SSR Genotyping and Marker–Trait Association with Yield Components in a Kazakh Germplasm Collection of Chickpea (Cicer arietinum L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Field Experiments
2.2. Molecular Analysis
2.3. Data Analysis
3. Results
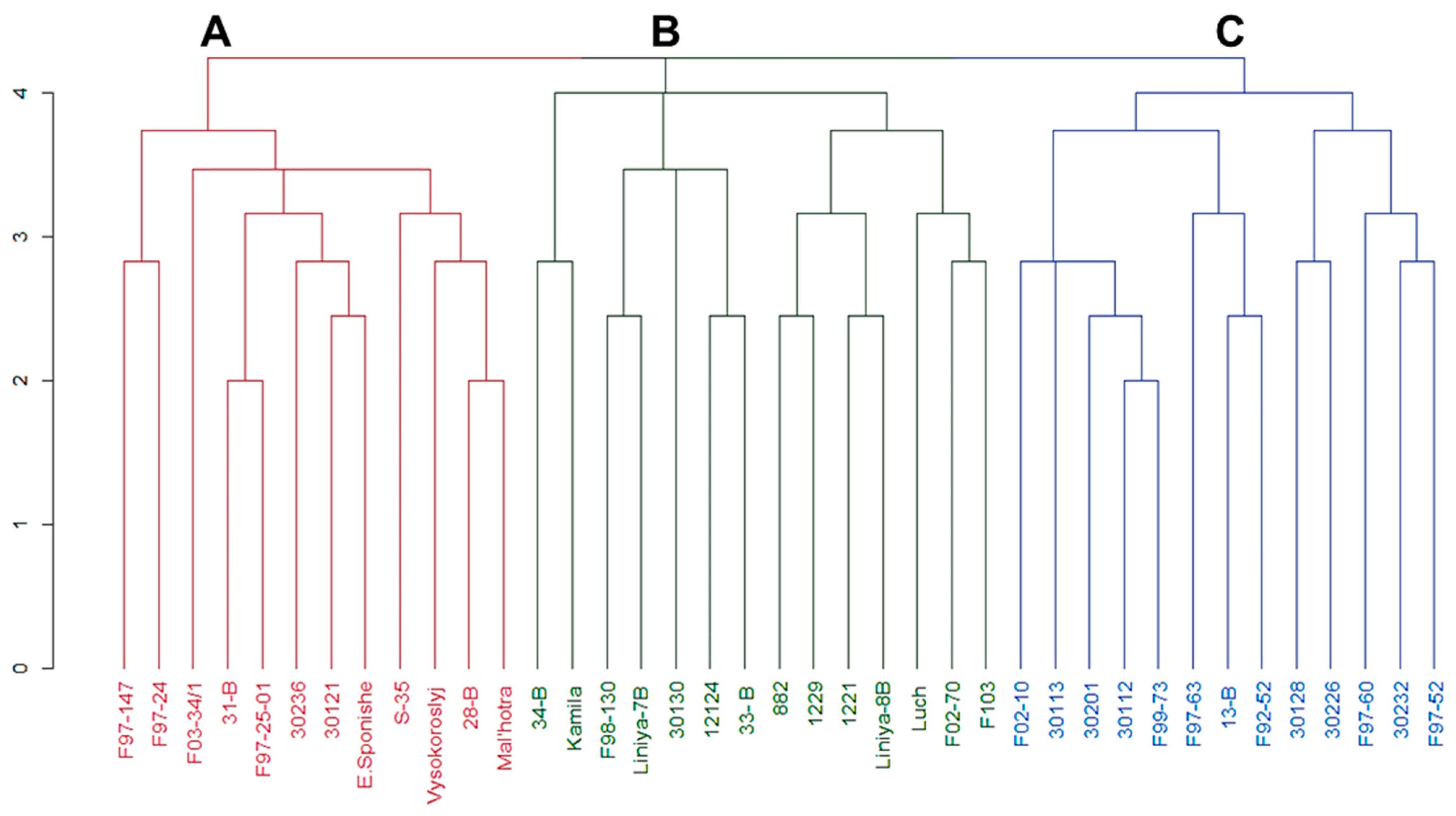
3.1. Genetic Diversity of Chickpea Germplasm Collection
3.2. Cluster Analysis
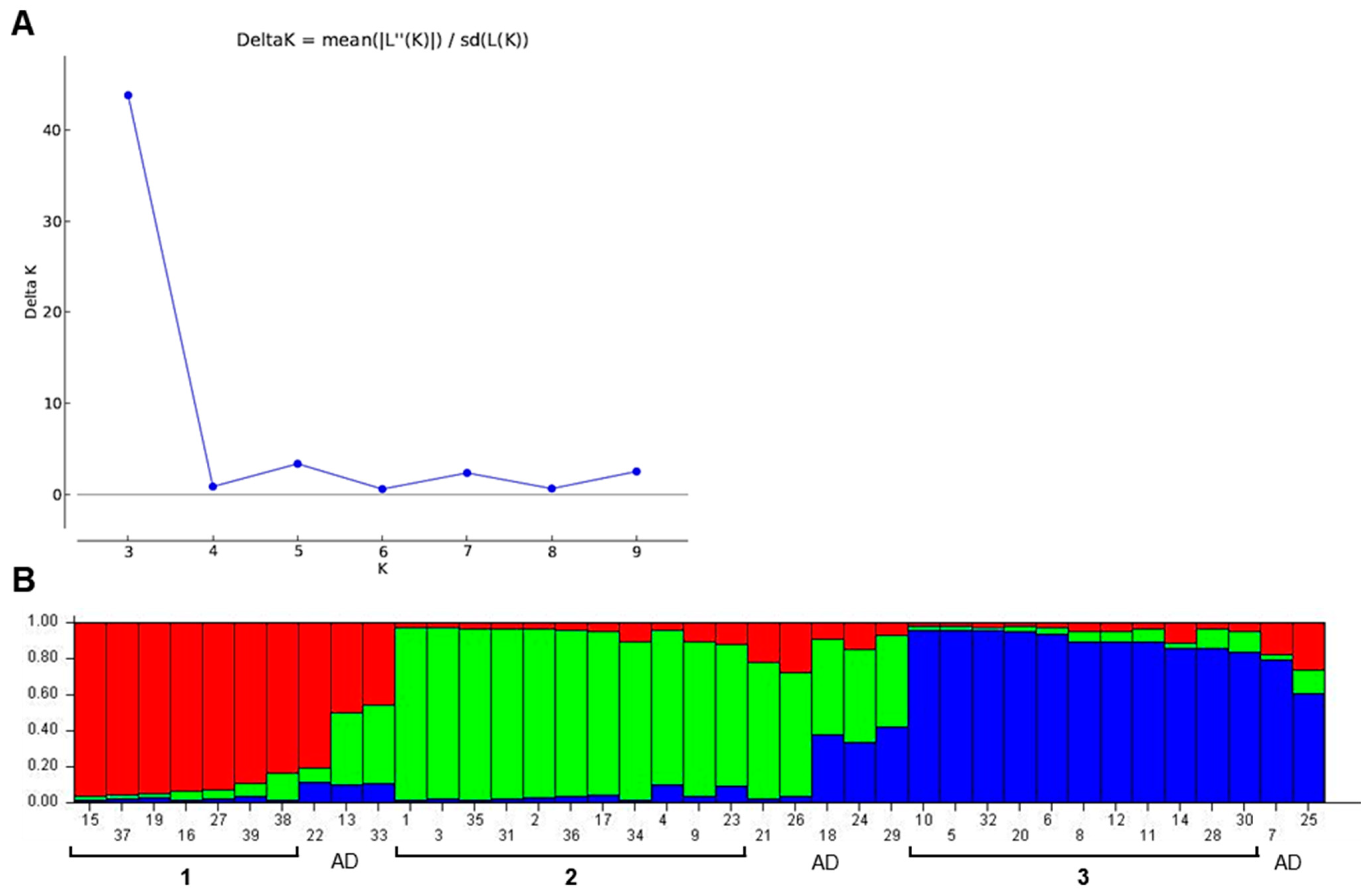
3.3. Population Structure Analysis
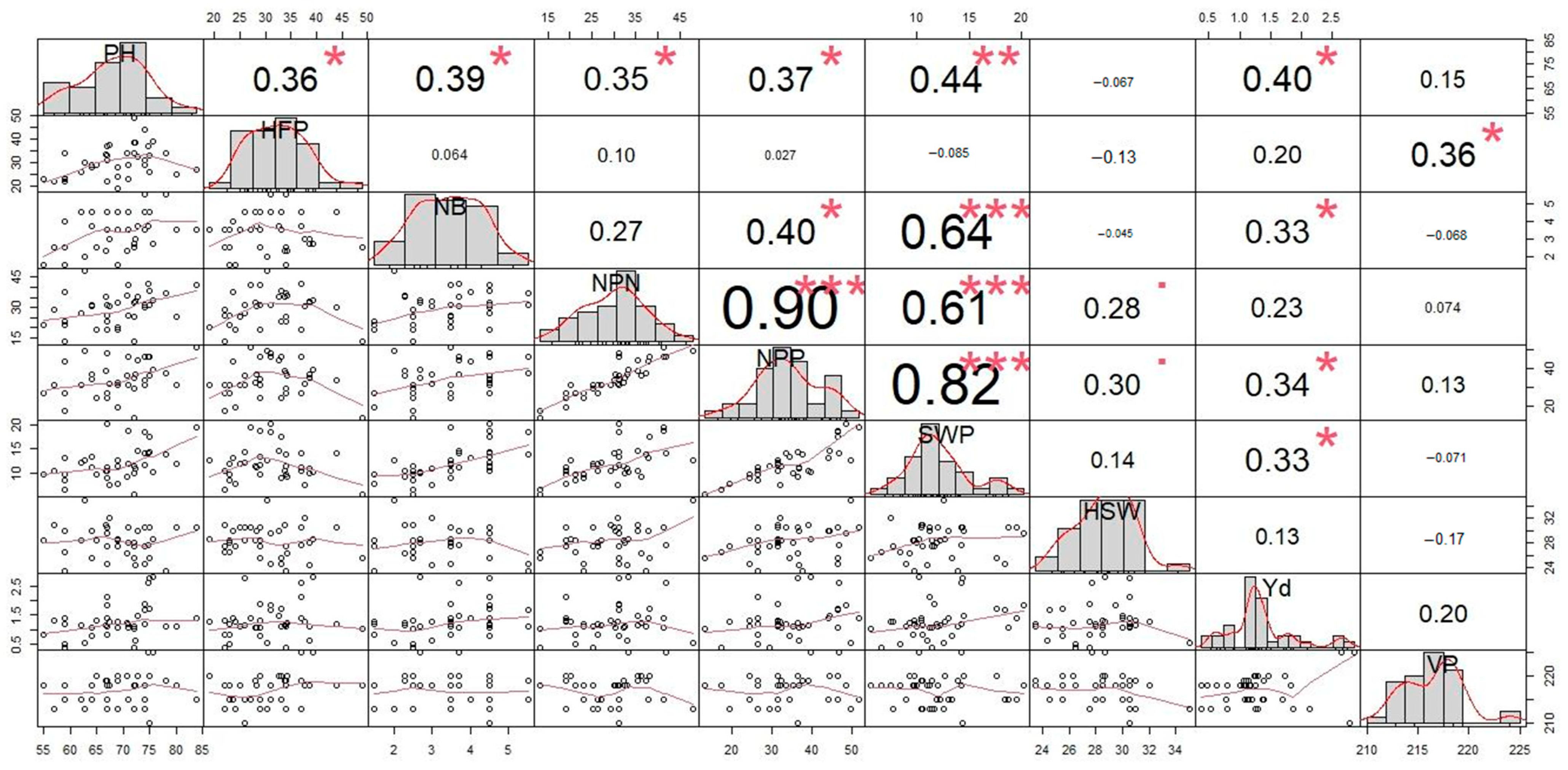
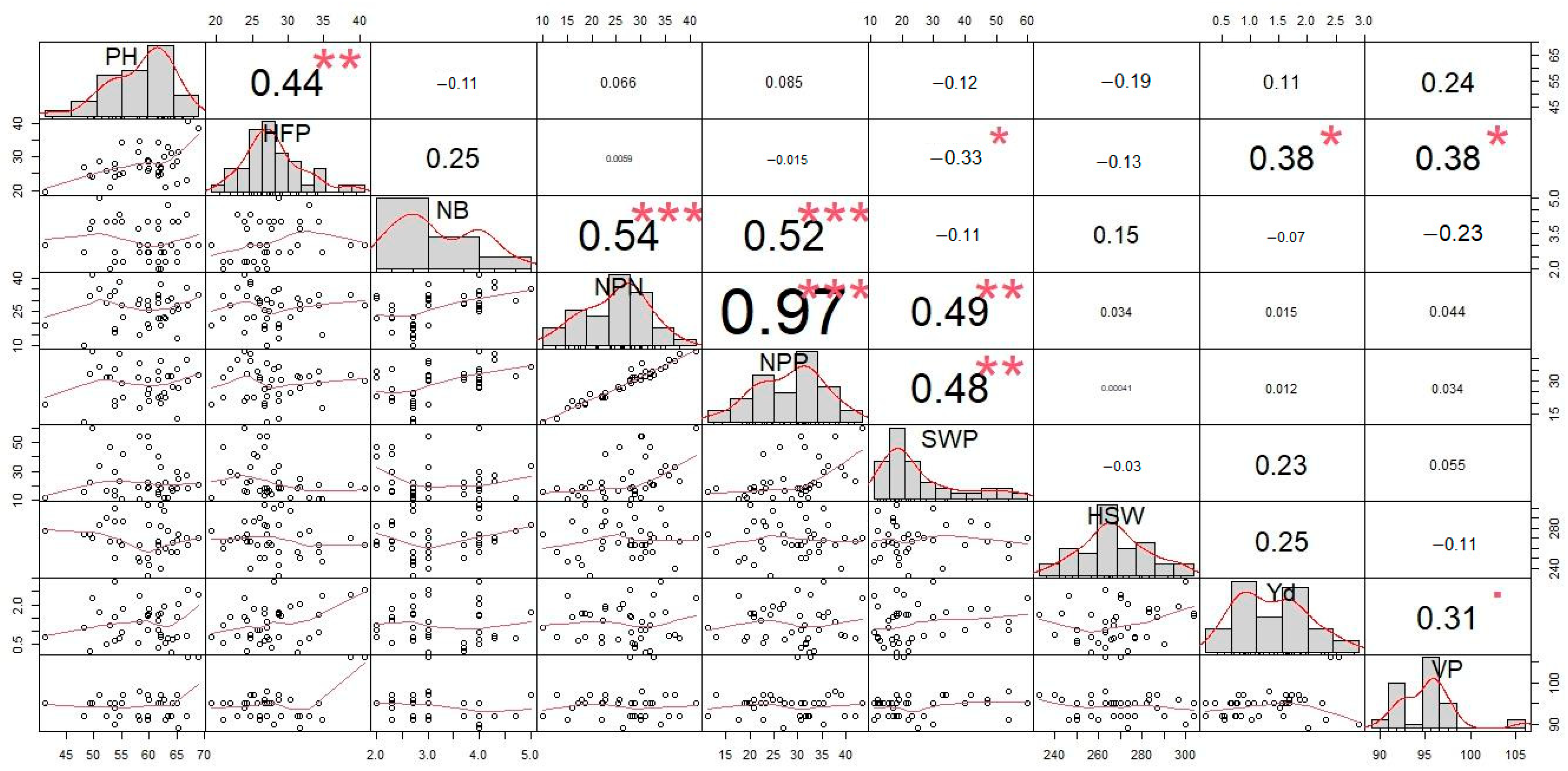
3.4. Correlation Analysis
3.5. Marker–Trait Association (MTA) Analysis between SSR Markers and Morphological Traits
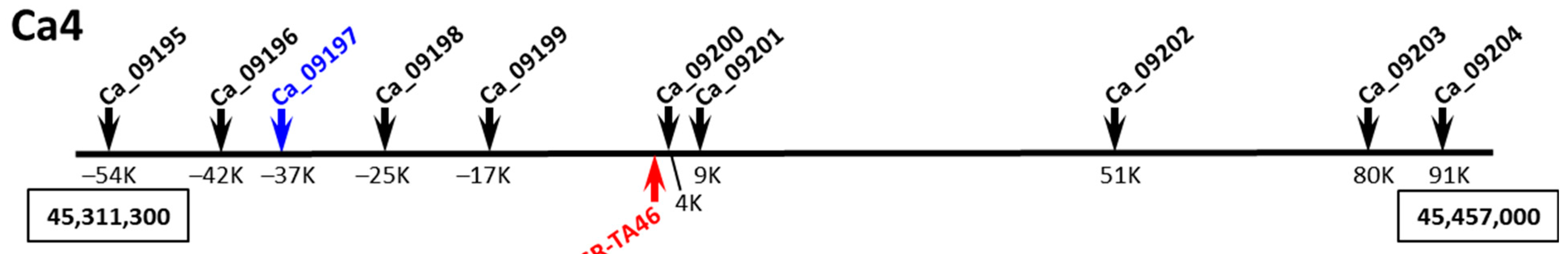
3.6. Potential Candidate Gene Identification
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Toker, C.; Berger, J.; Eker, T.; Sari, D.; Sari, H.; Gokturk, R.S.; Kahraman, A.; Aydin, B.; von Wettberg, E.J. Cicer turcicum: A new Cicer species and its potential to improve chickpea. Front. Plant Sci. 2021, 12, 662891. [Google Scholar] [CrossRef]
- Stat. Gov. Kazakhstan. Available online: https://stat.gov.kz/ru/industries/economy/national-/publications/5099 (accessed on 30 October 2023).
- Vinokurov, V.A. Technology of Chickpea Cultivation in Northern Kazakhstan; Ualikhan University: Kokshetau, Kazakhstan, 1999; 179p. (In Russian) [Google Scholar]
- Kadyrbekov, B.; Eshkebaev, K.; Kamzina, G. Selection of previously grown crops for chickpea cultivation for seed production. Bull. Shakarim Univ. Technol. Sci. 2020, 91, 319–322. (In Russian) [Google Scholar]
- Serekpayev, N.; Popov, V.; Stybayev, G.; Nogayev, A.; Ansabayeva, A. Agroecological aspects of chickpea growing in the dry steppe zone of Akmola region, Northern Kazakhstan. Biotechol. Res. Asia 2016, 13, 1341. [Google Scholar] [CrossRef]
- Shektybayeva, G.H.; Limaskaya, V.B.; Orynbayev, A.T.; Kasenova, A.S. Ecological variety testing of chickpeas in a changing climate in the West of Kazakhstan. E3S Web of Conf. 2023, 395, 02004. [Google Scholar] [CrossRef]
- Kudaybergenov, M.S.; Bulatova, K.M.; Baytarakova, K.; Mazkirat., S. Yield of chickpea collection samples at overwintering in the conditions of Southeastern Kazakhstan. Bull. Karaganda Univ. Biol. Med. Geogr. Ser. 2017, 88, 35–41. (In Russian) [Google Scholar]
- Singh, K.B.; Malhotra, R.S.; Saxena, M.C.; Bejiga, G. Superiority of winter sowing over traditional spring sowing of chickpea in the Mediterranean region. Agron. J. 1997, 89, 112–118. [Google Scholar] [CrossRef]
- Siddique, K.; Loss, S.; Regan, K.; Jettner, R. Adaptation and seed yield of cool season grain legumes in Mediterranean environments of south-western Australia. Aust. J. Agric. Res. 1999, 50, 375–388. [Google Scholar] [CrossRef]
- Iliadis, C. Evaluation of six chickpea varieties for seed yield under autumn and spring sowing. J. Agric. Sci. 2001, 137, 439–444. [Google Scholar] [CrossRef]
- Morison JI, L.; Butterfield, R.E. Cereal crop damage by frosts, spring 1990. Weather 1990, 45, 308–313. [Google Scholar] [CrossRef]
- O’Toole, N.; Stoddard, F.L.; O’Brien, L. Screening of chickpea for adaptation to autumn sowing. J. Agron. Crop Sci. 2001, 186, 193–207. [Google Scholar] [CrossRef]
- Valimohammai, F.; Tajbakhsh, M.; Saeid, A. Comparison winter and spring sowing dates and effect of plant density on yield, yield components and some quality, morphological traits of chickpea (Cicer arietinum L.) under environmental condition of Urmia, Iran. J. Agron. 2007, 6, 571–575. [Google Scholar] [CrossRef][Green Version]
- Mart, D. Investigation of the morphological characteristics of chickpea (Cicer arietinum L.) cultivars cultivated under irrigated and non-irrigated conditions sown in winter and early spring. Turk. J. Range Forage Sci. 2022, 3, 75–83. [Google Scholar] [CrossRef]
- Rubiales, D.; Moral, A.; Flores, F. Performance of winter-sown chickpea breeding lines with contrasting levels of resistance to Ascochyta blight. Agronomy 2022, 12, 2194. [Google Scholar] [CrossRef]
- Sharma, R. Planting Chickpea in October Shows Promise in the Cold Winter Dessert Climate of Uzbekistan; International Center for Agricultural Research in the Dry Areas (ICARDA): Beirut, Lebanon, 2020.
- Rani, A.; Devi, P.; Jha, U.C.; Sharma, K.D.; Siddique, K.H.M.; Nayyar, H. Developing climate-resilient chickpea involving physiological and molecular approaches with a focus on temperature and drought stresses. Front. Plant Sci. 2020, 10, 1759. [Google Scholar] [CrossRef]
- Hassaneian, K.H.; Lotfi, R. Advanced breeding approaches for cold-tolerant chickpea and lentil in dryland areas. In Legumes Research; Jimenez-Lopez, J.C., Clemente, A., Eds.; IntechOpen: London, UK, 2022. [Google Scholar] [CrossRef]
- Upadhyaya, H.D.; Dwivedi, S.L.; Baum, M.; Varshney, R.K.; Udupa, S.M.; Gowda, C.L.L. Genetic structure, diversity, and allelic richness in composite collection and reference set in chickpea (Cicer arietinum L.). BMC Plant Biol. 2008, 8, 106. [Google Scholar] [CrossRef]
- Singh, R.K.; Singh, C.; Ambika, C.B.S.; Mahto, R.K.; Patial, R.; Gupta, A.; Gahlaut, V.; Gayacharan, H.A.; Upadhyaya, H.D.; Kumar, R. Exploring chickpea germplasm diversity for broadening the genetic base utilizing genomic resources. Front. Genet. 2022, 13, 905771. [Google Scholar] [CrossRef]
- Hajibarat, Z.; Saidi, A.; Hajibarat, Z.; Talebi, R. Characterization of genetic diversity in chickpea using SSR markers, start codon targeted polymorphism (SCoT) and conserved DNA-derived polymorphism (CDDP). Physiol. Mol. Biol. Plants 2015, 21, 365–373. [Google Scholar] [CrossRef]
- Duhan, N.; Kaundal, R. LegumeSSRdb: A comprehensive microsatellite marker database of legumes for germplasm characterization and crop improvement. Int. J. Mol. Sci. 2021, 22, 11350. [Google Scholar] [CrossRef]
- Fayaz, H.; Mir, A.H.; Tyagi, S.; Wani, A.A.; Jan, N.; Yasin, M.; Mir, J.I.; Mondal, B.; Khan, M.A.; Mir, R.R. Assessment of molecular genetic diversity of 384 chickpea genotypes and development of core set of 192 genotypes for chickpea improvement programs. Genet. Resour. Crop Evol. 2021, 69, 1193–1205. [Google Scholar] [CrossRef]
- Jha, U.C.; Jha, R.; Thakro, V.; Kumar, A.; Gupta, S.; Nayyar, H.; Basu, P.; Parida, S.K.; Singh, N.P. Discerning molecular diversity and association mapping for phenological, physiological and yield traits under high temperature stress in chickpea (Cicer arietinum L.). J. Genet. 2021, 100, 4. [Google Scholar] [CrossRef]
- Zhou, R.; Yang, S.; Zhang, B.; Qi, Z.; Xin, D.; Su, A.; Li, S.; Cheng, P.; Bai, Y.; Yin, Z.; et al. Analysis of the genetic diversity of grain legume germplasm resources in China and the development of universal SSR primers. Biotechnol. Biotechnol. Equip. 2021, 35, 1706–1721. [Google Scholar] [CrossRef]
- Gore, P.G.; Gupta, V.; Singh, R.; Tripathi, K.; Kumar, R.; Kumari, G.; Madhavan, L.; Dikshit, H.K.; Venkateswaran, K.; Pandey, A.; et al. Insights into the genetic diversity of an underutilized Indian legume, Vigna stipulacea (Lam.) Kuntz., using morphological traits and microsatellite markers. PLoS ONE 2022, 17, e0262634. [Google Scholar] [CrossRef]
- Jannat, S.; Shah, A.H.; ul Hassan, M.; Sher, A.; Fiaz, S.; Elesawy, B.H.; Ismail, K.A.; El Askary, A.; Gharib, A.F.; Qayyum, A. Genetic diversity of common bean (Phaseolus vulgaris L.) ecotypes from Pakistan using Simple Sequence Repeats. Saudi J. Biol. Sci. 2022, 29, 103300. [Google Scholar] [CrossRef] [PubMed]
- Özkan, G.; Haliloğlu, K.; Türkoğlu, A.; Özturk, H.I.; Elkoca, E.; Poczai, P. Determining genetic diversity and population structure of common bean (Phaseolus vulgaris L.) landraces from Turkey using SSR markers. Genes 2022, 13, 1410. [Google Scholar] [CrossRef]
- Islam, M.A.; Alam, M.S.; Maniruzzaman, M.; Haque, M.S. Microsatellite marker-based genetic diversity assessment among exotic and native maize inbred lines of Bangladesh. Saudi J. Biol. Sci. 2023, 30, 103715. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Graner, A.; Sorrells, M.E. Genic microsatellite markers in plants: Features and applications. Trends Biotechnol. 2005, 23, 48–55. [Google Scholar] [CrossRef]
- Hüttel, B.; Winter, P.; Weising, K.; Choumane, W.; Weigand, F.; Kahl, G. Sequence-tagged microsatellite site markers for chickpea (Cicer arietinum L.). Genome 1999, 42, 210–217. [Google Scholar] [CrossRef]
- Winter, P.; Pfaff, T.; Udupa, S.M.; Hüttel, B.; Sharma, P.C.; Sahi, S.; Arreguin-Espinoza, R.; Weigand, F.; Muehlbauer, F.J.; Kahl, G. Characterization and mapping of sequence-tagged microsatellite sites in the chickpea (Cicer arietinum L.) genome. Mol. Gen. Genet. 1999, 262, 90–101. [Google Scholar] [CrossRef]
- Sethy, N.K.; Shokeen, B.; Bhatia, S. Isolation and characterization of sequence-tagged microsatellite sites markers in chickpea (Cicer arietinum L.). Mol. Ecol. Notes 2003, 3, 428–430. [Google Scholar] [CrossRef]
- Buhariwalla, H.K.; Eshwar, K.; Crouch, J.H. Development of ESTs from chickpea roots and their use in diversity analysis of the Cicer genus. BMC Plant Biol. 2005, 5, 16. [Google Scholar] [CrossRef]
- Naghavi, M.R.; Monfared, S.R.; Humberto, G. Genetic diversity in Iranian chickpea (Cicer arietinum L.) landraces as revealed by microsatellite markers. Czech J. Genet. Plant Breed. 2012, 48, 131–138. [Google Scholar] [CrossRef]
- Varshney, R.; Thudi, M.; Upadhyaya, H.; Dwivedi, S.; Udupa, S.; Furman, B.; Baum, M.; Hoisington, D. A SSR kit to study genetic diversity in chickpea (Cicer arietinum L.). Plant Genet. Resour. 2014, 12, S118–S120. [Google Scholar] [CrossRef]
- Bhattarai, G.; Shi, A.; Kandel, D.R.; Solis-Gracia, N.; Da Silva, J.A.; Avila, C.A. Genome-wide simple sequence repeats (SSR) markers discovered from whole-genome sequence comparisons of multiple spinach accessions. Sci. Rep. 2021, 11, 9999. [Google Scholar] [CrossRef]
- Sari, D.; Sari, H.; Ikten, C.; Toker, C. Genome-wide discovery of di-nucleotide SSR markers based on whole genome re-sequencing data of Cicer arietinum L. and Cicer reticulatum Ladiz. Sci. Rep. 2023, 13, 10351. [Google Scholar] [CrossRef]
- Nayak, S.N.; Zhu, H.; Varghese, N.; Datta, S.; Choi, H.K.; Horres, R.; Jüngling, R.; Singh, J.; Kishor, P.B.K.; Sivaramakrishnan, S.; et al. Integration of novel SSR and gene-based SNP marker loci in the chickpea genetic map and establishment of new anchor points with Medicago truncatula genome. Theor. Appl. Genet. 2010, 120, 1415–1441. [Google Scholar] [CrossRef]
- Mason, A.S. SSR genotyping. In Plant Genotyping: Methods and Protocols. Methods in Molecular Biology; Batley, J., Ed.; Humana: New York, NY, USA, 2015; Volume 1245, pp. 77–89. [Google Scholar] [CrossRef]
- UPOV. Available online: https://www.upov.int/edocs/tgdocs/en/tg143.pdf (accessed on 30 October 2023).
- Climate Classification. Wikipedia. Available online: https://en.wikipedia.org/wiki/K%C3%B6ppen_climate_classification (accessed on 30 October 2023).
- Climate in Kazakhstan. Climate Data. Available online: https://ru.climate-data.org (accessed on 30 October 2023).
- Vishniyakova, M.A.; Seferova, I.V.; Buravtseva, T.V.; Burlyaeva, M.O.; Semenova, E.V.; Filipenko, G.I.; Drugova, E.V. VIR Global Collection of Grain Legume Crop Genetic Resources: Replenishment, Conservation, and Study. Methodological Guidelines; VIR Publishers: St.-Petersburg, Russia, 2010. (In Russian) [Google Scholar]
- Lassner, M.W.; Peterson, P.; Yoder, J.I. Simultaneous amplification of multiple DNA fragments by polymerase chain reaction in the analysis of transgenic plants and their progeny. Plant Mol. Biol. Rep. 1989, 7, 116–128. [Google Scholar] [CrossRef]
- Kimura, M.; Crow, J.F. The number of alleles that can be maintained in a finite population. Genetics 1964, 49, 725. [Google Scholar] [CrossRef]
- Nei, M. Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA 1973, 70, 3321–3323. [Google Scholar] [CrossRef]
- Serrote, C.M.L.; Reiniger, L.R.S.; Silva, K.B.; Rabaiolli, S.M.D.S.; Stefanel, C.M. Determining the polymorphism information content of a molecular marker. Gene 2020, 726, 144175. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Muse, S.V. PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef] [PubMed]
- R_Core_Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2023; Available online: https://www.R-project.org (accessed on 30 October 2023).
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- LIS: Legume Information System. Available online: https://www.legumeinfo.org/tools (accessed on 30 October 2023).
- Mir, A.H.; Bhat, M.A.; Fayaz, H.; Wani, A.A.; Dar, S.A.; Maqbool, S.; Yasin, M.; Mir, J.I.; Khan, M.A.; Sofi, P.A.; et al. SSR markers in revealing extent of genetic diversity and phylogenetic relationships among chickpea core collection accessions for Western Himalayas. Mol. Biol. Rep. 2022, 49, 11469–11479. [Google Scholar] [CrossRef]
- Agrama, H.; Tuinstra, M. Phylogenetic diversity and relationships among sorghum accessions using SSRs and RAPDs. Afr. J. Biotechnol. 2003, 2, 334–340. [Google Scholar] [CrossRef]
- Zhou, H.F.; Xie, Z.W.; Ge, S. Microsatellite analysis of genetic diversity and population genetic structure of a wild rice (Oryza rufipogon Griff.) in China. Theor. Appl. Genet. 2003, 107, 332–339. [Google Scholar] [CrossRef] [PubMed]
- Segura-Alabart, N.; Serratosa, F.; Gómez, S.; Fernández, A. Nonunique UPGMA clusterings of microsatellite markers. Brief. Bioinform. 2022, 23, bbac312. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, N.A.; Mahajan, S.; Ramachandran, S.; Zhao, C.; Pritchard, J.K.; Feldman, M.W. Clines, clusters, and the effect of study design on the inference of human population structure. PLoS Genet. 2005, 1, e70. [Google Scholar] [CrossRef]
- Pocovi, M.I.; Mariotti, J.A. A Bayesian approach to inferring the genetic population structure of sugarcane accessions from INTA (Argentina). Chil. J. Agric. Res. 2015, 75, 152–159. [Google Scholar] [CrossRef]
- Stift, M.; Kolář, F.; Meirmans, P.G. Structure is more robust than other clustering methods in simulated mixed-ploidy populations. Heredity 2019, 123, 429–441. [Google Scholar] [CrossRef]
- Kumar, L.; Arora, P. Multivariate analysis in chickpea. Ind. J. Pulses Res. 1992, 5, 1–5. [Google Scholar]
- Lahiri, N.; Kumar, T.; Bharadwaj, C.; Sarker, A.; Rizvi, A.H.; Chauhan, S.K.; Verma, A.K.; Prasad, G. Diversity analysis among chickpea genetic stock as revealed through STMS marker analysis. Ind. J. Plant Genet. Resour. 2015, 28, 189–197. [Google Scholar] [CrossRef]
- Samyuktha, S.; Kannan, B.J.; Geethanjali, S. Molecular genetic diversity and population structure analysis in chickpea (Cicer arietinum L.) germplasm using SSR markers. Int. J. Curr. Microbiol. Appl. Sci. 2018, 7, 639–651. [Google Scholar] [CrossRef]
- Admas, S.; Tesfaye, K.; Haileselassie, T.; Shiferaw, E.; Flynn, K.C. Phenotypic variability of chickpea (Cicer arietinum L) germplasm with temporally varied collection from the Amhara Regional State, Ethiopia. Cogent Food Agric. 2021, 7, 1896117. [Google Scholar] [CrossRef]
- Sivasakthi, K.; Thudi, M.; Tharanya, M.; Kale, S.M.; Kholová, J.; Halime, M.H.; Jaganathan, D.; Baddam, R.; Thirunalasundari, T.; Gaur, P.M.; et al. Plant vigour QTLs co-map with an earlier reported QTL hotspot for drought tolerance while water saving QTLs map in other regions of the chickpea genome. BMC Plant Biol. 2018, 18, 29. [Google Scholar] [CrossRef] [PubMed]
- Astarak, H.; Sharifi, P.; Pouresmael, M. Correlation and path analysis for grain yield and yield components in chickpea (Cicer arietinum L.). Genetika 2017, 49, 273–284. [Google Scholar] [CrossRef]
- Shedge, P.J.; Patil, D.K.; Dawane, J.K. Correlation and path coefficient analysis of yield and yield components in chickpea (Cicer arietinum L.). Int. J. Curr. Microbiol. Appl. Sci. 2019, 8, 1326–1333. [Google Scholar] [CrossRef]
- Toker, C.; Cagirgan, M.I. The use of phenotypic correlations and factor analysis in determining characters for grain yield selection in chickpea (Cicer arietinum L.). Hereditas 2004, 140, 226–228. [Google Scholar] [CrossRef]
- Nezami, A.; Nabati, J.; Mirmiran, S.M.; Hasanfard, A.; Mohammadi, M. How does the freezing stress in the seedling stage affect the chickpea’s morpho-physiological and biochemical attributes? Gesunde Pflanz. 2023, 75, 1107–1119. [Google Scholar] [CrossRef]
- Kazhydromet. Republican State Enterprise. Review of the Climate Features on the Territory of Kazakhstan; Astana, Kazakhstan, 2023; 40p, (In Russian). Available online: https://www.kazhydromet.kz/ru/klimat/obzor-ob-osobennostyah-klimata-na-territorii-kazahstana (accessed on 30 October 2023).
- Al-Rifaee, M.; Al-Tawaha, A.; Ismael, F. Doubling chickpea yield by shifting from spring to winter sowing using Ascochyta blight resistant lines under typical Mediterranean climate. Biosci. Res. 2005, 2, 80–85. [Google Scholar]
- Özdemir, S.; Karadavut, U. Comparison of the performance of autumn and spring sowing of chickpeas in a temperate region. Turk. J. Agric. Forest. 2003, 27, 345–352. [Google Scholar]
- Gupta, S.; Kumar, T.; Verma, S.; Bharadwaj, C.; Bhatia, S. Development of gene-based markers for use in construction of the chickpea (Cicer arietinum L.) genetic linkage map and identification of QTLs associated with seed weight and plant height. Mol. Biol. Rep. 2015, 42, 1571–1580. [Google Scholar] [CrossRef] [PubMed]
- Radhika, P.; Gowda, S.J.M.; Kadoo, N.Y.; Mhase, L.B.; Jamadagni, B.M.; Sainani, M.N.; Chandra, S.; Gupta, V.S. Development of an integrated intraspecific map of chickpea (Cicer arietinum L.) using two recombinant inbred line populations. Theor. Appl. Genet. 2007, 115, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Sabbavarapu, M.M.; Sharma, M.; Chamarthi, S.K.; Swapna, N.; Rathore, A.; Thudi, M.; Gaur, P.M.; Pande, S.; Singh, S.; Kaur, L.; et al. Molecular mapping of QTLs for resistance to Fusarium wilt (race 1) and Ascochyta blight in chickpea (Cicer arietinum L.). Euphytica 2013, 193, 121–133. [Google Scholar] [CrossRef]
- Patil, B.S.; Ravikumar, R.L.; Bhat, J.S.; Soregaon, C.D. Molecular mapping of QTLs for resistance to early and late Fusarium wilt in chickpea. Czech J. Genet. Plant Breed. 2014, 50, 171–176. [Google Scholar] [CrossRef]


| Entry N | Name of Accessions | Origin | Morphological Characteristics | ||||
|---|---|---|---|---|---|---|---|
| Time to Flowering | Flower Color | Seed Color | Seed Shape | Seed Ribbing | |||
| 1 | 882 | Türkiye | medium | white | yellow | round | absent |
| 2 | 1221 | Russia | medium | white | grey-brown | round to angular | weak to medium |
| 3 | 1229 | Moldova | medium | white | grey-brown | round to angular | weak to medium |
| 4 | 12124 | Kazakhstan | medium | white | grey-brown | round to angular | weak |
| 5 | 30112 | ICARDA | early | white | grey-brown | round to angular | weak |
| 6 | 30113 | ICARDA | early | white | grey-brown | round to angular | medium |
| 7 | 30121 | ICARDA | early | white | grey-brown | round to angular | medium |
| 8 | 30128 | ICARDA | early | white | grey-brown | round to angular | medium |
| 9 | 30130 | ICARDA | early | white | grey-brown | round to angular | medium |
| 10 | 30201 | ICARDA | early | white | grey-brown | round to angular | medium |
| 11 | 30226 | ICARDA | early | white | grey-brown | round to angular | medium |
| 12 | 30232 | ICARDA | early | white | grey-brown | round to angular | medium |
| 13 | 30236 | ICARDA | early | white | grey-brown | round to angular | medium |
| 14 | 13-B | Syria | early | white | rose-brown | round to angular | weak to medium |
| 15 | 28-B | Ukraine | white | brown | angular | strong | |
| 16 | 31-B | Türkiye | medium | white | grey-brown | round to angular | medium |
| 17 | 33-B | Syria | medium | white | grey-brown | round to angular | weak to medium |
| 18 | 34-B | Morocco | medium | white | grey-brown | round to angular | medium |
| 19 | Ezbsen Sponishe | Germany | medium | white | grey-brown | round to angular | weak to medium |
| 20 | F02-10 | Kazakhstan | early | white | grey-brown | round to angular | medium |
| 21 | F02-70 | Kazakhstan | early | white | grey-brown | round to angular | medium |
| 22 | F03-34/1 | Kazakhstan | medium | white | grey-brown | round to angular | weak to medium |
| 23 | F103 | Kazakhstan | medium | white | rose-brown | round to angular | weak to medium |
| 24 | F92-52 | Kazakhstan | medium | white | rose-brown | round to angular | medium |
| 25 | F97-147 | Kazakhstan | medium | white | rose-brown | round to angular | medium |
| 26 | F97-24 | Kazakhstan | medium | white | grey-brown | round to angular | weak to medium |
| 27 | F97-25-01 | Kazakhstan | medium | white | rose-brown | round to angular | weak to medium |
| 28 | F97-52 | Kazakhstan | medium | white | grey-brown | round to angular | weak to medium |
| 29 | F97-60 | Kazakhstan | medium | white | grey-brown | round to angular | weak to medium |
| 30 | F97-63 | Kazakhstan | medium | white | grey-brown | round to angular | medium |
| 31 | F98-130 | Kazakhstan | early | white | grey-brown | round to angular | strong |
| 32 | F99-73 | Kazakhstan | early | white | rose-brown | round to angular | medium |
| 33 | Kamila | Kazakhstan | medium | white | yellow | round to angular | weak |
| 34 | Liniya-7B | Russia | medium | white | grey-brown | round | absent |
| 35 | Liniya-8B | Russia | medium | white | grey-brown | round | absent |
| 36 | Luch | Kazakhstan | medium | white | grey-brown | round | absent |
| 37 | Malhotra | Syria | late | purple-pink | brown | angular | strong |
| 38 | S-35 | Russia | medium | white | grey-brown | round to angular | medium |
| 39 | Vysokoroslyj | Azerbaijan | medium | white | grey-brown | round to angular | weak to medium |
| SSR Marker | Motif | Primer Sequence | LG | Amplicon (bp) |
|---|---|---|---|---|
| TA14 | (TAA)22 n(TAA)4 T(A)3 n(AAT)5 n(A)3 n(GAT)4 (TAA)5 | F: TGACTTGCTATTTAGGGAACA R: TGGCTAAAGACAATTAAAGTT | 6 | 144, 263–278 |
| TA22 | (ATT)40 | F: TCTCCAACCCTTTAGATTGA R: TCGTGTTTACTGAATGTGGA | 4 | 203–278 |
| TA46 | (TAA)22 | F: TTTATTGCAATAAAACTCATTTCTTATC R:TTCTTTTTGTGTGAAAAAAAAATATAGTGA | 6 | 69, 127–154 |
| TA71 | (AAT)32 | F: CGATTTAACACAAAACACAAA R: CCTATCCATTGTCATCTCGT | 5 | 138, 184–223 |
| TA76s | (AAT)7(AAT)4 [ACT(AAT)11]2 n(AAT)3 n(AAT)2 (ATT)5 | F: TCCTCTTCTTCGATATCATCA R: CCATTCTATCTTTGGTGCTT | 3 | 165, 203, 206, 212, 218 |
| TA142 | (TTA)15 | F: TGTTAACATTCCCTAATATCAATAACTT R: TTCCACAATGTTGTATGTTTTGTAAG | 7 | 84, 125–140 |
| NCPGR 4 | (CT)16 | F: TTACAGCTTGTGCTCAG R: AGTCAGATTCTTATCCGA | 6 | 180, 194, 196 |
| NCPGR 6 | (CA)12 | F: GACCAAGATTAGTAGAACCT R: TATGTCTACACCTATGCATC | 4 | 249, 251, 255 |
| NCPGR 7 | (CA)14 | F: GACCAAGATTAGTAGAACCT R: CTTGATAAGGATGAGTCATG | 4 | 217, 219, 223 |
| NCPGR 12 | (CT)35 | F: CCTTGTTAGTGTGTATAGGT R: GTAATGACCAAGTGAACA | 7 | 213–261 |
| NCPGR 19 | (GA)19 | F: TCCATTGTAGCTTAGCTTAG R: TCTTACTCTTAGCTTACCTCTT | 7 | 298–312 |
| SSR Marker | Amplicon (bp) in the Current Research | Amplicon (bp) in s | Reference |
|---|---|---|---|
| TA14 | 263, 278, 288, 300, 307 | 250 | [32] |
| 266, 272, 278 | [34] | ||
| 266, 272 | [31] | ||
| 263, 278 | [33] | ||
| 263, 266, 272, 278 | [36] | ||
| TA22 | 195, 203, 209, 218, 227, 236, 239, 245, 251, 263, 278 | 228 | [32] |
| 203, 209, 212 | [34] | ||
| 209, 278 | [31] | ||
| 206, 269 | [33] | ||
| 203, 206, 209, 212, 269, 278 | [36] | ||
| TA46 | 152, 155, 162, 164, 166, 171, 176, 186 | 152 | [32] |
| 142, 145 | [34] | ||
| 145, 148, 151 | [31] | ||
| 127, 154 | [33] | ||
| 127, 142, 145, 148, 151, 154 | [36] | ||
| TA71 | 184, 196, 202, 214, 223, 230, 237 | 225 | [32] |
| 196, 205, 214 | [34] | ||
| 196, 202, 223 | [31] | ||
| 184, 187, 202 | [33] | ||
| 184, 187, 196, 202, 205, 214, 223 | [36] | ||
| TA76s | 214, 218, 227, 230 | 206 | [32] |
| 203, 212, 218 | [34] | ||
| 212, 218 | [31] | ||
| 212, 218 | [33] | ||
| 203, 212, 218 | [36] | ||
| TA142 | 143, 147, 155, 174 | 135 | [32] |
| 131 | [33] | ||
| 125, 128, 137 | [31] | ||
| 134, 140 | [33] | ||
| 125, 128, 131, 134, 137, 140 | [36] | ||
| NCPGR4 | 174, 180, 186, 194, 198, 200 | 180, 194 | [34] |
| 194, 196 | [31] | ||
| 194, 195, 198 | [33] | ||
| 180, 194, 196 | [36] | ||
| NCPGR6 | 245 | 249, 251, 255 | [34] |
| 251, 255 | [31] | ||
| 245, 249, 251 | [33] | ||
| 249, 251, 255 | [36] | ||
| NCPGR7 | 211, 214, 217, 221 | 217, 219, 223 | [34] |
| 219, 223 | [31] | ||
| 217, 219, 222 | [33] | ||
| 217, 219,2 23 | [36] | ||
| NCPGR12 | 200 | 225, 259, 261 | [34] |
| 213, 253 | [31] | ||
| 235, 251, 255 | [33] | ||
| 213, 225, 253, 255, 259, 261 | [36] | ||
| NCPGR19 | 310, 312 | 298, 300, 308 | [34] |
| 298, 300, 308 | [31] | ||
| 306, 308, 312 | [33] | ||
| 298, 300, 308, 312 | [36] |
| SSR Marker | Major Allele Frequency | Observed Number of Alleles | Nei’s Gene Diversity (h) | Polymorphism Information Content, PIC |
|---|---|---|---|---|
| TA14 | 0.30 | 5 | 0.77 | 0.73 |
| TA22 | 0.18 | 11 | 0.87 | 0.86 |
| TA46 | 0.32 | 8 | 0.79 | 0.76 |
| TA71 | 0.24 | 7 | 0.83 | 0.80 |
| TA76s | 0.70 | 4 | 0.46 | 0.42 |
| TA142 | 0.48 | 4 | 0.63 | 0.57 |
| NCPGR4 | 0.36 | 6 | 0.76 | 0.72 |
| NCPGR7 | 0.52 | 4 | 0.64 | 0.58 |
| NCPGR19 | 0.62 | 2 | 0.47 | 0.36 |
| Mean | 0.41 | 5.7 | 0.69 | 0.65 |
| Traits | SSR Marker | Allele (Amplicon, bp) | Environments | p Value |
|---|---|---|---|---|
| PH | TA46 | 162 | Spring, 2016 | 0.042 * |
| TA46 | 162 | Spring, 2017 | 0.041 * | |
| HFP | TA142 | 147 | Autumn, 2016 | 0.0086 ** |
| TA71 | 223 | Autumn, 2017 | 0.038 * | |
| NB | TA14 | 278 | Autumn, 2017 | 0.029 * |
| NPN | TA142 | 155 | Autumn, 2016 | 0.01 * |
| TA142 | 155 | Autumn, 2017 | 0.0094 ** | |
| TA46 | 162 | Spring, 2016 | 0.04 * | |
| TA46 | 162 | Spring, 2017 | 0.04 * | |
| NPP | TA142 | 155 | Autumn, 2016 | 0.037 * |
| TA142 | 155 | Autumn, 2017 | 0.025 * | |
| TA46 | 162 | Spring, 2016 | 0.02 * | |
| TA46 | 162 | Spring, 2017 | 0.02 * | |
| NCPGR7 | 211 | Autumn, 2016 | 0.026 * | |
| SWP | TA71 | 230 | Spring, 2016 | 0.05 |
| TA71 | 230 | Spring, 2017 | 0.04 * | |
| NCPGR7 | 211 | Autumn, 2016 | 0.046 * | |
| HSW | TA71 | 237 | Autumn, 2016 | 0.0098 ** |
| TA71 | 237 | Autumn, 2017 | 0.01 * | |
| NCPGR7 | 221 | Spring, 2016 | 0.03 * | |
| NCPGR7 | 221 | Spring, 2017 | 0.04 * | |
| Yd | NCPGR7 | 211 | Autumn, 2016 | 0.034 * |
| NCPGR4 | 194 | Spring, 2016 | 0.033 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mazkirat, S.; Baitarakova, K.; Kudaybergenov, M.; Babissekova, D.; Bastaubayeva, S.; Bulatova, K.; Shavrukov, Y. SSR Genotyping and Marker–Trait Association with Yield Components in a Kazakh Germplasm Collection of Chickpea (Cicer arietinum L.). Biomolecules 2023, 13, 1722. https://doi.org/10.3390/biom13121722
Mazkirat S, Baitarakova K, Kudaybergenov M, Babissekova D, Bastaubayeva S, Bulatova K, Shavrukov Y. SSR Genotyping and Marker–Trait Association with Yield Components in a Kazakh Germplasm Collection of Chickpea (Cicer arietinum L.). Biomolecules. 2023; 13(12):1722. https://doi.org/10.3390/biom13121722
Chicago/Turabian StyleMazkirat, Shynar, Kuralay Baitarakova, Mukhtar Kudaybergenov, Dilyara Babissekova, Sholpan Bastaubayeva, Kulpash Bulatova, and Yuri Shavrukov. 2023. "SSR Genotyping and Marker–Trait Association with Yield Components in a Kazakh Germplasm Collection of Chickpea (Cicer arietinum L.)" Biomolecules 13, no. 12: 1722. https://doi.org/10.3390/biom13121722
APA StyleMazkirat, S., Baitarakova, K., Kudaybergenov, M., Babissekova, D., Bastaubayeva, S., Bulatova, K., & Shavrukov, Y. (2023). SSR Genotyping and Marker–Trait Association with Yield Components in a Kazakh Germplasm Collection of Chickpea (Cicer arietinum L.). Biomolecules, 13(12), 1722. https://doi.org/10.3390/biom13121722







