitination of p73. Cell Death Differ. 2007, 14, 743–751. [Google Scholar] [CrossRef] [PubMed]
- Strano, S.; Monti, O.; Pediconi, N.; Baccarini, A.; Fontemaggi, G.; Lapi, E.; Mantovani, F.; Damalas, A.; Citro, G.; Sacchi, A.; et al. The transcriptional coactivator Yes-associated protein drives p73 gene-target specificity in response to DNA Damage. Mol. Cell 2005, 18, 447–459. [Google Scholar] [CrossRef] [PubMed]
- Basei, F.L.; Meirelles, G.V.; Righetto, G.L.; Dos Santos Migueleti, D.L.; Smetana, J.H.; Kobarg, J. New interaction partners for Nek4.1 and Nek4.2 isoforms: From the DNA damage response to RNA splicing. Proteome Sci. 2015, 13, 11. [Google Scholar] [CrossRef] [PubMed]
- Sanz, M.M.; Proytcheva, M.; Ellis, N.A.; Holloman, W.K.; German, J. BLM, the Bloom’s syndrome protein, varies during the cell cycle in its amount, distribution, and co-localization with other nuclear proteins. Cytogenet. Cell Genet. 2000, 91, 217–223. [Google Scholar] [CrossRef]
- Luciani, J.J.; Depetris, D.; Usson, Y.; Metzler-Guillemain, C.; Mignon-Ravix, C.; Mitchell, M.J.; Megarbane, A.; Sarda, P.; Sirma, H.; Moncla, A.; et al. PML nuclear bodies are highly organised DNA-protein structures with a function in heterochromatin remodelling at the G2 phase. J. Cell Sci. 2006, 119, 2518–2531. [Google Scholar] [CrossRef]
- Zhang, Y.; Shao, A.W.; Tang, J.; Geng, Y. PML-II recruits ataxin-3 to PML-NBs and inhibits its deubiquitinating activity. Biochem. Biophys. Res. Commun. 2021, 554, 186–192. [Google Scholar] [CrossRef]
- Fu, C.; Ahmed, K.; Ding, H.; Ding, X.; Lan, J.; Yang, Z.; Miao, Y.; Zhu, Y.; Shi, Y.; Zhu, J.; et al. Stabilization of PML nuclear localization by conjugation and oligomerization of SUMO-3. Oncogene 2005, 24, 5401–5413. [Google Scholar] [CrossRef]
- Rabellino, A.; Carter, B.; Konstantinidou, G.; Wu, S.Y.; Rimessi, A.; Byers, L.A.; Heymach, J.V.; Girard, L.; Chiang, C.M.; Teruya-Feldstein, J.; et al. The SUMO E3-ligase PIAS1 regulates the tumor suppressor PML and its oncogenic counterpart PML-RARA. Cancer Res. 2012, 72, 2275–2284. [Google Scholar] [CrossRef]
- Duprez, E.; Saurin, A.J.; Desterro, J.M.; Lallemand-Breitenbach, V.; Howe, K.; Boddy, M.N.; Solomon, E.; de Thé, H.; Hay, R.T.; Freemont, P.S. SUMO-1 modification of the acute promyelocytic leukaemia protein PML: Implications for nuclear localisation. J. Cell Sci. 1999, 112 Pt 3, 381–393. [Google Scholar] [CrossRef]
- Barroso-Gomila, O.; Trulsson, F.; Muratore, V.; Canosa, I.; Merino-Cacho, L.; Cortazar, A.R.; Pérez, C.; Azkargorta, M.; Iloro, I.; Carracedo, A.; et al. Identification of proximal SUMO-dependent interactors using SUMO-ID. Nat. Commun. 2021, 12, 6671. [Google Scholar] [CrossRef] [PubMed]
- Häkli, M.; Karvonen, U.; Jänne, O.A.; Palvimo, J.J. SUMO-1 promotes association of SNURF (RNF4) with PML nuclear bodies. Exp. Cell Res. 2005, 304, 224–233. [Google Scholar] [CrossRef] [PubMed]
- Shastrula, P.K.; Sierra, I.; Deng, Z.; Keeney, F.; Hayden, J.E.; Lieberman, P.M.; Janicki, S.M. PML is recruited to heterochromatin during S phase and represses DAXX-mediated histone H3.3 chromatin assembly. J. Cell Sci. 2019, 132, jcs.220970. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Rogers, R.; Matunis, M.J.; Mayhew, C.N.; Goodson, M.L.; Park-Sarge, O.K.; Sarge, K.D. Regulation of heat shock transcription factor 1 by stress-induced SUMO-1 modification. J. Biol. Chem. 2001, 276, 40263–40267. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.S.; Xu, Z.X.; Hittelman, W.N.; Salomoni, P.; Pandolfi, P.P.; Chang, K.S. Promyelocytic leukemia protein sensitizes tumor necrosis factor alpha-induced apoptosis by inhibiting the NF-kappaB survival pathway. J. Biol. Chem. 2003, 278, 12294–12304. [Google Scholar] [CrossRef] [PubMed]
- Giogha, C.; Lung, T.W.; Mühlen, S.; Pearson, J.S.; Hartland, E.L. Substrate recognition by the zinc metalloprotease effector NleC from enteropathogenic Escherichia coli. Cell. Microbiol. 2015, 17, 1766–1778. [Google Scholar] [CrossRef] [PubMed]
- Hodgson, A.; Wier, E.M.; Fu, K.; Sun, X.; Yu, H.; Zheng, W.; Sham, H.P.; Johnson, K.; Bailey, S.; Vallance, B.A.; et al. Metalloprotease NleC suppresses host NF-κB/inflammatory responses by cleaving p65 and interfering with the p65/RPS3 interaction. PLoS Pathog. 2015, 11, e1004705. [Google Scholar] [CrossRef]
- Koken, M.H.; Reid, A.; Quignon, F.; Chelbi-Alix, M.K.; Davies, J.M.; Kabarowski, J.H.; Zhu, J.; Dong, S.; Chen, S.; Chen, Z.; et al. Leukemia-associated retinoic acid receptor alpha fusion partners, PML and PLZF, heterodimerize and colocalize to nuclear bodies. Proc. Natl. Acad. Sci. USA 1997, 94, 10255–10260. [Google Scholar] [CrossRef]
- Xu, D.; Holko, M.; Sadler, A.J.; Scott, B.; Higashiyama, S.; Berkofsky-Fessler, W.; McConnell, M.J.; Pandolfi, P.P.; Licht, J.D.; Williams, B.R. Promyelocytic leukemia zinc finger protein regulates interferon-mediated innate immunity. Immunity 2009, 30, 802–816. [Google Scholar] [CrossRef]
- Wang, X.; He, S.; Sun, J.M.; Delcuve, G.P.; Davie, J.R. Selective association of peroxiredoxin 1 with genomic DNA and COX-2 upstream promoter elements in estrogen receptor negative breast cancer cells. Mol. Biol. Cell 2010, 21, 2987–2995. [Google Scholar] [CrossRef]
- Sachini, N.; Arampatzi, P.; Klonizakis, A.; Nikolaou, C.; Makatounakis, T.; Lam, E.W.; Kretsovali, A.; Papamatheakis, J. Promyelocytic leukemia protein (PML) controls breast cancer cell proliferation by modulating Forkhead transcription factors. Mol. Oncol. 2019, 13, 1369–1387. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Liao, L.; Deng, X.; Chen, R.; Gray, N.S.; Yates, J.R., 3rd; Lee, J.D. BMK1 is involved in the regulation of p53 through disrupting the PML-MDM2 interaction. Oncogene 2013, 32, 3156–3164. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yoshida, N.; Murakami, N.; Kawata, K.; Ishizaki, H.; Tanaka-Okamoto, M.; Miyoshi, J.; Zinn, A.R.; Shime, H.; Inoue, N. Dynamic regulation of p53 subnuclear localization and senescence by MORC3. Mol. Biol. Cell 2007, 18, 1701–1709. [Google Scholar] [CrossRef] [PubMed]
- Sloan, E.; Orr, A.; Everett, R.D. MORC3, a Component of PML Nuclear Bodies, Has a Role in Restricting Herpes Simplex Virus 1 and Human Cytomegalovirus. J. Virol. 2016, 90, 8621–8633. [Google Scholar] [CrossRef] [PubMed]
- Mimura, Y.; Takahashi, K.; Kawata, K.; Akazawa, T.; Inoue, N. Two-step colocalization of MORC3 with PML nuclear bodies. J. Cell Sci. 2010, 123, 2014–2024. [Google Scholar] [CrossRef] [PubMed]
- Renner, F.; Moreno, R.; Schmitz, M.L. SUMOylation-dependent localization of IKKepsilon in PML nuclear bodies is essential for protection against DNA-damage-triggered cell death. Mol. Cell 2010, 37, 503–515. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Park, J.S.; Kang, Y.K. Dual functions of histone-lysine N-methyltransferase Setdb1 protein at promyelocytic leukemia-nuclear body (PML-NB): Maintaining PML-NB structure and regulating the expression of its associated genes. J. Biol. Chem. 2011, 286, 41115–41124. [Google Scholar] [CrossRef]
- Schmidt, J.A.; Danielson, K.G.; Duffner, E.R.; Radecki, S.G.; Walker, G.T.; Shelton, A.; Wang, T.; Knepper, J.E. Regulation of the oncogenic phenotype by the nuclear body protein ZC3H8. BMC Cancer 2018, 18, 759. [Google Scholar] [CrossRef]
- Kleijwegt, C.; Bressac, F.; Seurre, C.; Bouchereau, W.; Cohen, C.; Texier, P.; Simonet, T.; Schaeffer, L.; Lomonte, P.; Corpet, A. Interplay between PML NBs and HIRA for H3.3 dynamics following type I interferon stimulus. eLife 2023, 12, e80156. [Google Scholar] [CrossRef]
- Potts, P.R.; Yu, H. The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins. Nat. Struct. Mol. Biol. 2007, 14, 581–590. [Google Scholar] [CrossRef]
- Brouwer, A.K.; Schimmel, J.; Wiegant, J.C.; Vertegaal, A.C.; Tanke, H.J.; Dirks, R.W. Telomeric DNA mediates de novo PML body formation. Mol. Biol. Cell 2009, 20, 4804–4815. [Google Scholar] [CrossRef] [PubMed]
- González-Prieto, R.; Cuijpers, S.A.; Luijsterburg, M.S.; van Attikum, H.; Vertegaal, A.C. SUMOylation and PARylation cooperate to recruit and stabilize SLX4 at DNA damage sites. EMBO Rep. 2015, 16, 512–519. [Google Scholar] [CrossRef] [PubMed]
- Forlani, G.; Tosi, G.; Turrini, F.; Poli, G.; Vicenzi, E.; Accolla, R.S. Tripartite Motif-Containing Protein 22 Interacts with Class II Transactivator and Orchestrates Its Recruitment in Nuclear Bodies Containing TRIM19/PML and Cyclin T1. Front. Immunol. 2017, 8, 564. [Google Scholar] [CrossRef] [PubMed]
- Ivanschitz, L.; Takahashi, Y.; Jollivet, F.; Ayrault, O.; Le Bras, M.; de Thé, H. PML IV/ARF interaction enhances p53 SUMO-1 conjugation, activation, and senescence. Proc. Natl. Acad. Sci. USA 2015, 112, 14278–14283. [Google Scholar] [CrossRef] [PubMed]
- Oravcová, M.; Nie, M.; Zilio, N.; Maeda, S.; Jami-Alahmadi, Y.; Lazzerini-Denchi, E.; Wohlschlegel, J.A.; Ulrich, H.D.; Otomo, T.; Boddy, M.N. The Nse5/6-like SIMC1-SLF2 complex localizes SMC5/6 to viral replication centers. eLife 2022, 11, e79676. [Google Scholar] [CrossRef]
- Shiio, Y.; Rose, D.W.; Aur, R.; Donohoe, S.; Aebersold, R.; Eisenman, R.N. Identification and characterization of SAP25, a novel component of the mSin3 corepressor complex. Mol. Cell. Biol. 2006, 26, 1386–1397. [Google Scholar] [CrossRef]
- Pearson, M.; Carbone, R.; Sebastiani, C.; Cioce, M.; Fagioli, M.; Saito, S.; Higashimoto, Y.; Appella, E.; Minucci, S.; Pandolfi, P.P.; et al. PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature 2000, 406, 207–210. [Google Scholar] [CrossRef]
- Doucas, V. The promyelocytic (PML) nuclear compartment and transcription control. Biochem. Pharmacol. 2000, 60, 1197–1201. [Google Scholar] [CrossRef]
- Wu, Q.; Hu, H.; Lan, J.; Emenari, C.; Wang, Z.; Chang, K.S.; Huang, H.; Yao, X. PML3 Orchestrates the Nuclear Dynamics and Function of TIP60. J. Biol. Chem. 2009, 284, 8747–8759. [Google Scholar] [CrossRef]
- Everett, R.D.; Meredith, M.; Orr, A.; Cross, A.; Kathoria, M.; Parkinson, J. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 1997, 16, 1519–1530. [Google Scholar] [CrossRef]
- Song, M.S.; Salmena, L.; Carracedo, A.; Egia, A.; Lo-Coco, F.; Teruya-Feldstein, J.; Pandolfi, P.P. The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature 2008, 455, 813–817. [Google Scholar] [CrossRef] [PubMed]
- Langley, E.; Pearson, M.; Faretta, M.; Bauer, U.M.; Frye, R.A.; Minucci, S.; Pelicci, P.G.; Kouzarides, T. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 2002, 21, 2383–2396. [Google Scholar] [CrossRef] [PubMed]
- Hattersley, N.; Shen, L.; Jaffray, E.G.; Hay, R.T. The SUMO protease SENP6 is a direct regulator of PML nuclear bodies. Mol. Biol. Cell 2011, 22, 78–90. [Google Scholar] [CrossRef] [PubMed]
- Tatemichi, Y.; Shibazaki, M.; Yasuhira, S.; Kasai, S.; Tada, H.; Oikawa, H.; Suzuki, Y.; Takikawa, Y.; Masuda, T.; Maesawa, C. Nucleus accumbens associated 1 is recruited within the promyelocytic leukemia nuclear body through SUMO modification. Cancer Sci. 2015, 106, 848–856. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Wang, F.; Cao, W.B.; Lv, X.X.; Hua, F.; Cui, B.; Yu, J.J.; Zhang, X.W.; Shang, S.; Liu, S.S.; et al. TRIB3 Promotes APL Progression through Stabilization of the Oncoprotein PML-RARα and Inhibition of p53-Mediated Senescence. Cancer Cell 2017, 31, 697–710.e7. [Google Scholar] [CrossRef] [PubMed]
- Niu, C.; Livingston, C.M.; Li, L.; Beran, R.K.; Daffis, S.; Ramakrishnan, D.; Burdette, D.; Peiser, L.; Salas, E.; Ramos, H.; et al. The Smc5/6 Complex Restricts HBV when Localized to ND10 without Inducing an Innate Immune Response and Is Counteracted by the HBV X Protein Shortly after Infection. PLoS ONE 2017, 12, e0169648. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Park, Y.J.; Hwang, I.Y.; Youdim, M.B.; Park, K.S.; Oh, Y.J. Nuclear translocation of DJ-1 during oxidative stress-induced neuronal cell death. Free Radic. Biol. Med. 2012, 53, 936–950. [Google Scholar] [CrossRef]
- Suico, M.A.; Yoshida, H.; Seki, Y.; Uchikawa, T.; Lu, Z.; Shuto, T.; Matsuzaki, K.; Nakao, M.; Li, J.D.; Kai, H. Myeloid Elf-1-like factor, an ETS transcription factor, up-regulates lysozyme transcription in epithelial cells through interaction with promyelocytic leukemia protein. J. Biol. Chem. 2004, 279, 19091–19098. [Google Scholar] [CrossRef]
- Fukuyo, Y.; Mogi, K.; Tsunematsu, Y.; Nakajima, T. E2FBP1/hDril1 modulates cell growth through downregulation of promyelocytic leukemia bodies. Cell Death Differ. 2004, 11, 747–759. [Google Scholar] [CrossRef]
- Tatematsu, K.; Yoshimoto, N.; Koyanagi, T.; Tokunaga, C.; Tachibana, T.; Yoneda, Y.; Yoshida, M.; Okajima, T.; Tanizawa, K.; Kuroda, S. Nuclear-cytoplasmic shuttling of a RING-IBR protein RBCK1 and its functional interaction with nuclear body proteins. J. Biol. Chem. 2005, 280, 22937–22944. [Google Scholar] [CrossRef]
- He, W.; Hu, C.X.; Hou, J.K.; Fan, L.; Xu, Y.W.; Liu, M.H.; Yan, S.Y.; Chen, G.Q.; Huang, Y. Microtubule-associated protein 1 light chain 3 interacts with and contributes to growth inhibiting effect of PML. PLoS ONE 2014, 9, e113089. [Google Scholar] [CrossRef] [PubMed]
- Harhouri, K.; Navarro, C.; Depetris, D.; Mattei, M.G.; Nissan, X.; Cau, P.; De Sandre-Giovannoli, A.; Lévy, N. MG132-induced progerin clearance is mediated by autophagy activation and splicing regulation. EMBO Mol. Med. 2017, 9, 1294–1313. [Google Scholar] [CrossRef] [PubMed]
- Zannini, L.; Buscemi, G.; Fontanella, E.; Lisanti, S.; Delia, D. REGgamma/PA28gamma proteasome activator interacts with PML and Chk2 and affects PML nuclear bodies number. Cell Cycle 2009, 8, 2399–2407. [Google Scholar] [CrossRef] [PubMed]
- Park, H.J.; Kim, B.C.; Kim, S.J.; Choi, K.S. Role of MAP kinases and their cross-talk in TGF-beta1-induced apoptosis in FaO rat hepatoma cell line. Hepatology 2002, 35, 1360–1371. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, T.G.; Möller, A.; Sirma, H.; Zentgraf, H.; Taya, Y.; Dröge, W.; Will, H.; Schmitz, M.L. Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nat. Cell Biol. 2002, 4, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Banumathy, G.; Somaiah, N.; Zhang, R.; Tang, Y.; Hoffmann, J.; Andrake, M.; Ceulemans, H.; Schultz, D.; Marmorstein, R.; Adams, P.D. Human UBN1 is an ortholog of yeast Hpc2p and has an essential role in the HIRA/ASF1a chromatin-remodeling pathway in senescent cells. Mol. Cell. Biol. 2009, 29, 758–770. [Google Scholar] [CrossRef] [PubMed]
- Chakarova, C.F.; Papaioannou, M.G.; Khanna, H.; Lopez, I.; Waseem, N.; Shah, A.; Theis, T.; Friedman, J.; Maubaret, C.; Bujakowska, K.; et al. Mutations in TOPORS cause autosomal dominant retinitis pigmentosa with perivascular retinal pigment epithelium atrophy. Am. J. Hum. Genet. 2007, 81, 1098–1103. [Google Scholar] [CrossRef]
- Ji, L.; Huo, X.; Zhang, Y.; Yan, Z.; Wang, Q.; Wen, B. TOPORS, a tumor suppressor protein, contributes to the maintenance of higher-order chromatin architecture. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194518. [Google Scholar] [CrossRef]
- Hariharasudhan, G.; Jeong, S.Y.; Kim, M.J.; Jung, S.M.; Seo, G.; Moon, J.R.; Lee, S.; Chang, I.Y.; Kee, Y.; You, H.J.; et al. TOPORS-mediated RAD51 SUMOylation facilitates homologous recombination repair. Nucleic Acids Res. 2022, 50, 1501–1516. [Google Scholar] [CrossRef]
- Salsman, J.; Pinder, J.; Tse, B.; Corkery, D.; Dellaire, G. The translation initiation factor 3 subunit eIF3K interacts with PML and associates with PML nuclear bodies. Exp. Cell Res. 2013, 319, 2554–2565. [Google Scholar] [CrossRef]
- Kunapuli, P.; Kasyapa, C.S.; Chin, S.F.; Caldas, C.; Cowell, J.K. ZNF198, a zinc finger protein rearranged in myeloproliferative disease, localizes to the PML nuclear bodies and interacts with SUMO-1 and PML. Exp. Cell Res. 2006, 312, 3739–3751. [Google Scholar] [CrossRef] [PubMed]
- Shima, Y.; Shima, T.; Chiba, T.; Irimura, T.; Pandolfi, P.P.; Kitabayashi, I. PML activates transcription by protecting HIPK2 and p300 from SCFFbx3-mediated degradation. Mol. Cell. Biol. 2008, 28, 7126–7138. [Google Scholar] [CrossRef]
- Milovic-Holm, K.; Krieghoff, E.; Jensen, K.; Will, H.; Hofmann, T.G. FLASH links the CD95 signaling pathway to the cell nucleus and nuclear bodies. EMBO J. 2007, 26, 391–401. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Chen, Y.; Yan, K.; Shao, Y.; Zhang, Q.C.; Lin, Y.; Xi, Q. Recruitment of TRIM33 to cell-context specific PML nuclear bodies regulates nodal signaling in mESCs. EMBO J. 2023, 42, e112058. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.Q.; Nguyen, A.; Cao, Y.; Chang, A.C.; Reddel, R.R. HP1-mediated formation of alternative lengthening of telomeres-associated PML bodies requires HIRA but not ASF1a. PLoS ONE 2011, 6, e17036. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.; Block, S.; Scribano, C.M.; Thiry, R.; Esbona, K.; Audhya, A.; Weaver, B.A. Mad1 destabilizes p53 by preventing PML from sequestering MDM2. Nat. Commun. 2019, 10, 1540. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Seo, T.; Kim, H.; Choe, J. Sumoylation of the novel protein hRIP{beta} is involved in replication protein A deposition in PML nuclear bodies. Mol. Cell. Biol. 2005, 25, 8202–8214. [Google Scholar] [CrossRef]
- Sohn, S.Y.; Hearing, P. Adenoviral strategies to overcome innate cellular responses to infection. FEBS Lett. 2019, 593, 3484–3495. [Google Scholar] [CrossRef] [PubMed]
- Barr, S.M.; Leung, C.G.; Chang, E.E.; Cimprich, K.A. ATR kinase activity regulates the intranuclear translocation of ATR and RPA following ionizing radiation. Curr. Biol. 2003, 13, 1047–1051. [Google Scholar] [CrossRef]
- Bloch, D.B.; Chiche, J.D.; Orth, D.; de la Monte, S.M.; Rosenzweig, A.; Bloch, K.D. Structural and functional heterogeneity of nuclear bodies. Mol. Cell. Biol. 1999, 19, 4423–4430. [Google Scholar] [CrossRef]
- Bloch, D.B.; de la Monte, S.M.; Guigaouri, P.; Filippov, A.; Bloch, K.D. Identification and characterization of a leukocyte-specific component of the nuclear body. J. Biol. Chem. 1996, 271, 29198–29204. [Google Scholar] [CrossRef] [PubMed]
- Bloch, D.B.; Nakajima, A.; Gulick, T.; Chiche, J.D.; Orth, D.; de La Monte, S.M.; Bloch, K.D. Sp110 localizes to the PML-Sp100 nuclear body and may function as a nuclear hormone receptor transcriptional coactivator. Mol. Cell. Biol. 2000, 20, 6138–6146. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Chu, J.; Yucer, N.; Leng, M.; Wang, S.Y.; Chen, B.P.; Hittelman, W.N.; Wang, Y. RING finger and WD repeat domain 3 (RFWD3) associates with replication protein A (RPA) and facilitates RPA-mediated DNA damage response. J. Biol. Chem. 2011, 286, 22314–22322. [Google Scholar] [CrossRef] [PubMed]
- Lunardi, A.; Chiacchiera, F.; D’Este, E.; Carotti, M.; Dal Ferro, M.; Di Minin, G.; Del Sal, G.; Collavin, L. The evolutionary conserved gene C16orf35 encodes a nucleo-cytoplasmic protein that interacts with p73. Biochem. Biophys. Res. Commun. 2009, 388, 428–433. [Google Scholar] [CrossRef] [PubMed]
- Deiss, K.; Lockwood, N.; Howell, M.; Segeren, H.A.; Saunders, R.E.; Chakravarty, P.; Soliman, T.N.; Martini, S.; Rocha, N.; Semple, R.; et al. A genome-wide RNAi screen identifies the SMC5/6 complex as a non-redundant regulator of a Topo2a-dependent G2 arrest. Nucleic Acids Res. 2019, 47, 2906–2921. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; He, M.; Gong, R.; Wang, Z.; Lu, L.; Peng, S.; Duan, Z.; Feng, Y.; Liu, Y.; Gao, B. Forkhead O Transcription Factor 4 Restricts HBV Covalently Closed Circular DNA Transcription and HBV Replication through Genetic Downregulation of Hepatocyte Nuclear Factor 4 Alpha and Epigenetic Suppression of Covalently Closed Circular DNA via Interacting with Promyelocytic Leukemia Protein. J. Virol. 2022, 96, e0054622. [Google Scholar] [CrossRef] [PubMed]
- Borden, K.L.; Campbelldwyer, E.J.; Carlile, G.W.; Djavani, M.; Salvato, M.S. Two RING finger proteins, the oncoprotein PML and the arenavirus Z protein, colocalize with the nuclear fraction of the ribosomal P proteins. J. Virol. 1998, 72, 3819–3826. [Google Scholar] [CrossRef]
- Bernassola, F.; Salomoni, P.; Oberst, A.; Di Como, C.J.; Pagano, M.; Melino, G.; Pandolfi, P.P. Ubiquitin-dependent degradation of p73 is inhibited by PML. J. Exp. Med. 2004, 199, 1545–1557. [Google Scholar] [CrossRef]
- Lee, Y.R.; Yuan, W.C.; Ho, H.C.; Chen, C.H.; Shih, H.M.; Chen, R.H. The Cullin 3 substrate adaptor KLHL20 mediates DAPK ubiquitination to control interferon responses. EMBO J. 2010, 29, 1748–1761. [Google Scholar] [CrossRef]
- Martin, N.; Benhamed, M.; Nacerddine, K.; Demarque, M.D.; van Lohuizen, M.; Dejean, A.; Bischof, O. Physical and functional interaction between PML and TBX2 in the establishment of cellular senescence. EMBO J. 2012, 31, 95–109. [Google Scholar] [CrossRef]
- Oh, W.; Ghim, J.; Lee, E.W.; Yang, M.R.; Kim, E.T.; Ahn, J.H.; Song, J. PML-IV functions as a negative regulator of telomerase by interacting with TERT. J. Cell Sci. 2009, 122, 2613–2622. [Google Scholar] [CrossRef] [PubMed]
- Salomoni, P.; Bernardi, R.; Bergmann, S.; Changou, A.; Tuttle, S.; Pandolfi, P.P. The promyelocytic leukemia protein PML regulates c-Jun function in response to DNA damage. Blood 2005, 105, 3686–3690. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, J.; Zou, W.X.; Chang, K.S. Inhibition of Sp1 functions by its sequestration into PML nuclear bodies. PLoS ONE 2014, 9, e94450. [Google Scholar] [CrossRef] [PubMed]
- Kordon, M.M.; Szczurek, A.; Berniak, K.; Szelest, O.; Solarczyk, K.; Tworzydło, M.; Wachsmann-Hogiu, S.; Vaahtokari, A.; Cremer, C.; Pederson, T.; et al. PML-like subnuclear bodies, containing XRCC1, juxtaposed to DNA replication-based single-strand breaks. FASEB J. 2019, 33, 2301–2313. [Google Scholar] [CrossRef]
- Wu, J.; Zhou, L.Q.; Yu, W.; Zhao, Z.G.; Xie, X.M.; Wang, W.T.; Xiong, J.; Li, M.; Xue, Z.; Wang, X.; et al. PML4 facilitates erythroid differentiation by enhancing the transcriptional activity of GATA-1. Blood 2014, 123, 261–270. [Google Scholar] [CrossRef]
- Rao, V.A.; Fan, A.M.; Meng, L.; Doe, C.F.; North, P.S.; Hickson, I.D.; Pommier, Y. Phosphorylation of BLM, dissociation from topoisomerase IIIalpha, and colocalization with gamma-H2AX after topoisomerase I-induced replication damage. Mol. Cell. Biol. 2005, 25, 8925–8937. [Google Scholar] [CrossRef]
- Carbone, R.; Pearson, M.; Minucci, S.; Pelicci, P.G. PML NBs associate with the hMre11 complex and p53 at sites of irradiation induced DNA damage. Oncogene 2002, 21, 1633–1640. [Google Scholar] [CrossRef]
- Gibb, S.L.; Boston-Howes, W.; Lavina, Z.S.; Gustincich, S.; Brown, R.H., Jr.; Pasinelli, P.; Trotti, D. A caspase-3-cleaved fragment of the glial glutamate transporter EAAT2 is sumoylated and targeted to promyelocytic leukemia nuclear bodies in mutant SOD1-linked amyotrophic lateral sclerosis. J. Biol. Chem. 2007, 282, 32480–32490. [Google Scholar] [CrossRef]
- Alsheich-Bartok, O.; Haupt, S.; Alkalay-Snir, I.; Saito, S.; Appella, E.; Haupt, Y. PML enhances the regulation of p53 by CK1 in response to DNA damage. Oncogene 2008, 27, 3653–3661. [Google Scholar] [CrossRef]
- Gialitakis, M.; Arampatzi, P.; Makatounakis, T.; Papamatheakis, J. Gamma interferon-dependent transcriptional memory via relocalization of a gene locus to PML nuclear bodies. Mol. Cell. Biol. 2010, 30, 2046–2056. [Google Scholar] [CrossRef]
- Kurki, S.; Latonen, L.; Laiho, M. Cellular stress and DNA damage invoke temporally distinct Mdm2, p53 and PML complexes and damage-specific nuclear relocalization. J. Cell Sci. 2003, 116, 3917–3925. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.A.; Sun, Y.; Song, J.; Chen, Y.; Krontiris, T.G.; Durrin, L.K. SUMO conjugation to the matrix attachment region-binding protein, special AT-rich sequence-binding protein-1 (SATB1), targets SATB1 to promyelocytic nuclear bodies where it undergoes caspase cleavage. J. Biol. Chem. 2008, 283, 18124–18134. [Google Scholar] [CrossRef] [PubMed]
- Sapetschnig, A.; Rischitor, G.; Braun, H.; Doll, A.; Schergaut, M.; Melchior, F.; Suske, G. Transcription factor Sp3 is silenced through SUMO modification by PIAS1. EMBO J. 2002, 21, 5206–5215. [Google Scholar] [CrossRef] [PubMed]
- Ross, S.; Best, J.L.; Zon, L.I.; Gill, G. SUMO-1 modification represses Sp3 transcriptional activation and modulates its subnuclear localization. Mol. Cell 2002, 10, 831–842. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.J.; Meloni, A.R.; Nevins, J.R. The Rb-related p130 protein controls telomere lengthening through an interaction with a Rad50-interacting protein, RINT-1. Mol. Cell 2006, 22, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, Y.I.; Kitamura, T.; Kruse, J.P.; Raum, J.C.; Stein, R.; Gu, W.; Accili, D. FoxO1 protects against pancreatic beta cell failure through NeuroD and MafA induction. Cell Metab. 2005, 2, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Voisset, E.; Moravcsik, E.; Stratford, E.W.; Jaye, A.; Palgrave, C.J.; Hills, R.K.; Salomoni, P.; Kogan, S.C.; Solomon, E.; Grimwade, D. Pml nuclear body disruption cooperates in APL pathogenesis and impairs DNA damage repair pathways in mice. Blood 2018, 131, 636–648. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Aster, J.C.; Blacklow, S.C.; Lake, R.; Artavanis-Tsakonas, S.; Griffin, J.D. MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat. Genet. 2000, 26, 484–489. [Google Scholar] [CrossRef]
- Takahashi, H.; Hatakeyama, S.; Saitoh, H.; Nakayama, K.I. Noncovalent SUMO-1 binding activity of thymine DNA glycosylase (TDG) is required for its SUMO-1 modification and colocalization with the promyelocytic leukemia protein. J. Biol. Chem. 2005, 280, 5611–5621. [Google Scholar] [CrossRef]
- Blander, G.; Zalle, N.; Daniely, Y.; Taplick, J.; Gray, M.D.; Oren, M. DNA damage-induced translocation of the Werner helicase is regulated by acetylation. J. Biol. Chem. 2002, 277, 50934–50940. [Google Scholar] [CrossRef]
- El-Asmi, F.; El-Mchichi, B.; Maroui, M.A.; Dianoux, L.; Chelbi-Alix, M.K. TGF-β induces PML SUMOylation, degradation and PML nuclear body disruption. Cytokine 2019, 120, 264–272. [Google Scholar] [CrossRef] [PubMed]
- Osterwald, S.; Deeg, K.I.; Chung, I.; Parisotto, D.; Wörz, S.; Rohr, K.; Erfle, H.; Rippe, K. PML induces compaction, TRF2 depletion and DNA damage signaling at telomeres and promotes their alternative lengthening. J. Cell Sci. 2015, 128, 1887–1900. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Park, B.; Cho, S.; Lee, S.; Kim, Y.; Lee, S.O.; Cho, K.; Lee, S.; Jin, B.S.; Ahn, J.H.; et al. Promyelocytic leukemia is a direct inhibitor of SAPK2/p38 mitogen-activated protein kinase. J. Biol. Chem. 2004, 279, 40994–41003. [Google Scholar] [CrossRef] [PubMed]
- Merkl, P.E.; Orzalli, M.H.; Knipe, D.M. Mechanisms of Host IFI16, PML, and Daxx Protein Restriction of Herpes Simplex Virus 1 Replication. J. Virol. 2018, 92, e00057-18. [Google Scholar] [CrossRef] [PubMed]
- Sasai, N.; Saitoh, N.; Saitoh, H.; Nakao, M. The transcriptional cofactor MCAF1/ATF7IP is involved in histone gene expression and cellular senescence. PLoS ONE 2013, 8, e68478. [Google Scholar] [CrossRef]
- Sun, H.; Liu, Y.; Hunter, T. Multiple Arkadia/RNF111 structures coordinate its Polycomb body association and transcriptional control. Mol. Cell. Biol. 2014, 34, 2981–2995. [Google Scholar] [CrossRef]
- Schmitz, M.L.; Grishina, I. Regulation of the tumor suppressor PML by sequential post-translational modifications. Front. Oncol. 2012, 2, 204. [Google Scholar] [CrossRef][Green Version]
- Rauth, S.; Karmakar, S.; Shah, A.; Seshacharyulu, P.; Nimmakayala, R.K.; Ganguly, K.; Bhatia, R.; Muniyan, S.; Kumar, S.; Dutta, S.; et al. SUMO Modification of PAF1/PD2 Enables PML Interaction and Promotes Radiation Resistance in Pancreatic Ductal Adenocarcinoma. Mol. Cell. Biol. 2021, 41, e0013521. [Google Scholar] [CrossRef]
- Xu, Z.X.; Timanova-Atanasova, A.; Zhao, R.X.; Chang, K.S. PML colocalizes with and stabilizes the DNA damage response protein TopBP1. Mol. Cell. Biol. 2003, 23, 4247–4256. [Google Scholar] [CrossRef]
- Rokudai, S.; Laptenko, O.; Arnal, S.M.; Taya, Y.; Kitabayashi, I.; Prives, C. MOZ increases p53 acetylation and premature senescence through its complex formation with PML. Proc. Natl. Acad. Sci. USA 2013, 110, 3895–3900. [Google Scholar] [CrossRef]
- Guan, D.; Factor, D.; Liu, Y.; Wang, Z.; Kao, H.Y. The epigenetic regulator UHRF1 promotes ubiquitination-mediated degradation of the tumor-suppressor protein promyelocytic leukemia protein. Oncogene 2013, 32, 3819–3828. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Li, J.; Dong, F.N.; Dong, J.T. Characterization of nuclear localization and SUMOylation of the ATBF1 transcription factor in epithelial cells. PLoS ONE 2014, 9, e92746. [Google Scholar] [CrossRef] [PubMed]
- Georges, A.; Benayoun, B.A.; Marongiu, M.; Dipietromaria, A.; L’Hôte, D.; Todeschini, A.L.; Auer, J.; Crisponi, L.; Veitia, R.A. SUMOylation of the Forkhead transcription factor FOXL2 promotes its stabilization/activation through transient recruitment to PML bodies. PLoS ONE 2011, 6, e25463. [Google Scholar] [CrossRef] [PubMed]
- Shire, K.; Wong, A.I.; Tatham, M.H.; Anderson, O.F.; Ripsman, D.; Gulstene, S.; Moffat, J.; Hay, R.T.; Frappier, L. Identification of RNF168 as a PML nuclear body regulator. J. Cell Sci. 2016, 129, 580–591. [Google Scholar] [CrossRef] [PubMed]
- Imrichova, T.; Hubackova, S.; Kucerova, A.; Kosla, J.; Bartek, J.; Hodny, Z.; Vasicova, P. Dynamic PML protein nucleolar associations with persistent DNA damage lesions in response to nucleolar stress and senescence-inducing stimuli. Aging 2019, 11, 7206–7235. [Google Scholar] [CrossRef] [PubMed]
- Lång, A.; Eriksson, J.; Schink, K.O.; Lång, E.; Blicher, P.; Połeć, A.; Brech, A.; Dalhus, B.; Bøe, S.O. Visualization of PML nuclear import complexes reveals FG-repeat nucleoporins at cargo retrieval sites. Nucleus 2017, 8, 404–420. [Google Scholar] [CrossRef] [PubMed]
- Lallemand-Breitenbach, V.; Zhu, J.; Puvion, F.; Koken, M.; Honoré, N.; Doubeikovsky, A.; Duprez, E.; Pandolfi, P.P.; Puvion, E.; Freemont, P.; et al. Role of promyelocytic leukemia (PML) sumolation in nuclear body formation, 11S proteasome recruitment, and As2O3-induced PML or PML/retinoic acid receptor alpha degradation. J. Exp. Med. 2001, 193, 1361–1371. [Google Scholar] [CrossRef]
- Tang, J.; Xie, W.; Yang, X. Association of caspase-2 with the promyelocytic leukemia protein nuclear bodies. Cancer Biol. Ther. 2005, 4, 645–649. [Google Scholar] [CrossRef]
- Scheper, G.C.; Parra, J.L.; Wilson, M.; Van Kollenburg, B.; Vertegaal, A.C.; Han, Z.G.; Proud, C.G. The N and C termini of the splice variants of the human mitogen-activated protein kinase-interacting kinase Mnk2 determine activity and localization. Mol. Cell. Biol. 2003, 23, 5692–5705. [Google Scholar] [CrossRef]
- Sabò, A.; Lusic, M.; Cereseto, A.; Giacca, M. Acetylation of conserved lysines in the catalytic core of cyclin-dependent kinase 9 inhibits kinase activity and regulates transcription. Mol. Cell. Biol. 2008, 28, 2201–2212. [Google Scholar] [CrossRef]
- Tomasini, R.; Samir, A.A.; Carrier, A.; Isnardon, D.; Cecchinelli, B.; Soddu, S.; Malissen, B.; Dagorn, J.C.; Iovanna, J.L.; Dusetti, N.J. TP53INP1s and homeodomain-interacting protein kinase-2 (HIPK2) are partners in regulating p53 activity. J. Biol. Chem. 2003, 278, 37722–37729. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Kim, D.U.; Kehrl, J.H. RGS14 is a centrosomal and nuclear cytoplasmic shuttling protein that traffics to promyelocytic leukemia nuclear bodies following heat shock. J. Biol. Chem. 2005, 280, 805–814. [Google Scholar] [CrossRef]
- Messaoudi, L.; Yang, Y.G.; Kinomura, A.; Stavreva, D.A.; Yan, G.; Bortolin-Cavaillé, M.L.; Arakawa, H.; Buerstedde, J.M.; Hainaut, P.; Cavaillé, J.; et al. Subcellular distribution of human RDM1 protein isoforms and their nucleolar accumulation in response to heat shock and proteotoxic stress. Nucleic Acids Res. 2007, 35, 6571–6587. [Google Scholar] [CrossRef] [PubMed]
- Gongora, C.; David, G.; Pintard, L.; Tissot, C.; Hua, T.D.; Dejean, A.; Mechti, N. Molecular cloning of a new interferon-induced PML nuclear body-associated protein. J. Biol. Chem. 1997, 272, 19457–19463. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Akira, S.; Reed, J.C. ZIP kinase triggers apoptosis from nuclear PML oncogenic domains. Mol. Cell. Biol. 2003, 23, 6174–6186. [Google Scholar] [CrossRef]
- Sharma, P.; Murillas, R.; Zhang, H.; Kuehn, M.R. N4BP1 is a newly identified nucleolar protein that undergoes SUMO-regulated polyubiquitylation and proteasomal turnover at promyelocytic leukemia nuclear bodies. J. Cell Sci. 2010, 123, 1227–1234. [Google Scholar] [CrossRef]
- Oughtred, R.; Rust, J.; Chang, C.; Breitkreutz, B.J.; Stark, C.; Willems, A.; Boucher, L.; Leung, G.; Kolas, N.; Zhang, F.; et al. The BioGRID database: A comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 2021, 30, 187–200. [Google Scholar] [CrossRef]
- Xu, P.; Roizman, B. The SP100 component of ND10 enhances accumulation of PML and suppresses replication and the assembly of HSV replication compartments. Proc. Natl. Acad. Sci. USA 2017, 114, E3823–E3829. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Wang, P.; Benhenda, S.; Wu, H.; Lallemand-Breitenbach, V.; Zhen, T.; Jollivet, F.; Peres, L.; Li, Y.; Chen, S.J.; Chen, Z.; et al. RING tetramerization is required for nuclear body biogenesis and PML sumoylation. Nat. Commun. 2018, 9, 1277. [Google Scholar] [CrossRef]
- Lang, M.; Jegou, T.; Chung, I.; Richter, K.; Munch, S.; Udvarhelyi, A.; Cremer, C.; Hemmerich, P.; Engelhardt, J.; Hell, S.W.; et al. Three-dimensional organization of promyelocytic leukemia nuclear bodies. J. Cell Sci. 2010, 123, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Liebl, M.C.; Hofmann, T.G. Regulating the p53 Tumor Suppressor Network at PML Biomolecular Condensates. Cancers 2022, 14, 4549. [Google Scholar] [CrossRef] [PubMed]
- Willms, A.; Schupp, H.; Poelker, M.; Adawy, A.; Debus, J.F.; Hartwig, T.; Krichel, T.; Fritsch, J.; Singh, S.; Walczak, H.; et al. TRAIL-receptor 2-a novel negative regulator of p53. Cell Death Dis. 2021, 12, 757. [Google Scholar] [CrossRef] [PubMed]
- Kwek, S.S.; Derry, J.; Tyner, A.L.; Shen, Z.; Gudkov, A.V. Functional analysis and intracellular localization of p53 modified by SUMO-1. Oncogene 2001, 20, 2587–2599. [Google Scholar] [CrossRef] [PubMed]
- Sha, Z.; Blyszcz, T.; Gonzalez-Prieto, R.; Vertegaal, A.C.O.; Goldberg, A.L. Inhibiting ubiquitination causes an accumulation of SUMOylated newly synthesized nuclear proteins at PML bodies. J. Biol. Chem. 2019, 294, 15218–15234. [Google Scholar] [CrossRef] [PubMed]
- Erker, Y.; Neyret-Kahn, H.; Seeler, J.S.; Dejean, A.; Atfi, A.; Levy, L. Arkadia, a novel SUMO-targeted ubiquitin ligase involved in PML degradation. Mol. Cell. Biol. 2013, 33, 2163–2177. [Google Scholar] [CrossRef] [PubMed]
- Bregnard, T.; Ahmed, A.; Semenova, I.V.; Weller, S.K.; Bezsonova, I. The B-box1 domain of PML mediates SUMO E2-E3 complex formation through an atypical interaction with UBC9. Biophys. Chem. 2022, 287, 106827. [Google Scholar] [CrossRef] [PubMed]
- Matt, S.; Hofmann, T.G. Crosstalk between p53 modifiers at PML bodies. Mol. Cell. Oncol. 2018, 5, e1074335. [Google Scholar] [CrossRef]
- Ye, M.; Tang, Y.; Tang, S.; Liu, J.; Wu, K.; Yao, S.; Sun, Y.; Zhou, L.; Deng, T.; Chen, Y.; et al. STIP is a critical nuclear scaffolding protein linking USP7 to p53-Mdm2 pathway regulation. Oncotarget 2015, 6, 34718–34731. [Google Scholar] [CrossRef]
- Nagasaka, M.; Miyajima, C.; Aoki, H.; Aoyama, M.; Morishita, D.; Inoue, Y.; Hayashi, H. Insights into Regulators of p53 Acetylation. Cells 2022, 11, 3825. [Google Scholar] [CrossRef]
- Moller, A.; Sirma, H.; Hofmann, T.G.; Staege, H.; Gresko, E.; Ludi, K.S.; Klimczak, E.; Droge, W.; Will, H.; Schmitz, M.L. Sp100 is important for the stimulatory effect of homeodomain-interacting protein kinase-2 on p53-dependent gene expression. Oncogene 2003, 22, 8731–8737. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mohammed, A.S.; Uversky, V.N. Intrinsic Disorder as a Natural Preservative: High Levels of Intrinsic Disorder in Proteins Found in the 2600-Year-Old Human Brain. Biology 2022, 11, 1704. [Google Scholar] [CrossRef] [PubMed]
- Eskiw, C.H.; Dellaire, G.; Bazett-Jones, D.P. Chromatin contributes to structural integrity of promyelocytic leukemia bodies through a SUMO-1-independent mechanism. J. Biol. Chem. 2004, 279, 9577–9585. [Google Scholar] [CrossRef] [PubMed]
- Torok, D.; Ching, R.W.; Bazett-Jones, D.P. PML nuclear bodies as sites of epigenetic regulation. Front. Biosci. 2009, 14, 1325–1336. [Google Scholar] [CrossRef]
- Delbarre, E.; Ivanauskiene, K.; Spirkoski, J.; Shah, A.; Vekterud, K.; Moskaug, J.O.; Boe, S.O.; Wong, L.H.; Kuntziger, T.; Collas, P. PML protein organizes heterochromatin domains where it regulates histone H3.3 deposition by ATRX/DAXX. Genome Res. 2017, 27, 913–921. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).