When Dad’s Stress Gets under Kid’s Skin—Impacts of Stress on Germline Cargo and Embryonic Development
Abstract
1. Epigenetic Inheritance
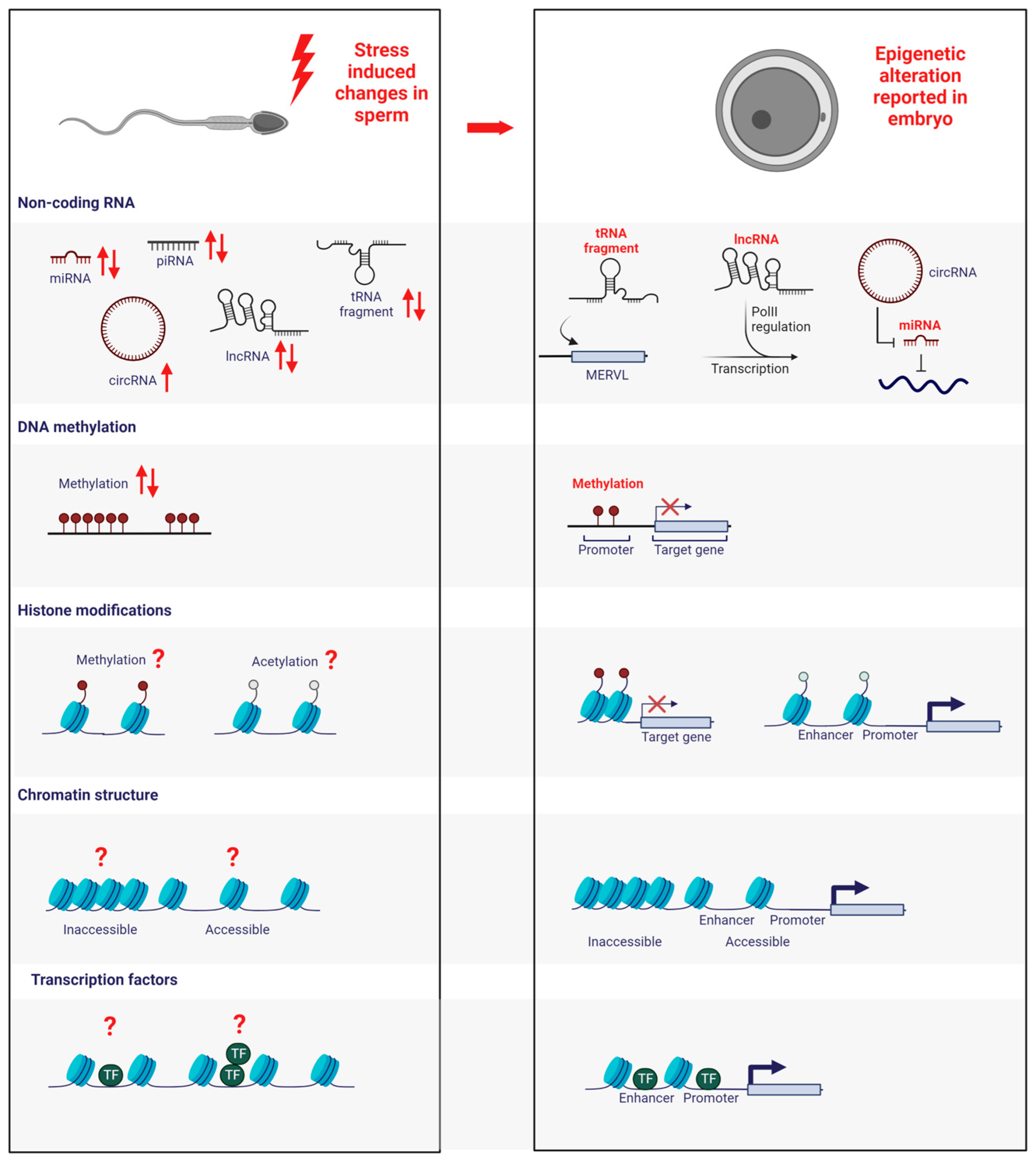
2. Vertical Information Carriers of Epigenetic Inheritance in the Paternal Germline?
2.1. Non-Coding RNAs
2.2. DNA Methylation
2.3. Histone Modifications
2.4. Chromatin Structure
2.5. Transcription Factors
3. Epigenetic Regulators in the Early Embryo—How Do Stress-Induced Changes in the Paternal Epigenome Impact Them?
3.1. Non-Coding RNA
3.2. DNA Methylation
3.3. Histone Modifications
3.4. Chromatin Structure
3.5. Transcription Factors
4. Stress Sensitivity as a Converging Effect of Distinct Types of Exposures
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Santomauro, D.F.; Mantilla Herrera, A.M.; Shadid, J.; Zheng, P.; Ashbaugh, C.; Pigott, D.M.; Abbafati, C.; Adolph, C.; Amlag, J.O.; Aravkin, A.Y.; et al. Global prevalence and burden of depressive and anxiety disorders in 204 countries and territories in 2020 due to the COVID-19 pandemic. Lancet 2021, 398, 1700–1712. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, C. Genetics in psychiatry: Methods, clinical applications and future perspectives. Psychiatry Clin. Neurosci. Rep. 2022, 1, e6. [Google Scholar] [CrossRef]
- Yehuda, R.; Bierer, L.M. Transgenerational Transmission of Cortisol and PTSD Risk. Prog. Brain Res. 2007, 167, 121–135. [Google Scholar] [CrossRef]
- Yehuda, R.; Daskalakis, N.P.; Lehrner, A.; Desarnaud, F.; Bader, H.N.; Makotkine, I.; Flory, J.D.; Bierer, L.M.; Meaney, M.J. Influences of Maternal and Paternal PTSD on Epigenetic Regulation of the Glucocorticoid Receptor Gene in Holocaust Survivor Offspring. Am. J. Psychiatry 2014, 171, 872–880. [Google Scholar] [CrossRef] [PubMed]
- Bohacek, J.; Mansuy, I.M. Molecular insights into transgenerational non-genetic inheritance of acquired behaviours. Nat. Rev. Genet. 2015, 16, 641–652. [Google Scholar] [CrossRef] [PubMed]
- Fraser, R.; Lin, C.-J. Epigenetic reprogramming of the zygote in mice and men: On your marks, get set, go! Reproduction 2016, 152, R211–R222. [Google Scholar] [CrossRef] [PubMed]
- Morgan, H.D.; Sutherland, H.G.E.; Martin, D.I.K.; Whitelaw, E. Epigenetic inheritance at the agouti locus in the mouse. Nat. Genet. 1999, 23, 314–318. [Google Scholar] [CrossRef]
- Rakyan, V.K.; Chong, S.; Champ, M.E.; Cuthbert, P.C.; Morgan, H.D.; Luu, K.V.K.; Whitelaw, E. Transgenerational inheritance of epigenetic states at the murine AxinFu allele occurs after maternal and paternal transmission. Proc. Natl. Acad. Sci. USA 2003, 100, 2538–2543. [Google Scholar] [CrossRef]
- Rassoulzadegan, M.; Grandjean, V.; Gounon, P.; Vincent, S.; Gillot, I.; Cuzin, F. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature 2006, 441, 469–474. [Google Scholar] [CrossRef]
- Krawetz, S.A. Paternal contribution: New insights and future challenges. Nat. Rev. Genet. 2005, 6, 633–642. [Google Scholar] [CrossRef]
- Ostermeier, G.C.; Miller, D.; Huntriss, J.D.; Diamond, M.P.; Krawetz, S.A. Delivering spermatozoan RNA to the oocyte. Nature 2004, 429, 154. [Google Scholar] [CrossRef] [PubMed]
- Clarke, H.J.; Vieux, K.-F. Epigenetic inheritance through the female germ-line: The known, the unknown, and the possible. Semin. Cell Dev. Biol. 2015, 43, 106–116. [Google Scholar] [CrossRef]
- Stäubli, A.; Peters, A.H. Mechanisms of maternal intergenerational epigenetic inheritance. Curr. Opin. Genet. Dev. 2021, 67, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Anway, M.D.; Cupp, A.S.; Uzumcu, M.; Skinner, M.K. Epigenetic Transgenerational Actions of Endocrine Disruptors and Male Fertility. Science 2005, 308, 1466–1469. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, E.E.; Skinner, M.K. Environmentally induced epigenetic transgenerational inheritance of disease susceptibility. Transl. Res. 2015, 165, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Radford, E.J. Exploring the extent and scope of epigenetic inheritance. Nat. Rev. Endocrinol. 2018, 14, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Sirard, M.-A. Epigenetic inheritance of acquired traits through DNA methylation. Anim. Front. 2021, 11, 19–27. [Google Scholar] [CrossRef]
- Mattick, J.S.; Makunin, I.V. Non-Coding RNA. Hum. Mol. Genet. 2006, 15, R17–R29. [Google Scholar] [CrossRef]
- Takemata, N.; Ohta, K. Role of non-coding RNA transcription around gene regulatory elements in transcription factor recruitment. RNA Biol. 2017, 14, 1–5. [Google Scholar] [CrossRef]
- Kotaja, N. MicroRNAs and spermatogenesis. Fertil. Steril. 2014, 101, 1552–1562. [Google Scholar] [CrossRef]
- Felden, B.; Paillard, L. When eukaryotes and prokaryotes look alike: The case of regulatory RNAs. FEMS Microbiol. Rev. 2017, 41, 624–639. [Google Scholar] [CrossRef] [PubMed]
- Xie, R.; Lin, X.; Du, T.; Xu, K.; Shen, H.; Wei, F.; Hao, W.; Lin, T.; Lin, X.; Qin, Y.; et al. Targeted Disruption of miR-17-92 Impairs Mouse Spermatogenesis by Activating mTOR Signaling Pathway. Medicine 2016, 95, e2713. [Google Scholar] [CrossRef] [PubMed]
- Chuma, S.; Nakano, T. piRNA and spermatogenesis in mice. Philos. Trans. R. Soc. B Biol. Sci. 2013, 368, 20110338. [Google Scholar] [CrossRef] [PubMed]
- Goh, W.S.S.; Falciatori, I.; Tam, O.H.; Burgess, R.; Meikar, O.; Kotaja, N.; Hammell, M.; Hannon, G.J. piRNA-directed cleavage of meiotic transcripts regulates spermatogenesis. Genes Dev. 2015, 29, 1032–1044. [Google Scholar] [CrossRef] [PubMed]
- Gou, L.-T.; Dai, P.; Yang, J.-H.; Xue, Y.; Hu, Y.-P.; Zhou, Y.; Kang, J.-Y.; Wang, X.; Li, H.; Hua, M.-M.; et al. Pachytene piRNAs instruct massive mRNA elimination during late spermiogenesis. Cell Res. 2014, 24, 680–700. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Shi, J.; Zhang, Y.; Zhang, H.; Liao, S.; Li, W.; Lei, L.; Han, C.; Ning, L.; Cao, Y.; et al. A novel class of tRNA-derived small RNAs extremely enriched in mature mouse sperm. Cell Res. 2012, 22, 1609–1612. [Google Scholar] [CrossRef] [PubMed]
- Sharma, U.; Conine, C.C.; Shea, J.M.; Boskovic, A.; Derr, A.G.; Bing, X.Y.; Belleannee, C.; Kucukural, A.; Serra, R.W.; Sun, F.; et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 2016, 351, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Yin, Q.; Yang, C.-H.; Strelkova, O.S.; Wu, J.; Sun, Y.; Gopalan, S.; Yang, L.; Dekker, J.; Fazzio, T.G.; Li, X.Z.; et al. Revisiting chromatin packaging in mouse sperm. bioRxiv 2023. bioRxiv:2022.12.26.521943. [Google Scholar]
- Gustafsson, H.T.; Galan, C.; Yu, T.; Upton, H.E.; Ferguson, L.; Kaymak, E.; Weng, Z.; Collins, K.; Rando, O.J. Deep sequencing of yeast and mouse tRNAs and tRNA fragments using OTTR. bioRxiv 2022. bioRxiv:2022.02.04.479139. [Google Scholar]
- Magee, R.; Rigoutsos, I. On the expanding roles of tRNA fragments in modulating cell behavior. Nucleic Acids Res. 2020, 48, 9433–9448. [Google Scholar] [CrossRef]
- Kigami, D.; Minami, N.; Takayama, H.; Imai, H. MuERV-L Is One of the Earliest Transcribed Genes in Mouse One-Cell Embryos1. Biol. Reprod. 2003, 68, 651–654. [Google Scholar] [CrossRef] [PubMed]
- Macfarlan, T.S.; Gifford, W.D.; Driscoll, S.; Lettieri, K.; Rowe, H.M.; Bonanomi, D.; Firth, A.; Singer, O.; Trono, D.; Pfaff, S.L. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 2012, 487, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Svoboda, P.; Stein, P.; Anger, M.; Bernstein, E.; Hannon, G.J.; Schultz, R.M. RNAi and expression of retrotransposons MuERV-L and IAP in preimplantation mouse embryos. Dev. Biol. 2004, 269, 276–285. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Huang, B.; Chen, H.; Yin, Q.; Liu, Y.; Xiang, Y.; Zhang, B.; Liu, B.; Wang, Q.; Xia, W.; et al. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature 2016, 534, 652–657. [Google Scholar] [CrossRef] [PubMed]
- Joshi, M.; Rajender, S. Long non-coding RNAs (lncRNAs) in spermatogenesis and male infertility. Reprod. Biol. Endocrinol. 2020, 18, 103. [Google Scholar] [CrossRef]
- Qin, T.; Li, J.; Zhang, K.-Q. Structure, Regulation, and Function of Linear and Circular Long Non-Coding RNAs. Front. Genet. 2020, 11, 150. [Google Scholar] [CrossRef]
- Barrett, S.P.; Salzman, J. Circular RNAs: Analysis, expression and potential functions. Development 2016, 143, 1838–1847. [Google Scholar] [CrossRef] [PubMed]
- Ji, P.; Wu, W.; Chen, S.; Zheng, Y.; Zhou, L.; Zhang, J.; Cheng, H.; Yan, J.; Zhang, S.; Yang, P.; et al. Expanded Expression Landscape and Prioritization of Circular RNAs in Mammals. Cell Rep. 2019, 26, 3444–3460.e5. [Google Scholar] [CrossRef]
- Tang, C.; Xie, Y.; Yu, T.; Liu, N.; Wang, Z.; Woolsey, R.J.; Tang, Y.; Zhang, X.; Qin, W.; Zhang, Y.; et al. m6A-dependent biogenesis of circular RNAs in male germ cells. Cell Res. 2020, 30, 211–228. [Google Scholar] [CrossRef]
- Das, A.; Sinha, T.; Shyamal, S.; Panda, A.C. Emerging Role of Circular RNA–Protein Interactions. Non-Coding RNA 2021, 7, 48. [Google Scholar] [CrossRef]
- Panda, A.C. Circular RNAs Act as miRNA Sponges. Circ. RNAs. Adv. Exp. Med. Biol. 2018, 1087, 67–79. [Google Scholar] [CrossRef]
- Chen, Q.; Yan, W.; Duan, E. Epigenetic inheritance of acquired traits through sperm RNAs and sperm RNA modifications. Nat. Rev. Genet. 2016, 17, 733–743. [Google Scholar] [CrossRef] [PubMed]
- Fullston, T.; Teague, E.M.C.O.; Palmer, N.O.; DeBlasio, M.J.; Mitchell, M.; Corbett, M.; Print, C.G.; Owens, J.A.; Lane, M. Paternal obesity initiates metabolic disturbances in two generations of mice with incomplete penetrance to the F 2 generation and alters the transcriptional profile of testis and sperm microRNA content. FASEB J. 2013, 27, 4226–4243. [Google Scholar] [CrossRef] [PubMed]
- Rodgers, A.B.; Morgan, C.P.; Bronson, S.L.; Revello, S.; Bale, T.L. Paternal Stress Exposure Alters Sperm MicroRNA Content and Reprograms Offspring HPA Stress Axis Regulation. J. Neurosci. 2013, 33, 9003–9012. [Google Scholar] [CrossRef] [PubMed]
- Pegtel, D.M.; Cosmopoulos, K.; Thorley-Lawson, D.A.; van Eijndhoven, M.A.J.; Hopmans, E.S.; Lindenberg, J.L.; de Gruijl, T.D.; Würdinger, T.; Middeldorp, J.M. Functional delivery of viral miRNAs via exosomes. Proc. Natl. Acad. Sci. USA 2010, 107, 6328–6333. [Google Scholar] [CrossRef] [PubMed]
- Vickers, K.C.; Palmisano, B.T.; Shoucri, B.M.; Shamburek, R.D.; Remaley, A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011, 13, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Gapp, K.; Jawaid, A.; Sarkies, P.; Bohacek, J.; Pelczar, P.; Prados, J.; Farinelli, L.; Miska, E.; Mansuy, I.M. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat. Neurosci. 2014, 17, 667–669. [Google Scholar] [CrossRef]
- Sharma, U.; Sun, F.; Conine, C.C.; Reichholf, B.; Kukreja, S.; Herzog, V.A.; Ameres, S.L.; Rando, O.J. Small RNAs Are Trafficked from the Epididymis to Developing Mammalian Sperm. Dev. Cell 2018, 46, 481–494.e6. [Google Scholar] [CrossRef]
- Chan, J.C.; Morgan, C.P.; Adrian Leu, N.; Shetty, A.; Cisse, Y.M.; Nugent, B.M.; Morrison, K.E.; Jašarević, E.; Huang, W.; Kanyuch, N.; et al. Reproductive tract extracellular vesicles are sufficient to transmit intergenerational stress and program neurodevelopment. Nat. Commun. 2020, 11, 1499. [Google Scholar] [CrossRef]
- Morgan, C.P.; Meadows, V.E.; Marx-Rattner, R.; Cisse, Y.M.; Bale, T.L. HA-tag CD63 is a novel conditional transgenic approach to track extracellular vesicle interactions with sperm and their transfer at conception. Sci. Rep. 2023, 13, 707. [Google Scholar] [CrossRef]
- Wang, H.; Wang, Z.; Zhou, T.; Morris, D.; Chen, S.; Li, M.; Wang, Y.; Zheng, H.; Fu, W.; Yan, W. Small RNA shuffling between murine sperm and their cytoplasmic droplets during epididymal maturation. Dev. Cell 2023, 58, 779–790.e4. [Google Scholar] [CrossRef]
- Zhang, Y.; Shi, J.; Rassoulzadegan, M.; Tuorto, F.; Chen, Q. Sperm RNA code programmes the metabolic health of offspring. Nat. Rev. Endocrinol. 2019, 15, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Bohacek, J.; Rassoulzadegan, M. Sperm RNA: Quo vadis? Semin. Cell Dev. Biol. 2020, 97, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Bošković, A.; Rando, O.J. Transgenerational Epigenetic Inheritance. Annu. Rev. Genet. 2018, 52, 21–41. [Google Scholar] [CrossRef] [PubMed]
- Gapp, K.; Bohacek, J. Epigenetic germline inheritance in mammals: Looking to the past to understand the future. Genes Brain Behav. 2018, 17, e12407. [Google Scholar] [CrossRef]
- Kretschmer, M.; Gapp, K. Deciphering the RNA universe in sperm in its role as a vertical information carrier. Environ. Epigenet. 2022, 8, dvac011. [Google Scholar] [CrossRef]
- Skvortsova, K.; Iovino, N.; Bogdanović, O. Functions and mechanisms of epigenetic inheritance in animals. Nat. Rev. Mol. Cell Biol. 2018, 19, 774–790. [Google Scholar] [CrossRef]
- Franklin, T.B.; Russig, H.; Weiss, I.C.; Gräff, J.; Linder, N.; Michalon, A.; Vizi, S.; Mansuy, I.M. Epigenetic Transmission of the Impact of Early Stress Across Generations. Biol. Psychiatry 2010, 68, 408–415. [Google Scholar] [CrossRef]
- Rodgers, A.B.; Morgan, C.P.; Leu, N.A.; Bale, T.L. Transgenerational epigenetic programming via sperm microRNA recapitulates effects of paternal stress. Proc. Natl. Acad. Sci. USA 2015, 112, 13699–13704. [Google Scholar] [CrossRef]
- Gapp, K.; Bohacek, J.; Grossmann, J.; Brunner, A.M.; Manuella, F.; Nanni, P.; Mansuy, I.M. Potential of Environmental Enrichment to Prevent Transgenerational Effects of Paternal Trauma. Neuropsychopharmacology 2016, 41, 2749–2758. [Google Scholar] [CrossRef]
- Wu, L.; Lu, Y.; Jiao, Y.; Liu, B.; Li, S.; Li, Y.; Xing, F.; Chen, D.; Liu, X.; Zhao, J.; et al. Paternal Psychological Stress Reprograms Hepatic Gluconeogenesis in Offspring. Cell Metab. 2016, 23, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Dickson, D.A.; Paulus, J.K.; Mensah, V.; Lem, J.; Saavedra-Rodriguez, L.; Gentry, A.; Pagidas, K.; Feig, L.A. Reduced levels of miRNAs 449 and 34 in sperm of mice and men exposed to early life stress. Transl. Psychiatry 2018, 8, 101. [Google Scholar] [CrossRef] [PubMed]
- Gapp, K.; van Steenwyk, G.; Germain, P.L.; Matsushima, W.; Rudolph, K.L.M.; Manuella, F.; Roszkowski, M.; Vernaz, G.; Ghosh, T.; Pelczar, P.; et al. Alterations in sperm long RNA contribute to the epigenetic inheritance of the effects of postnatal trauma. Mol. Psychiatry 2020, 25, 2162–2174. [Google Scholar] [CrossRef] [PubMed]
- Gapp, K.; Parada, G.E.; Gross, F.; Corcoba, A.; Kaur, J.; Grau, E.; Hemberg, M.; Bohacek, J.; Miska, E.A. Single paternal dexamethasone challenge programs offspring metabolism and reveals multiple candidates in RNA-mediated inheritance. IScience 2021, 24, 102870. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, A.M.; Walker, D.M.; Ramakrishnan, A.; Doyle, M.A.; Bagot, R.C.; Cates, H.M.; Peña, C.J.; Issler, O.; Lardner, C.K.; Browne, C.; et al. Sperm Transcriptional State Associated with Paternal Transmission of Stress Phenotypes. J. Neurosci. 2021, 41, 6202–6216. [Google Scholar] [CrossRef] [PubMed]
- Kong, Q.-Q.; Tian, X.-D.; Wang, J.; Yuan, H.-J.; Ning, S.-F.; Luo, M.-J.; Tan, J.-H. Author Correction: A next-generation sequencing study on mechanisms by which restraint and social instability stresses of male mice alter offspring anxiety-like behavior. Sci. Rep. 2022, 12, 11342. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, Z.-P.; Hu, H.; Lei, J.; Zhou, Z.; Yao, B.; Chen, L.; Liang, G.; Zhan, S.; Zhu, X.; et al. Sperm microRNAs confer depression susceptibility to offspring. Sci. Adv. 2021, 7, eabd7605c. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Li, Z.; Wang, G.; Wang, H.; Zhou, Y.; Zhao, X.; Cheng, C.Y.; Qiao, Y.; Sun, F. Sperm epigenetic alterations contribute to inter- and transgenerational effects of paternal exposure to long-term psychological stress via evading offspring embryonic reprogramming. Cell Discov. 2021, 7, 101. [Google Scholar] [CrossRef]
- Bestor, T.H. The DNA methyltransferases of mammals. Hum. Mol. Genet. 2000, 9, 2395–2402. [Google Scholar] [CrossRef]
- Jurkowska, R.Z.; Jurkowski, T.P.; Jeltsch, A. Structure and Function of Mammalian DNA Methyltransferases. ChemBioChem 2011, 12, 206–222. [Google Scholar] [CrossRef]
- Adalsteinsson, B.; Ferguson-Smith, A. Epigenetic Control of the Genome—Lessons from Genomic Imprinting. Genes 2014, 5, 635–655. [Google Scholar] [CrossRef] [PubMed]
- Ferguson-Smith, A.C. Genomic imprinting: The emergence of an epigenetic paradigm. Nat. Rev. Genet. 2011, 12, 565–575. [Google Scholar] [CrossRef] [PubMed]
- SanMiguel, J.M.; Bartolomei, M.S. DNA methylation dynamics of genomic imprinting in mouse development. Biol. Reprod. 2018, 99, 252–262. [Google Scholar] [CrossRef]
- Shirane, K.; Lorincz, M. Epigenetic Mechanisms Governing Female and Male Germline Development in Mammals. Sex. Dev. 2022, 16, 365–387. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Hore, T.A.; Reik, W. Reprogramming the Methylome: Erasing Memory and Creating Diversity. Cell Stem Cell 2014, 14, 710–719. [Google Scholar] [CrossRef] [PubMed]
- Wossidlo, M.; Nakamura, T.; Lepikhov, K.; Marques, C.J.; Zakhartchenko, V.; Boiani, M.; Arand, J.; Nakano, T.; Reik, W.; Walter, J. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat. Commun. 2011, 2, 241. [Google Scholar] [CrossRef]
- Dahlet, T.; Argüeso Lleida, A.; Al Adhami, H.; Dumas, M.; Bender, A.; Ngondo, R.P.; Tanguy, M.; Vallet, J.; Auclair, G.; Bardet, A.F.; et al. Genome-wide analysis in the mouse embryo reveals the importance of DNA methylation for transcription integrity. Nat. Commun. 2020, 11, 3153. [Google Scholar] [CrossRef]
- Greenberg, M.V.C.; Bourc’his, D. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 2019, 20, 590–607. [Google Scholar] [CrossRef]
- Hackett, J.A.; Sengupta, R.; Zylicz, J.J.; Murakami, K.; Lee, C.; Down, T.A.; Surani, M.A. Germline DNA Demethylation Dynamics and Imprint Erasure Through 5-Hydroxymethylcytosine. Science 2013, 339, 448–452. [Google Scholar] [CrossRef]
- Reik, W.; Dean, W.; Walter, J. Epigenetic Reprogramming in Mammalian Development. Science 2001, 293, 1089–1093. [Google Scholar] [CrossRef]
- Seah, M.K.Y.; Messerschmidt, D.M. From Germline to Soma: Epigenetic Dynamics in the Mouse Preimplantation Embryo. Curr. Top. Dev. Biol. 2018, 128, 203–235. [Google Scholar] [CrossRef] [PubMed]
- Bohacek, J.; Farinelli, M.; Mirante, O.; Steiner, G.; Gapp, K.; Coiret, G.; Ebeling, M.; Durán-Pacheco, G.; Iniguez, A.L.; Manuella, F.; et al. Pathological brain plasticity and cognition in the offspring of males subjected to postnatal traumatic stress. Mol. Psychiatry 2015, 20, 621–631. [Google Scholar] [CrossRef] [PubMed]
- Lismer, A.; Kimmins, S. Emerging evidence that the mammalian sperm epigenome serves as a template for embryo development. Nat. Commun. 2023, 14, 2142. [Google Scholar] [CrossRef] [PubMed]
- Ramaswamy, A.; Ioshikhes, I. Chapter Four-Dynamics of Modeled Oligonucleosomes and the Role of Histone Variant Proteins in Nucleosome Organization. Adv. Protein Chem. Struct. Biol. 2013, 90, 119–149. [Google Scholar] [CrossRef] [PubMed]
- Kurumizaka, H.; Kujirai, T.; Takizawa, Y. Contributions of Histone Variants in Nucleosome Structure and Function. J. Mol. Biol. 2021, 433, 166678. [Google Scholar] [CrossRef] [PubMed]
- Tvardovskiy, A.; Schwämmle, V.; Kempf, S.J.; Rogowska-Wrzesinska, A.; Jensen, O.N. Accumulation of histone variant H3.3 with age is associated with profound changes in the histone methylation landscape. Nucleic Acids Res. 2017, 45, 9272–9289. [Google Scholar] [CrossRef]
- Farrelly, L.A.; Thompson, R.E.; Zhao, S.; Lepack, A.E.; Lyu, Y.; Bhanu, N.V.; Zhang, B.; Loh, Y.-H.E.; Ramakrishnan, A.; Vadodaria, K.C.; et al. Histone serotonylation is a permissive modification that enhances TFIID binding to H3K4me3. Nature 2019, 567, 535–539. [Google Scholar] [CrossRef] [PubMed]
- Fulton, S.L.; Mitra, S.; Lepack, A.E.; Martin, J.A.; Stewart, A.F.; Converse, J.; Hochstetler, M.; Dietz, D.M.; Maze, I. Histone H3 dopaminylation in ventral tegmental area underlies heroin-induced transcriptional and behavioral plasticity in male rats. Neuropsychopharmacology 2022, 47, 1776–1783. [Google Scholar] [CrossRef]
- Peterson, C.L.; Laniel, M.-A. Histones and histone modifications. Curr. Biol. 2004, 14, R546–R551. [Google Scholar] [CrossRef]
- Yun, M.; Wu, J.; Workman, J.L.; Li, B. Readers of histone modifications. Cell Res. 2011, 21, 564–578. [Google Scholar] [CrossRef]
- Clapier, C.R.; Cairns, B.R. The Biology of Chromatin Remodeling Complexes. Annu. Rev. Biochem. 2009, 78, 273–304. [Google Scholar] [CrossRef] [PubMed]
- Klemm, S.L.; Shipony, Z.; Greenleaf, W.J. Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 2019, 20, 207–220. [Google Scholar] [CrossRef] [PubMed]
- Gates, L.A.; Foulds, C.E.; O’Malley, B.W. Histone Marks in the ‘Driver’s Seat’: Functional Roles in Steering the Transcription Cycle. Trends Biochem. Sci. 2017, 42, 977–989. [Google Scholar] [CrossRef] [PubMed]
- Brykczynska, U.; Hisano, M.; Erkek, S.; Ramos, L.; Oakeley, E.J.; Roloff, T.C.; Beisel, C.; Schübeler, D.; Stadler, M.B.; Peters, A.H.F.M. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat. Struct. Mol. Biol. 2010, 17, 679–687. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Alves, M.B.R.; Belleannée, C. Contribution of epididymal epithelial cell functions to sperm epigenetic changes and the health of progeny. Hum. Reprod. Update 2021, 28, 51–66. [Google Scholar] [CrossRef]
- Odroniec, A.; Olszewska, M.; Kurpisz, M. Epigenetic markers in the embryonal germ cell development and spermatogenesis. Basic Clin. Androl. 2023, 33, 6. [Google Scholar] [CrossRef]
- Wang, T.; Gao, H.; Li, W.; Liu, C. Essential Role of Histone Replacement and Modifications in Male Fertility. Front. Genet. 2019, 10, 962. [Google Scholar] [CrossRef]
- Govin, J.; Escoffier, E.; Rousseaux, S.; Kuhn, L.; Ferro, M.; Thévenon, J.; Catena, R.; Davidson, I.; Garin, J.; Khochbin, S.; et al. Pericentric heterochromatin reprogramming by new histone variants during mouse spermiogenesis. J. Cell Biol. 2007, 176, 283–294. [Google Scholar] [CrossRef]
- Tang, M.C.W.; Jacobs, S.A.; Mattiske, D.M.; Soh, Y.M.; Graham, A.N.; Tran, A.; Lim, S.L.; Hudson, D.F.; Kalitsis, P.; O’Bryan, M.K.; et al. Contribution of the Two Genes Encoding Histone Variant H3.3 to Viability and Fertility in Mice. PLoS Genet. 2015, 11, e1004964. [Google Scholar] [CrossRef]
- Yuen, B.T.K.; Bush, K.M.; Barrilleaux, B.L.; Cotterman, R.; Knoepfler, P.S. Histone H3.3 regulates dynamic chromatin states during spermatogenesis. Development 2014, 141, 3483–3494. [Google Scholar] [CrossRef]
- Wen, D.; Noh, K.; Goldberg, A.D.; Allis, C.D.; Rosenwaks, Z.; Rafii, S.; Banaszynski, L.A. Genome editing a mouse locus encoding a variant histone, H3.3B, to report on its expression in live animals. Genesis 2014, 52, 959–966. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.H.; Sauria, M.E.G.; Lyu, X.; Cheema, M.S.; Ausio, J.; Taylor, J.; Corces, V.G. Chromatin States in Mouse Sperm Correlate with Embryonic and Adult Regulatory Landscapes. Cell Rep. 2017, 18, 1366–1382. [Google Scholar] [CrossRef] [PubMed]
- Samans, B.; Yang, Y.; Krebs, S.; Sarode, G.V.; Blum, H.; Reichenbach, M.; Wolf, E.; Steger, K.; Dansranjavin, T.; Schagdarsurengin, U. Uniformity of Nucleosome Preservation Pattern in Mammalian Sperm and Its Connection to Repetitive DNA Elements. Dev. Cell 2014, 30, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.H.; Kremsky, I.; Gold, H.B.; Rowley, M.J.; Punyawai, K.; Buonanotte, A.; Lyu, X.; Bixler, B.J.; Chan, A.W.S.; Corces, V.G. Maintenance of CTCF- and Transcription Factor-Mediated Interactions from the Gametes to the Early Mouse Embryo. Mol. Cell 2019, 75, 154–171.e5. [Google Scholar] [CrossRef] [PubMed]
- Bedi, Y.S.; Wang, H.; Thomas, K.N.; Basel, A.; Prunier, J.; Robert, C.; Golding, M.C. Alcohol induced increases in sperm Histone H3 lysine 4 trimethylation correlate with increased placental CTCF occupancy and altered developmental programming. Sci. Rep. 2022, 12, 8839. [Google Scholar] [CrossRef]
- Cambiasso, M.Y.; Gotfryd, L.; Stinson, M.G.; Birolo, S.; Salamone, G.; Romanato, M.; Calvo, J.C.; Fontana, V.A. Paternal alcohol consumption has intergenerational consequences in male offspring. J. Assist. Reprod. Genet. 2022, 39, 441–459. [Google Scholar] [CrossRef]
- Lismer, A.; Dumeaux, V.; Lafleur, C.; Lambrot, R.; Brind’Amour, J.; Lorincz, M.C.; Kimmins, S. Histone H3 lysine 4 trimethylation in sperm is transmitted to the embryo and associated with diet-induced phenotypes in the offspring. Dev. Cell 2021, 56, 671–686.e6. [Google Scholar] [CrossRef]
- Pepin, A.-S.; Lafleur, C.; Lambrot, R.; Dumeaux, V.; Kimmins, S. Sperm histone H3 lysine 4 tri-methylation serves as a metabolic sensor of paternal obesity and is associated with the inheritance of metabolic dysfunction. Mol. Metab. 2022, 59, 101463. [Google Scholar] [CrossRef]
- Sims, R.J.; Millhouse, S.; Chen, C.-F.; Lewis, B.A.; Erdjument-Bromage, H.; Tempst, P.; Manley, J.L.; Reinberg, D. Recognition of Trimethylated Histone H3 Lysine 4 Facilitates the Recruitment of Transcription Postinitiation Factors and Pre-mRNA Splicing. Mol. Cell 2007, 28, 665–676. [Google Scholar] [CrossRef]
- Wang, H.; Fan, Z.; Shliaha, P.V.; Miele, M.; Hendrickson, R.C.; Jiang, X.; Helin, K. Publisher Correction: H3K4me3 regulates RNA polymerase II promoter-proximal pause-release. Nature 2023, 616, E7. [Google Scholar] [CrossRef]
- Dekker, J.; Marti-Renom, M.A.; Mirny, L.A. Exploring the three-dimensional organization of genomes: Interpreting chromatin interaction data. Nat. Rev. Genet. 2013, 14, 390–403. [Google Scholar] [CrossRef] [PubMed]
- Belton, J.-M.; McCord, R.P.; Gibcus, J.H.; Naumova, N.; Zhan, Y.; Dekker, J. Hi–C: A comprehensive technique to capture the conformation of genomes. Methods 2012, 58, 268–276. [Google Scholar] [CrossRef]
- Lieberman-Aiden, E.; van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Amit, I.; Lajoie, B.R.; Sabo, P.J.; Dorschner, M.O.; et al. Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science 2009, 326, 289–293. [Google Scholar] [CrossRef] [PubMed]
- McArthur, E.; Capra, J.A. Topologically associating domain boundaries that are stable across diverse cell types are evolutionarily constrained and enriched for heritability. Am. J. Hum. Genet. 2021, 108, 269–283. [Google Scholar] [CrossRef] [PubMed]
- Battulin, N.; Fishman, V.S.; Mazur, A.M.; Pomaznoy, M.; Khabarova, A.A.; Afonnikov, D.A.; Prokhortchouk, E.B.; Serov, O.L. Comparison of the three-dimensional organization of sperm and fibroblast genomes using the Hi-C approach. Genome Biol. 2015, 16, 77. [Google Scholar] [CrossRef] [PubMed]
- Ke, Y.; Xu, Y.; Chen, X.; Feng, S.; Liu, Z.; Sun, Y.; Yao, X.; Li, F.; Zhu, W.; Gao, L.; et al. 3D Chromatin Structures of Mature Gametes and Structural Reprogramming during Mammalian Embryogenesis. Cell 2017, 170, 367–381.e20. [Google Scholar] [CrossRef]
- Agarwal, A.; Virk, G.; Ong, C.; du Plessis, S.S. Effect of Oxidative Stress on Male Reproduction. World J. Men’s Health 2014, 32, 1–17. [Google Scholar] [CrossRef]
- Meng, X.; Peng, L.; Xu, J.; Guo, D.; Cao, W.; Xu, Y.; Li, S. Betaine attenuate chronic restraint stress-induced changes in testicular damage and oxidative stress in male mice. Reprod. Biol. Endocrinol. 2022, 20, 80. [Google Scholar] [CrossRef]
- Sabeti, P.; Pourmasumi, S.; Rahiminia, T.; Akyash, F.; Talebi, A.R. Etiologies of sperm oxidative stress. Int. J. Reprod. Biomed. 2016, 14, 231–240. [Google Scholar] [CrossRef]
- Sanders, J.T.; Freeman, T.F.; Xu, Y.; Golloshi, R.; Stallard, M.A.; Hill, A.M.; San Martin, R.; Balajee, A.S.; McCord, R.P. Radiation-induced DNA damage and repair effects on 3D genome organization. Nat. Commun. 2020, 11, 6178. [Google Scholar] [CrossRef]
- Xiang, J.-F.; Corces, V.G. Regulation of 3D chromatin organization by CTCF. Curr. Opin. Genet. Dev. 2021, 67, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Marahrens, Y.; Panning, B.; Dausman, J.; Strauss, W.; Jaenisch, R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 1997, 11, 156–166. [Google Scholar] [CrossRef] [PubMed]
- Andergassen, D.; Rinn, J.L. From genotype to phenotype: Genetics of mammalian long non-coding RNAs in vivo. Nat. Rev. Genet. 2022, 23, 229–243. [Google Scholar] [CrossRef] [PubMed]
- Barlow, D.P.; Bartolomei, M.S. Genomic Imprinting in Mammals. Cold Spring Harb. Perspect. Biol. 2014, 6, a018382. [Google Scholar] [CrossRef]
- Fatica, A.; Bozzoni, I. Long non-coding RNAs: New players in cell differentiation and development. Nat. Rev. Genet. 2014, 15, 7–21. [Google Scholar] [CrossRef]
- Kamikawa, Y.; Donohoe, M.E. The dynamics of X-chromosome inactivation in mouse development. Mol. Reprod. Dev. 2014, 81, 141–147. [Google Scholar] [CrossRef]
- Sahakyan, A.; Yang, Y.; Plath, K. The Role of Xist in X-Chromosome Dosage Compensation. Trends Cell Biol. 2018, 28, 999–1013. [Google Scholar] [CrossRef]
- Tucci, V.; Isles, A.R.; Kelsey, G.; Ferguson-Smith, A.C.; Tucci, V.; Bartolomei, M.S.; Benvenisty, N.; Bourc’his, D.; Charalambous, M.; Dulac, C.; et al. Genomic Imprinting and Physiological Processes in Mammals. Cell 2019, 176, 952–965. [Google Scholar] [CrossRef]
- Hupalowska, A.; Jedrusik, A.; Zhu, M.; Bedford, M.T.; Glover, D.M.; Zernicka-Goetz, M. CARM1 and Paraspeckles Regulate Pre-implantation Mouse Embryo Development. Cell 2018, 175, 1902–1916.e13. [Google Scholar] [CrossRef]
- Torres-Padilla, M.-E.; Parfitt, D.-E.; Kouzarides, T.; Zernicka-Goetz, M. Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature 2007, 445, 214–218. [Google Scholar] [CrossRef]
- Wang, J.; Wang, L.; Feng, G.; Wang, Y.; Li, Y.; Li, X.; Liu, C.; Jiao, G.; Huang, C.; Shi, J.; et al. Asymmetric Expression of LincGET Biases Cell Fate in Two-Cell Mouse Embryos. Cell 2018, 175, 1887–1901.e18. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Duan, E. LncRNAs and paraspeckles predict cell fate in early mouse embryo†. Biol. Reprod. 2019, 100, 1129–1131. [Google Scholar] [CrossRef] [PubMed]
- Ohnishi, Y.; Totoki, Y.; Toyoda, A.; Watanabe, T.; Yamamoto, Y.; Tokunaga, K.; Sakaki, Y.; Sasaki, H.; Hohjoh, H. Small RNA class transition from siRNA/piRNA to miRNA during pre-implantation mouse development. Nucleic Acids Res. 2010, 38, 5141–5151. [Google Scholar] [CrossRef]
- Yang, Q.; Lin, J.; Liu, M.; Li, R.; Tian, B.; Zhang, X.; Xu, B.; Liu, M.; Zhang, X.; Li, Y.; et al. Highly sensitive sequencing reveals dynamic modifications and activities of small RNAs in mouse oocytes and early embryos. Sci. Adv. 2016, 2, e1501482. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.; Kaneda, M.; O’Carroll, D.; Hajkova, P.; Barton, S.C.; Sun, Y.A.; Lee, C.; Tarakhovsky, A.; Lao, K.; Surani, M.A. Maternal microRNAs are essential for mouse zygotic development. Genes Dev. 2007, 21, 644–648. [Google Scholar] [CrossRef] [PubMed]
- Suh, N.; Baehner, L.; Moltzahn, F.; Melton, C.; Shenoy, A.; Chen, J.; Blelloch, R. MicroRNA Function Is Globally Suppressed in Mouse Oocytes and Early Embryos. Curr. Biol. 2010, 20, 271–277. [Google Scholar] [CrossRef]
- Murchison, E.P.; Stein, P.; Xuan, Z.; Pan, H.; Zhang, M.Q.; Schultz, R.M.; Hannon, G.J. Critical roles for Dicer in the female germline. Genes Dev. 2007, 21, 682–693. [Google Scholar] [CrossRef] [PubMed]
- Lykke-Andersen, K.; Gilchrist, M.J.; Grabarek, J.B.; Das, P.; Miska, E.; Zernicka-Goetz, M. Maternal Argonaute 2 Is Essential for Early Mouse Development at the Maternal-Zygotic Transition. Mol. Biol. Cell 2008, 19, 4383–4392. [Google Scholar] [CrossRef]
- Yuan, S.; Tang, C.; Schuster, A.; Zhang, Y.; Zheng, H.; Yan, W. Paternal pachytene piRNAs are not required for fertilization, embryonic development and sperm-mediated epigenetic inheritance in mice. Environ. Epigenetics 2016, 2, dvw021. [Google Scholar] [CrossRef]
- Mineno, J. The expression profile of microRNAs in mouse embryos. Nucleic Acids Res. 2006, 34, 1765–1771. [Google Scholar] [CrossRef]
- Yang, Y.; Bai, W.; Zhang, L.; Yin, G.; Wang, X.; Wang, J.; Zhao, H.; Han, Y.; Yao, Y. Determination of microRNAs in mouse preimplantation embryos by microarray. Dev. Dyn. 2008, 237, 2315–2327. [Google Scholar] [CrossRef] [PubMed]
- Perillo, G.; Shibata, K.; Wu, P.-H. piRNAs in sperm function and embryo viability. Reproduction 2022, 165, R91–R102. [Google Scholar] [CrossRef] [PubMed]
- García-López, J.; Hourcade, J.d.D.; Alonso, L.; Cárdenas, D.B.; del Mazo, J. Global characterization and target identification of piRNAs and endo-siRNAs in mouse gametes and zygotes. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2014, 1839, 463–475. [Google Scholar] [CrossRef] [PubMed]
- Flemr, M.; Malik, R.; Franke, V.; Nejepinska, J.; Sedlacek, R.; Vlahovicek, K.; Svoboda, P. A Retrotransposon-Driven Dicer Isoform Directs Endogenous Small Interfering RNA Production in Mouse Oocytes. Cell 2013, 155, 807–816. [Google Scholar] [CrossRef] [PubMed]
- Lim, A.K.; Lorthongpanich, C.; Chew, T.G.; Tan, C.W.G.; Shue, Y.T.; Balu, S.; Gounko, N.; Kuramochi-Miyagawa, S.; Matzuk, M.M.; Chuma, S.; et al. The nuage mediates retrotransposon silencing in mouse primordial ovarian follicles. Development 2013, 140, 3819–3825. [Google Scholar] [CrossRef]
- Watanabe, T.; Totoki, Y.; Toyoda, A.; Kaneda, M.; Kuramochi-Miyagawa, S.; Obata, Y.; Chiba, H.; Kohara, Y.; Kono, T.; Nakano, T.; et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 2008, 453, 539–543. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Chao, S.-B.; Xiao, L.; Wang, Z.-B.; Meng, T.-G.; Li, Y.-Y.; Han, Z.-M.; Ouyang, Y.-C.; Hou, Y.; Sun, Q.-Y.; et al. Sperm-carried RNAs play critical roles in mouse embryonic development. Oncotarget 2017, 8, 67394–67405. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Lu, X.; Dean, J. The maternal to zygotic transition in mammals. Mol. Asp. Med. 2013, 34, 919–938. [Google Scholar] [CrossRef]
- Li, E.; Zhang, Y. DNA Methylation in Mammals. Cold Spring Harb. Perspect. Biol. 2014, 6, a019133. [Google Scholar] [CrossRef]
- Messerschmidt, D.M.; Knowles, B.B.; Solter, D. DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes Dev. 2014, 28, 812–828. [Google Scholar] [CrossRef]
- Zeng, Y.; Chen, T. DNA Methylation Reprogramming during Mammalian Development. Genes 2019, 10, 257. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.; Tang, Q.; Wan, M.; Cui, K.; Zhang, Y.; Ren, G.; Ni, B.; Sklar, J.; Przytycka, T.M.; Childs, R.; et al. Genome-wide detection of DNase I hypersensitive sites in single cells and FFPE tissue samples. Nature 2015, 528, 142–146. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, H.; Sakurai, T.; Imai, M.; Takahashi, N.; Fukuda, A.; Yayoi, O.; Sato, S.; Nakabayashi, K.; Hata, K.; Sotomaru, Y.; et al. Contribution of Intragenic DNA Methylation in Mouse Gametic DNA Methylomes to Establish Oocyte-Specific Heritable Marks. PLoS Genet. 2012, 8, e1002440. [Google Scholar] [CrossRef] [PubMed]
- Lu, F.; Liu, Y.; Inoue, A.; Suzuki, T.; Zhao, K.; Zhang, Y. Establishing Chromatin Regulatory Landscape during Mouse Preimplantation Development. Cell 2016, 165, 1375–1388. [Google Scholar] [CrossRef] [PubMed]
- Mayer, W.; Niveleau, A.; Walter, J.; Fundele, R.; Haaf, T. Demethylation of the zygotic paternal genome. Nature 2000, 403, 501–502. [Google Scholar] [CrossRef] [PubMed]
- Oswald, J.; Engemann, S.; Lane, N.; Mayer, W.; Olek, A.; Fundele, R.; Dean, W.; Reik, W.; Walter, J. Active demethylation of the paternal genome in the mouse zygote. Curr. Biol. 2000, 10, 475–478. [Google Scholar] [CrossRef] [PubMed]
- Santos, F.; Hendrich, B.; Reik, W.; Dean, W. Dynamic Reprogramming of DNA Methylation in the Early Mouse Embryo. Dev. Biol. 2002, 241, 172–182. [Google Scholar] [CrossRef]
- Smith, Z.D.; Chan, M.M.; Mikkelsen, T.S.; Gu, H.; Gnirke, A.; Regev, A.; Meissner, A. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 2012, 484, 339–344. [Google Scholar] [CrossRef]
- Nichols, J.; Smith, A. Naive and Primed Pluripotent States. Cell Stem Cell 2009, 4, 487–492. [Google Scholar] [CrossRef]
- Peng, G.; Suo, S.; Cui, G.; Yu, F.; Wang, R.; Chen, J.; Chen, S.; Liu, Z.; Chen, G.; Qian, Y.; et al. Molecular architecture of lineage allocation and tissue organization in early mouse embryo. Nature 2019, 572, 528–532. [Google Scholar] [CrossRef]
- Theunissen, T.W.; Friedli, M.; He, Y.; Planet, E.; O’Neil, R.C.; Markoulaki, S.; Pontis, J.; Wang, H.; Iouranova, A.; Imbeault, M.; et al. Molecular Criteria for Defining the Naive Human Pluripotent State. Cell Stem Cell 2016, 19, 502–515. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xiang, Y.; Yin, Q.; Du, Z.; Peng, X.; Wang, Q.; Fidalgo, M.; Xia, W.; Li, Y.; Zhao, Z.; et al. Dynamic epigenomic landscapes during early lineage specification in mouse embryos. Nat. Genet. 2018, 50, 96–105. [Google Scholar] [CrossRef] [PubMed]
- Rulands, S.; Lee, H.J.; Clark, S.J.; Angermueller, C.; Smallwood, S.A.; Krueger, F.; Mohammed, H.; Dean, W.; Nichols, J.; Rugg-Gunn, P.; et al. Genome-Scale Oscillations in DNA Methylation during Exit from Pluripotency. Cell Syst. 2018, 7, 63–76.e12. [Google Scholar] [CrossRef] [PubMed]
- Smith, Z.D.; Shi, J.; Gu, H.; Donaghey, J.; Clement, K.; Cacchiarelli, D.; Gnirke, A.; Michor, F.; Meissner, A. Epigenetic restriction of extraembryonic lineages mirrors the somatic transition to cancer. Nature 2017, 549, 543–547. [Google Scholar] [CrossRef] [PubMed]
- Hackett, J.A.; Reddington, J.P.; Nestor, C.E.; Dunican, D.S.; Branco, M.R.; Reichmann, J.; Reik, W.; Surani, M.A.; Adams, I.R.; Meehan, R.R. Promoter DNA methylation couples genome-defence mechanisms to epigenetic reprogramming in the mouse germline. Development 2012, 139, 3623–3632. [Google Scholar] [CrossRef] [PubMed]
- Li, E.; Beard, C.; Jaenisch, R. Role for DNA methylation in genomic imprinting. Nature 1993, 366, 362–365. [Google Scholar] [CrossRef]
- Weaver, J.R.; Sarkisian, G.; Krapp, C.; Mager, J.; Mann, M.R.W.; Bartolomei, M.S. Domain-Specific Response of Imprinted Genes to Reduced DNMT1. Mol. Cell. Biol. 2010, 30, 3916–3928. [Google Scholar] [CrossRef]
- Bertozzi, T.M.; Takahashi, N.; Hanin, G.; Kazachenka, A.; Ferguson-Smith, A.C. A spontaneous genetically induced epiallele at a retrotransposon shapes host genome function. eLife 2021, 10, e65233. [Google Scholar] [CrossRef]
- Walsh, C.P.; Chaillet, J.R.; Bestor, T.H. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 1998, 20, 116–117. [Google Scholar] [CrossRef]
- Oda, M.; Yamagiwa, A.; Yamamoto, S.; Nakayama, T.; Tsumura, A.; Sasaki, H.; Nakao, K.; Li, E.; Okano, M. DNA methylation regulates long-range gene silencing of an X-linked homeobox gene cluster in a lineage-specific manner. Genes Dev. 2006, 20, 3382–3394. [Google Scholar] [CrossRef][Green Version]
- Borgel, J.; Guibert, S.; Li, Y.; Chiba, H.; Schübeler, D.; Sasaki, H.; Forné, T.; Weber, M. Targets and dynamics of promoter DNA methylation during early mouse development. Nat. Genet. 2010, 42, 1093–1100. [Google Scholar] [CrossRef] [PubMed]
- Peters, J. The role of genomic imprinting in biology and disease: An expanding view. Nat. Rev. Genet. 2014, 15, 517–530. [Google Scholar] [CrossRef] [PubMed]
- Kazachenka, A.; Bertozzi, T.M.; Sjoberg-Herrera, M.K.; Walker, N.; Gardner, J.; Gunning, R.; Pahita, E.; Adams, S.; Adams, D.; Ferguson-Smith, A.C. Identification, Characterization, and Heritability of Murine Metastable Epialleles: Implications for Non-genetic Inheritance. Cell 2018, 175, 1259–1271.e13. [Google Scholar] [CrossRef] [PubMed]
- Tsompana, M.; Buck, M.J. Chromatin accessibility: A window into the genome. Epigenetics Chromatin 2014, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Kremsky, I.; Corces, V.G. Protection from DNA re-methylation by transcription factors in primordial germ cells and pre-implantation embryos can explain trans-generational epigenetic inheritance. Genome Biol. 2020, 21, 118. [Google Scholar] [CrossRef] [PubMed]
- Fadloun, A.; Le Gras, S.; Jost, B.; Ziegler-Birling, C.; Takahashi, H.; Gorab, E.; Carninci, P.; Torres-Padilla, M.-E. Chromatin signatures and retrotransposon profiling in mouse embryos reveal regulation of LINE-1 by RNA. Nat. Struct. Mol. Biol. 2013, 20, 332–338. [Google Scholar] [CrossRef]
- Jachowicz, J.W.; Bing, X.; Pontabry, J.; Bošković, A.; Rando, O.J.; Torres-Padilla, M.-E. LINE-1 activation after fertilization regulates global chromatin accessibility in the early mouse embryo. Nat. Genet. 2017, 49, 1502–1510. [Google Scholar] [CrossRef]
- Bourque, G. Transposable elements in gene regulation and in the evolution of vertebrate genomes. Curr. Opin. Genet. Dev. 2009, 19, 607–612. [Google Scholar] [CrossRef]
- Peaston, A.E.; Evsikov, A.V.; Graber, J.H.; de Vries, W.N.; Holbrook, A.E.; Solter, D.; Knowles, B.B. Retrotransposons Regulate Host Genes in Mouse Oocytes and Preimplantation Embryos. Dev. Cell 2004, 7, 597–606. [Google Scholar] [CrossRef]
- Hatanaka, Y.; Inoue, K.; Oikawa, M.; Kamimura, S.; Ogonuki, N.; Kodama, E.N.; Ohkawa, Y.; Tsukada, Y.; Ogura, A. Histone chaperone CAF-1 mediates repressive histone modifications to protect preimplantation mouse embryos from endogenous retrotransposons. Proc. Natl. Acad. Sci. USA 2015, 112, 14641–14646. [Google Scholar] [CrossRef]
- Wang, C.; Liu, X.; Gao, Y.; Yang, L.; Li, C.; Liu, W.; Chen, C.; Kou, X.; Zhao, Y.; Chen, J.; et al. Reprogramming of H3K9me3-dependent heterochromatin during mammalian embryo development. Nat. Cell Biol. 2018, 20, 620–631. [Google Scholar] [CrossRef] [PubMed]
- Percharde, M.; Lin, C.-J.; Yin, Y.; Guan, J.; Peixoto, G.A.; Bulut-Karslioglu, A.; Biechele, S.; Huang, B.; Shen, X.; Ramalho-Santos, M. A LINE1-Nucleolin Partnership Regulates Early Development and ESC Identity. Cell 2018, 174, 391–405.e19. [Google Scholar] [CrossRef] [PubMed]
- Gapp, K.; Soldado-Magraner, S.; Alvarez-Sánchez, M.; Bohacek, J.; Vernaz, G.; Shu, H.; Franklin, T.B.; Wolfer, D.; Mansuy, I.M. Early life stress in fathers improves behavioural flexibility in their offspring. Nat. Commun. 2014, 5, 5466. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Morales Valencia, M.; Yu, Y.; Ouchi, Y.; Takahashi, K.; Shokhirev, M.N.; Lande, K.; Williams, A.E.; Fresia, C.; Kurita, M.; et al. Transgenerational inheritance of acquired epigenetic signatures at CpG islands in mice. Cell 2023, 186, 715–731.e19. [Google Scholar] [CrossRef]
- Stewart, K.R.; Veselovska, L.; Kim, J.; Huang, J.; Saadeh, H.; Tomizawa, S.; Smallwood, S.A.; Chen, T.; Kelsey, G. Dynamic changes in histone modifications precede de novo DNA methylation in oocytes. Genes Dev. 2015, 29, 2449–2462. [Google Scholar] [CrossRef]
- Xu, Q.; Xie, W. Epigenome in Early Mammalian Development: Inheritance, Reprogramming and Establishment. Trends Cell Biol. 2018, 28, 237–253. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Zhang, C.; Zhang, Y. Epigenetic regulation of mouse preimplantation embryo development. Curr. Opin. Genet. Dev. 2020, 64, 13–20. [Google Scholar] [CrossRef]
- Xu, R.; Li, C.; Liu, X.; Gao, S. Insights into epigenetic patterns in mammalian early embryos. Protein Cell 2021, 12, 7–28. [Google Scholar] [CrossRef]
- Dahl, J.A.; Jung, I.; Aanes, H.; Greggains, G.D.; Manaf, A.; Lerdrup, M.; Li, G.; Kuan, S.; Li, B.; Lee, A.Y.; et al. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature 2016, 537, 548–552. [Google Scholar] [CrossRef]
- Inoue, A.; Jiang, L.; Lu, F.; Suzuki, T.; Zhang, Y. Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature 2017, 547, 419–424. [Google Scholar] [CrossRef]
- Lepikhov, K.; Walter, J. Differential dynamics of histone H3 methylation at positions K4 and K9 in the mouse zygote. BMC Dev. Biol. 2004, 4, 12. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, C.; Liu, W.; Li, J.; Li, C.; Kou, X.; Chen, J.; Zhao, Y.; Gao, H.; Wang, H.; et al. Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature 2016, 537, 558–562. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Zheng, H.; Huang, B.; Li, W.; Xiang, Y.; Peng, X.; Ming, J.; Wu, X.; Zhang, Y.; Xu, Q.; et al. Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature 2016, 537, 553–557. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Huang, B.; Zhang, B.; Xiang, Y.; Du, Z.; Xu, Q.; Li, Y.; Wang, Q.; Ma, J.; Peng, X.; et al. Resetting Epigenetic Memory by Reprogramming of Histone Modifications in Mammals. Mol. Cell 2016, 63, 1066–1079. [Google Scholar] [CrossRef] [PubMed]
- Lismer, A.; Siklenka, K.; Lafleur, C.; Dumeaux, V.; Kimmins, S. Sperm histone H3 lysine 4 trimethylation is altered in a genetic mouse model of transgenerational epigenetic inheritance. Nucleic Acids Res. 2020, 48, 11380–11393. [Google Scholar] [CrossRef] [PubMed]
- Aoshima, K.; Inoue, E.; Sawa, H.; Okada, Y. Paternal H3K4 methylation is required for minor zygotic gene activation and early mouse embryonic development. EMBO Rep. 2015, 16, 803–812. [Google Scholar] [CrossRef] [PubMed]
- Bošković, A.; Bender, A.; Gall, L.; Ziegler-Birling, C.; Beaujean, N.; Torres-Padilla, M.-E. Analysis of active chromatin modifications in early mammalian embryos reveals uncoupling of H2A.Z acetylation and H3K36 trimethylation from embryonic genome activation. Epigenetics 2012, 7, 747–757. [Google Scholar] [CrossRef] [PubMed]
- Ziegler-Birling, C.; Daujat, S.; Schneider, R.; Torres-Padilla, M.-E. Dynamics of histone H3 acetylation in the nucleosome core during mouse pre-implantation development. Epigenetics 2016, 11, 553–562. [Google Scholar] [CrossRef]
- Bogliotti, Y.S.; Ross, P.J. Mechanisms of histone H3 lysine 27 trimethylation remodeling during early mammalian development. Epigenetics 2012, 7, 976–981. [Google Scholar] [CrossRef]
- Dahl, J.A.; Reiner, A.H.; Klungland, A.; Wakayama, T.; Collas, P. Histone H3 Lysine 27 Methylation Asymmetry on Developmentally-Regulated Promoters Distinguish the First Two Lineages in Mouse Preimplantation Embryos. PLoS ONE 2010, 5, e9150. [Google Scholar] [CrossRef]
- Chen, Z.; Djekidel, M.N.; Zhang, Y. Distinct dynamics and functions of H2AK119ub1 and H3K27me3 in mouse preimplantation embryos. Nat. Genet. 2021, 53, 551–563. [Google Scholar] [CrossRef] [PubMed]
- Santenard, A.; Ziegler-Birling, C.; Koch, M.; Tora, L.; Bannister, A.J.; Torres-Padilla, M.-E. Heterochromatin formation in the mouse embryo requires critical residues of the histone variant H3.3. Nat. Cell Biol. 2010, 12, 853–862. [Google Scholar] [CrossRef] [PubMed]
- Santos, F.; Peters, A.H.; Otte, A.P.; Reik, W.; Dean, W. Dynamic Chromatin Modifications Characterise the First Cell Cycle in Mouse Embryos. Dev. Biol. 2005, 280, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Ishiuchi, T.; Abe, S.; Inoue, K.; Yeung, W.K.A.; Miki, Y.; Ogura, A.; Sasaki, H. Reprogramming of the histone H3.3 landscape in the early mouse embryo. Nat. Struct. Mol. Biol. 2021, 28, 38–49. [Google Scholar] [CrossRef] [PubMed]
- Kaya-Okur, H.S.; Wu, S.J.; Codomo, C.A.; Pledger, E.S.; Bryson, T.D.; Henikoff, J.G.; Ahmad, K.; Henikoff, S. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 2019, 10, 1930. [Google Scholar] [CrossRef] [PubMed]
- Lismer, A.; Lambrot, R.; Lafleur, C.; Dumeaux, V.; Kimmins, S. ChIP-seq protocol for sperm cells and embryos to assess environmental impacts and epigenetic inheritance. STAR Protoc. 2021, 2, 100602. [Google Scholar] [CrossRef] [PubMed]
- Swain, J.E.; Pool, T.B. ART failure: Oocyte contributions to unsuccessful fertilization. Hum. Reprod. Update 2008, 14, 431–446. [Google Scholar] [CrossRef] [PubMed]
- Gou, L.-T.; Lim, D.-H.; Ma, W.; Aubol, B.E.; Hao, Y.; Wang, X.; Zhao, J.; Liang, Z.; Shao, C.; Zhang, X.; et al. Initiation of Parental Genome Reprogramming in Fertilized Oocyte by Splicing Kinase SRPK1-Catalyzed Protamine Phosphorylation. Cell 2020, 180, 1212–1227.e14. [Google Scholar] [CrossRef]
- Inoue, A.; Zhang, Y. Nucleosome assembly is required for nuclear pore complex assembly in mouse zygotes. Nat. Struct. Mol. Biol. 2014, 21, 609–616. [Google Scholar] [CrossRef]
- Lin, C.-J.; Koh, F.M.; Wong, P.; Conti, M.; Ramalho-Santos, M. Hira-Mediated H3.3 Incorporation Is Required for DNA Replication and Ribosomal RNA Transcription in the Mouse Zygote. Dev. Cell 2014, 30, 268–279. [Google Scholar] [CrossRef]
- Nashun, B.; Hill, P.W.S.; Smallwood, S.A.; Dharmalingam, G.; Amouroux, R.; Clark, S.J.; Sharma, V.; Ndjetehe, E.; Pelczar, P.; Festenstein, R.J.; et al. Continuous Histone Replacement by Hira Is Essential for Normal Transcriptional Regulation and De Novo DNA Methylation during Mouse Oogenesis. Mol. Cell 2015, 60, 611–625. [Google Scholar] [CrossRef] [PubMed]
- Buenrostro, J.D.; Wu, B.; Litzenburger, U.M.; Ruff, D.; Gonzales, M.L.; Snyder, M.P.; Chang, H.Y.; Greenleaf, W.J. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 2015, 523, 486–490. [Google Scholar] [CrossRef] [PubMed]
- Cusanovich, D.A.; Daza, R.; Adey, A.; Pliner, H.A.; Christiansen, L.; Gunderson, K.L.; Steemers, F.J.; Trapnell, C.; Shendure, J. Multiplex single-cell profiling of chromatin accessibility by combinatorial cellular indexing. Science 2015, 348, 910–914. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zibetti, C.; Wan, J.; Wang, G.; Blackshaw, S.; Qian, J. Assessing the model transferability for prediction of transcription factor binding sites based on chromatin accessibility. BMC Bioinform. 2017, 18, 355. [Google Scholar] [CrossRef]
- Bultman, S.; Gebuhr, T.; Yee, D.; La Mantia, C.; Nicholson, J.; Gilliam, A.; Randazzo, F.; Metzger, D.; Chambon, P.; Crabtree, G.; et al. A Brg1 Null Mutation in the Mouse Reveals Functional Differences among Mammalian SWI/SNF Complexes. Mol. Cell 2000, 6, 1287–1295. [Google Scholar] [CrossRef] [PubMed]
- de la Serna, I.L.; Ohkawa, Y.; Imbalzano, A.N. Chromatin remodelling in mammalian differentiation: Lessons from ATP-dependent remodellers. Nat. Rev. Genet. 2006, 7, 461–473. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Hyttel, P.; Hall, V.J. Regulation of H3K27me3 and H3K4me3 during early porcine embryonic development. Mol. Reprod. Dev. 2010, 77, 540–549. [Google Scholar] [CrossRef]
- Lessard, J.A.; Crabtree, G.R. Chromatin Regulatory Mechanisms in Pluripotency. Annu. Rev. Cell Dev. Biol. 2010, 26, 503–532. [Google Scholar] [CrossRef]
- Panamarova, M.; Cox, A.; Wicher, K.; Butler, R.; Bulgakova, N.; Jeon, S.; Rosen, B.; Seong, R.H.; Skarnes, W.; Crabtree, G.; et al. BAF chromatin remodelling complex is an epigenetic regulator of lineage specification in the early mouse embryo. Development 2016, 143, 1271–1283. [Google Scholar] [CrossRef]
- Xu, F.; Flowers, S.; Moran, E. Essential Role of ARID2 Protein-containing SWI/SNF Complex in Tissue-specific Gene Expression. J. Biol. Chem. 2012, 287, 5033–5041. [Google Scholar] [CrossRef]
- Lu, X.; Kovalev, G.I.; Chang, H.; Kallin, E.; Knudsen, G.; Xia, L.; Mishra, N.; Ruiz, P.; Li, E.; Su, L.; et al. Inactivation of NuRD Component Mta2 Causes Abnormal T Cell Activation and Lupus-like Autoimmune Disease in Mice. J. Biol. Chem. 2008, 283, 13825–13833. [Google Scholar] [CrossRef] [PubMed]
- Snider, A.C.; Leong, D.; Wang, Q.T.; Wysocka, J.; Yao, M.W.M.; Scott, M.P. The chromatin remodeling factor Chd1l is required in the preimplantation embryo. Biol. Open 2013, 2, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Nozawa, Y.; Tsukamoto, S.; Kaneko, T.; Manabe, I.; Imai, H.; Minami, N. CHD1 acts via the Hmgpi pathway to regulate mouse early embryogenesis. Development 2015, 142, 2375–2384. [Google Scholar] [CrossRef] [PubMed]
- Landry, J.; Sharov, A.A.; Piao, Y.; Sharova, L.V.; Xiao, H.; Southon, E.; Matta, J.; Tessarollo, L.; Zhang, Y.E.; Ko, M.S.H.; et al. Essential Role of Chromatin Remodeling Protein Bptf in Early Mouse Embryos and Embryonic Stem Cells. PLoS Genet. 2008, 4, e1000241. [Google Scholar] [CrossRef] [PubMed]
- Stopka, T.; Skoultchi, A.I. The ISWI ATPase Snf2h is required for early mouse development. Proc. Natl. Acad. Sci. USA 2003, 100, 14097–14102. [Google Scholar] [CrossRef] [PubMed]
- Yip, D.J.; Corcoran, C.P.; Alvarez-Saavedra, M.; DeMaria, A.; Rennick, S.; Mears, A.J.; Rudnicki, M.A.; Messier, C.; Picketts, D.J. Snf2l Regulates Foxg1-Dependent Progenitor Cell Expansion in the Developing Brain. Dev. Cell 2012, 22, 871–878. [Google Scholar] [CrossRef] [PubMed]
- Gopinathan, G.; Diekwisch, T.G.H. Epigenetics and Early Development. J. Dev. Biol. 2022, 10, 26. [Google Scholar] [CrossRef]
- Hota, S.K.; Bruneau, B.G. ATP-dependent chromatin remodeling during mammalian development. Development 2016, 143, 2882–2897. [Google Scholar] [CrossRef]
- Du, Z.; Zheng, H.; Huang, B.; Ma, R.; Wu, J.; Zhang, X.; He, J.; Xiang, Y.; Wang, Q.; Li, Y.; et al. Allelic reprogramming of 3D chromatin architecture during early mammalian development. Nature 2017, 547, 232–235. [Google Scholar] [CrossRef]
- Borsos, M.; Perricone, S.M.; Schauer, T.; Pontabry, J.; de Luca, K.L.; de Vries, S.S.; Ruiz-Morales, E.R.; Torres-Padilla, M.-E.; Kind, J. Genome–lamina interactions are established de novo in the early mouse embryo. Nature 2019, 569, 729–733. [Google Scholar] [CrossRef]
- Collombet, S.; Ranisavljevic, N.; Nagano, T.; Varnai, C.; Shisode, T.; Leung, W.; Piolot, T.; Galupa, R.; Borensztein, M.; Servant, N.; et al. Parental-to-embryo switch of chromosome organization in early embryogenesis. Nature 2020, 580, 142–146. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Zheng, H.; Kawamura, Y.K.; Zhang, K.; Gassler, J.; Powell, S.; Xu, Q.; Lin, Z.; Xu, K.; Zhou, Q.; et al. Polycomb Group Proteins Regulate Chromatin Architecture in Mouse Oocytes and Early Embryos. Mol. Cell 2020, 77, 825–839.e7. [Google Scholar] [CrossRef] [PubMed]
- Barbero, G.; de Sousa Serro, M.G.; Perez Lujan, C.; Vitullo, A.D.; González, C.R.; González, B. Transcriptome profiling of histone writers/erasers enzymes across spermatogenesis, mature sperm and pre-cleavage embryo: Implications in paternal epigenome transitions and inheritance mechanisms. Front. Cell Dev. Biol. 2023, 11, 1086573. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.H.; Wang, H.-L.V.; Ruiz, D.; Bixler, B.J.; Linsenbaum, H.; Xiang, J.-F.; Forestier, S.; Shafik, A.M.; Jin, P.; Corces, V.G. Recruitment of CTCF to an Fto enhancer is responsible for transgenerational inheritance of BPA-induced obesity. Proc. Natl. Acad. Sci. USA 2022, 119, e2214988119. [Google Scholar] [CrossRef] [PubMed]
- Aoki, F.; Worrad, D.M.; Schultz, R.M. Regulation of Transcriptional Activity during the First and Second Cell Cycles in the Preimplantation Mouse Embryo. Dev. Biol. 1997, 181, 296–307. [Google Scholar] [CrossRef] [PubMed]
- Worrad, D.M.; Ram, P.T.; Schultz, R.M. Regulation of gene expression in the mouse oocyte and early preimplantation embryo: Developmental changes in Sp1 and TATA box-binding protein, TBP. Development 1994, 120, 2347–2357. [Google Scholar] [CrossRef] [PubMed]
- Schultz, R.M. The molecular foundations of the maternal to zygotic transition in the preimplantation embryo. Hum. Reprod. Update 2002, 8, 323–331. [Google Scholar] [CrossRef]
- De Iaco, A.; Planet, E.; Coluccio, A.; Verp, S.; Duc, J.; Trono, D. DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat. Genet. 2017, 49, 941–945. [Google Scholar] [CrossRef]
- Kaltschmidt, C.; Greiner, J.F.W.; Kaltschmidt, B. The Transcription Factor NF-κB in Stem Cells and Development. Cells 2021, 10, 2042. [Google Scholar] [CrossRef]
- Martello, G.; Smith, A. The Nature of Embryonic Stem Cells. Annu. Rev. Cell Dev. Biol. 2014, 30, 647–675. [Google Scholar] [CrossRef]
- Schulz, W.A.; Hoffmann, M.J. Transcription Factor Networks in Embryonic Stem Cells and Testicular Cancer and the Definition of Epigenetics. Epigenetics 2007, 2, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Oldfield, A.J.; Yang, P.; Conway, A.E.; Cinghu, S.; Freudenberg, J.M.; Yellaboina, S.; Jothi, R. Histone-Fold Domain Protein NF-Y Promotes Chromatin Accessibility for Cell Type-Specific Master Transcription Factors. Mol. Cell 2014, 55, 708–722. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Ji, S.-Y.; Dang, Y.-J.; Sha, Q.-Q.; Yuan, Y.-F.; Zhou, J.-J.; Yan, L.-Y.; Qiao, J.; Tang, F.; Fan, H.-Y. Oocyte-expressed yes-associated protein is a key activator of the early zygotic genome in mouse. Cell Res. 2016, 26, 275–287. [Google Scholar] [CrossRef] [PubMed]
- Whiddon, J.L.; Langford, A.T.; Wong, C.-J.; Zhong, J.W.; Tapscott, S.J. Conservation and innovation in the DUX4-family gene network. Nat. Genet. 2017, 49, 935–940. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhang, Y. Loss of DUX causes minor defects in zygotic genome activation and is compatible with mouse development. Nat. Genet. 2019, 51, 947–951. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Zhang, Y.; Zhou, J.; Bi, Y.; Xu, J.; Xu, C.; Kou, X.; Zhao, Y.; Li, Y.; Tu, Z.; et al. Precise temporal regulation of Dux is important for embryo development. Cell Res. 2019, 29, 956–959. [Google Scholar] [CrossRef] [PubMed]
- Eckersley-Maslin, M.; Alda-Catalinas, C.; Blotenburg, M.; Kreibich, E.; Krueger, C.; Reik, W. Dppa2 and Dppa4 directly regulate the Dux-driven zygotic transcriptional program. Genes Dev. 2019, 33, 194–208. [Google Scholar] [CrossRef]
- Zhang, W.; Chen, F.; Chen, R.; Xie, D.; Yang, J.; Zhao, X.; Guo, R.; Zhang, Y.; Shen, Y.; Göke, J.; et al. Zscan4c activates endogenous retrovirus MERVL and cleavage embryo genes. Nucleic Acids Res. 2019, 47, 8485–8501. [Google Scholar] [CrossRef]
- Bulut-Karslioglu, A.; De La Rosa-Velázquez, I.A.; Ramirez, F.; Barenboim, M.; Onishi-Seebacher, M.; Arand, J.; Galán, C.; Winter, G.E.; Engist, B.; Gerle, B.; et al. Suv39h-Dependent H3K9me3 Marks Intact Retrotransposons and Silences LINE Elements in Mouse Embryonic Stem Cells. Mol. Cell 2014, 55, 277–290. [Google Scholar] [CrossRef]
- Rebollo, R.; Miceli-Royer, K.; Zhang, Y.; Farivar, S.; Gagnier, L.; Mager, D.L. Epigenetic interplay between mouse endogenous retroviruses and host genes. Genome Biol. 2012, 13, R89. [Google Scholar] [CrossRef]
- Andreu, M.J.; Alvarez-Franco, A.; Portela, M.; Gimenez-Llorente, D.; Cuadrado, A.; Badia-Careaga, C.; Tiana, M.; Losada, A.; Manzanares, M. Establishment of 3D chromatin structure after fertilization and the metabolic switch at the morula-to-blastocyst transition require CTCF. Cell Rep. 2022, 41, 111501. [Google Scholar] [CrossRef] [PubMed]
- Patty, B.J.; Hainer, S.J. Transcription factor chromatin profiling genome-wide using uliCUT&RUN in single cells and individual blastocysts. Nat. Protoc. 2021, 16, 2633–2666. [Google Scholar] [CrossRef] [PubMed]
- Dixon, J.R.; Selvaraj, S.; Yue, F.; Kim, A.; Li, Y.; Shen, Y.; Hu, M.; Liu, J.S.; Ren, B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012, 485, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Phillips-Cremins, J.E.; Sauria, M.E.G.; Sanyal, A.; Gerasimova, T.I.; Lajoie, B.R.; Bell, J.S.K.; Ong, C.-T.; Hookway, T.A.; Guo, C.; Sun, Y.; et al. Architectural Protein Subclasses Shape 3D Organization of Genomes during Lineage Commitment. Cell 2013, 153, 1281–1295. [Google Scholar] [CrossRef] [PubMed]
- Bell, A.C.; Felsenfeld, G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 2000, 405, 482–485. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.M.; Rabaia, N.A.; Smith, L.E.; Fagerlie, S.; Gurley, K.; Loukinov, D.; Disteche, C.M.; Collins, S.J.; Kemp, C.J.; Lobanenkov, V.V.; et al. Loss of Maternal CTCF Is Associated with Peri-Implantation Lethality of Ctcf Null Embryos. PLoS ONE 2012, 7, e34915. [Google Scholar] [CrossRef] [PubMed]
- Wan, L.-B.; Pan, H.; Hannenhalli, S.; Cheng, Y.; Ma, J.; Fedoriw, A.; Lobanenkov, V.; Latham, K.E.; Schultz, R.M.; Bartolomei, M.S. Maternal depletion of CTCF reveals multiple functions during oocyte and preimplantation embryo development. Development 2008, 135, 2729–2738. [Google Scholar] [CrossRef]
- Skene, P.J.; Henikoff, S. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife 2017, 6, e21856. [Google Scholar] [CrossRef]
- Crews, D.; Gillette, R.; Scarpino, S.V.; Manikkam, M.; Savenkova, M.I.; Skinner, M.K. Epigenetic transgenerational inheritance of altered stress responses. Proc. Natl. Acad. Sci. USA 2012, 109, 9143–9148. [Google Scholar] [CrossRef]
- Wolstenholme, J.T.; Drobná, Z.; Henriksen, A.D.; Goldsby, J.A.; Stevenson, R.; Irvin, J.W.; Flaws, J.A.; Rissman, E.F. Transgenerational Bisphenol A Causes Deficits in Social Recognition and Alters Postsynaptic Density Genes in Mice. Endocrinology 2019, 160, 1854–1867. [Google Scholar] [CrossRef]
- Sabzevari, S.; Rohbani, K.; Sadat-Shirazi, M.-S.; Babhadi-Ashar, N.; Shakeri, A.; Ashabi, G.; Khalifeh, S.; Ale-Ebrahim, M.; Zarrindast, M.-R. Morphine exposure before conception affects anxiety-like behavior and CRF level (in the CSF and plasma) in the adult male offspring. Brain Res. Bull. 2019, 144, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Xu, N.; Lei, L.; Lin, Y.; Ju, L.-S.; Morey, T.E.; Gravenstein, N.; Yang, J.; Martynyuk, A.E. A Methyltransferase Inhibitor (Decitabine) Alleviates Intergenerational Effects of Paternal Neonatal Exposure to Anesthesia With Sevoflurane. Anesth. Analg. 2020, 131, 1291–1299. [Google Scholar] [CrossRef] [PubMed]
- Rompala, G.R.; Finegersh, A.; Homanics, G.E. Paternal preconception ethanol exposure blunts hypothalamic-pituitary-adrenal axis responsivity and stress-induced excessive fluid intake in male mice. Alcohol 2016, 53, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Farah Naquiah, M.Z.; James, R.J.; Suratman, S.; Lee, L.S.; Mohd Hafidz, M.I.; Salleh, M.Z.; Teh, L.K. Transgenerational effects of paternal heroin addiction on anxiety and aggression behavior in male offspring. Behav. Brain Funct. 2016, 12, 23. [Google Scholar] [CrossRef]
- Lassi, M.; Tomar, A.; Comas-Armangué, G.; Vogtmann, R.; Dijkstra, D.J.; Corujo, D.; Gerlini, R.; Darr, J.; Scheid, F.; Rozman, J.; et al. Disruption of paternal circadian rhythm affects metabolic health in male offspring via nongerm cell factors. Sci. Adv. 2021, 7, eabg6424. [Google Scholar] [CrossRef]
- Mashoodh, R.; Habrylo, I.B.; Gudsnuk, K.; Champagne, F.A. Sex-specific effects of chronic paternal stress on offspring development are partially mediated via mothers. Horm. Behav. 2023, 152, 105357. [Google Scholar] [CrossRef]
| Study | Stressor | Epimodification Affected in Sperm | Epimodification Affected in Offspring (Organ) | Paternal Phenotype | Offspring Phenotype | Causality Checked | ||
|---|---|---|---|---|---|---|---|---|
| Behavioural | Metabolic | Behavioural | Metabolic | |||||
| Franklin et al., 2010 [58] | MSUS | DNA methylation | DNA methylation (sperm) | + | N.A. | + | N.A. | N.A. |
| Rodgers et al., 2013 [44] | chronic variable stress | miRNA | N.A. | N.A. | N.A. | + | + | N.A. |
| Gapp et al., 2014 [47] | MSUS | miRNA, tRF, piRNA, DNA methylation | miRNA (serum) | + | + | + | + | RNA injection |
| Rodgers et al., 2015 [59] | chronic variable stress | miRNA | − | N.A. | N.A. | N.A. | + | RNA injection |
| Gapp et al., 2016 [60] | MSUS | DNA methylation | DNA methylation (brain) | + | N.A. | + | N.A. | N.A. |
| Wu et al., 2016 [61] | chronic restraint | DNA methylation | DNA methylation (brain) | N.A. | + | N.A. | + | N.A. |
| Dickson et al., 2018 [62] | chronic social instability | miRNA | miRNA (embryo) | N.A. | N.A. | N.A. | N.A. | embryonic miRNA |
| Gapp et al., 2020 [63] | MSUS | lncRNA | lncRNA (zygote) | + | N.A. | + | + | RNA injection |
| Gapp et al., 2021 [64] | Dexamethasone | miRNA, tRF, rRNA, circRNA | tRF | N.A. | N.A. | N.A. | + | embryonic tRF |
| Cunningham et al., 2021 [65] | chronic social defeat | lncRNA | N.A. | + | N.A. | + | N.A. | N.A. |
| Kong et al., 2021 [66] | chronic restraint | DNA methylation | DNA methylation (brain) | − | − | + | N.A. | N.A. |
| Y. Wang et al., 2021 [67] | chronic variable stress | miRNA, piRNA | − | + | + | + | + | RNA injection, antisense strand RNA injection |
| X. Zheng et al., 2021 [68] | chronic restraint | DNA methylation | DNA methylation (embryo) | + | + | + | + | N.A. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kretschmer, M.; Fischer, V.; Gapp, K. When Dad’s Stress Gets under Kid’s Skin—Impacts of Stress on Germline Cargo and Embryonic Development. Biomolecules 2023, 13, 1750. https://doi.org/10.3390/biom13121750
Kretschmer M, Fischer V, Gapp K. When Dad’s Stress Gets under Kid’s Skin—Impacts of Stress on Germline Cargo and Embryonic Development. Biomolecules. 2023; 13(12):1750. https://doi.org/10.3390/biom13121750
Chicago/Turabian StyleKretschmer, Miriam, Vincent Fischer, and Katharina Gapp. 2023. "When Dad’s Stress Gets under Kid’s Skin—Impacts of Stress on Germline Cargo and Embryonic Development" Biomolecules 13, no. 12: 1750. https://doi.org/10.3390/biom13121750
APA StyleKretschmer, M., Fischer, V., & Gapp, K. (2023). When Dad’s Stress Gets under Kid’s Skin—Impacts of Stress on Germline Cargo and Embryonic Development. Biomolecules, 13(12), 1750. https://doi.org/10.3390/biom13121750






