The Characterization of R2R3-MYB Genes in Ammopiptanthus nanus Uncovers That the miR858-AnaMYB87 Module Mediates the Accumulation of Anthocyanin under Osmotic Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Stress Treatment
2.2. Identification of R2R3-MYB Genes
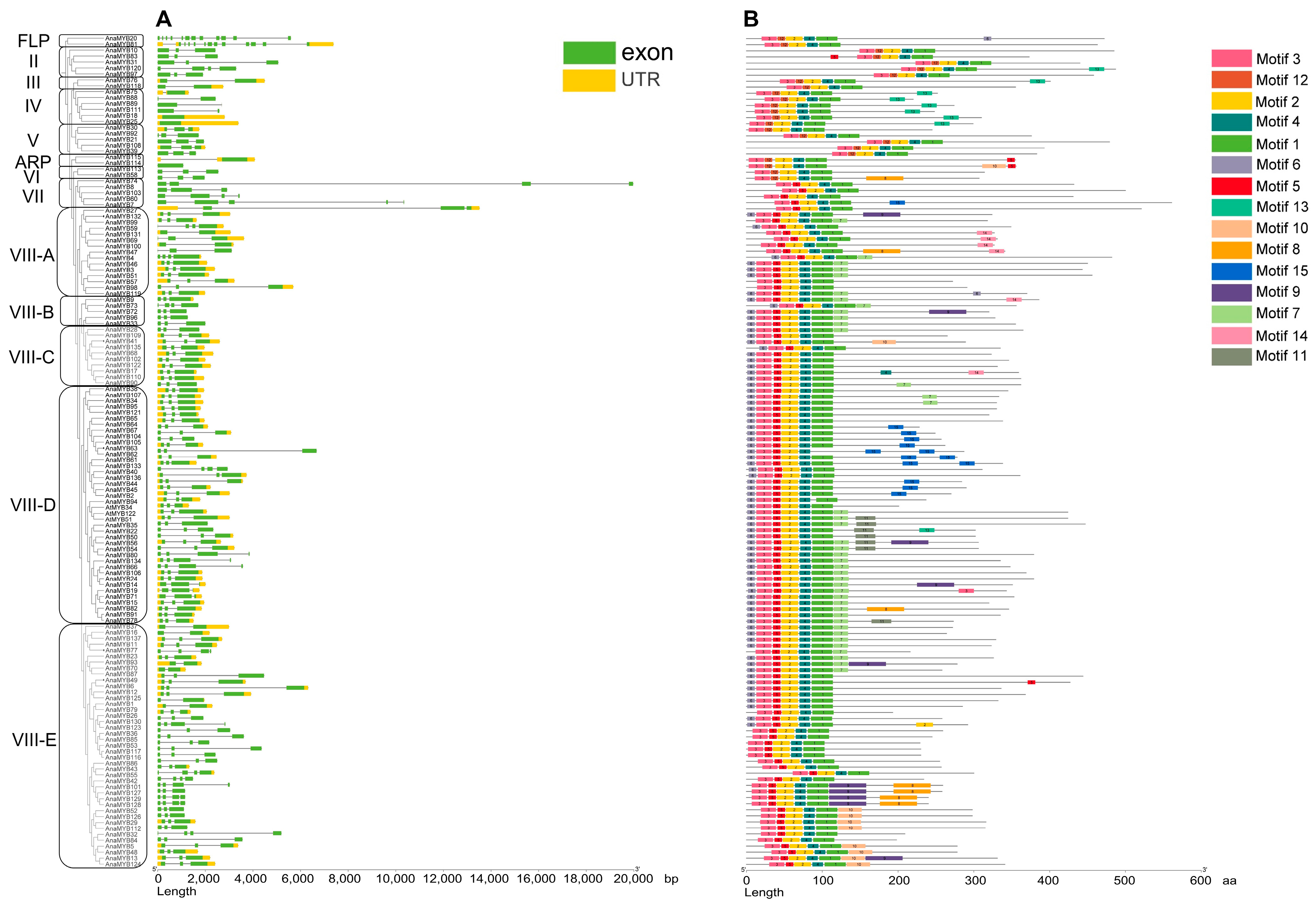
2.3. Structure Analysis of R2R3-MYB
2.4. Phylogenetic Analysis of R2R3-MYB
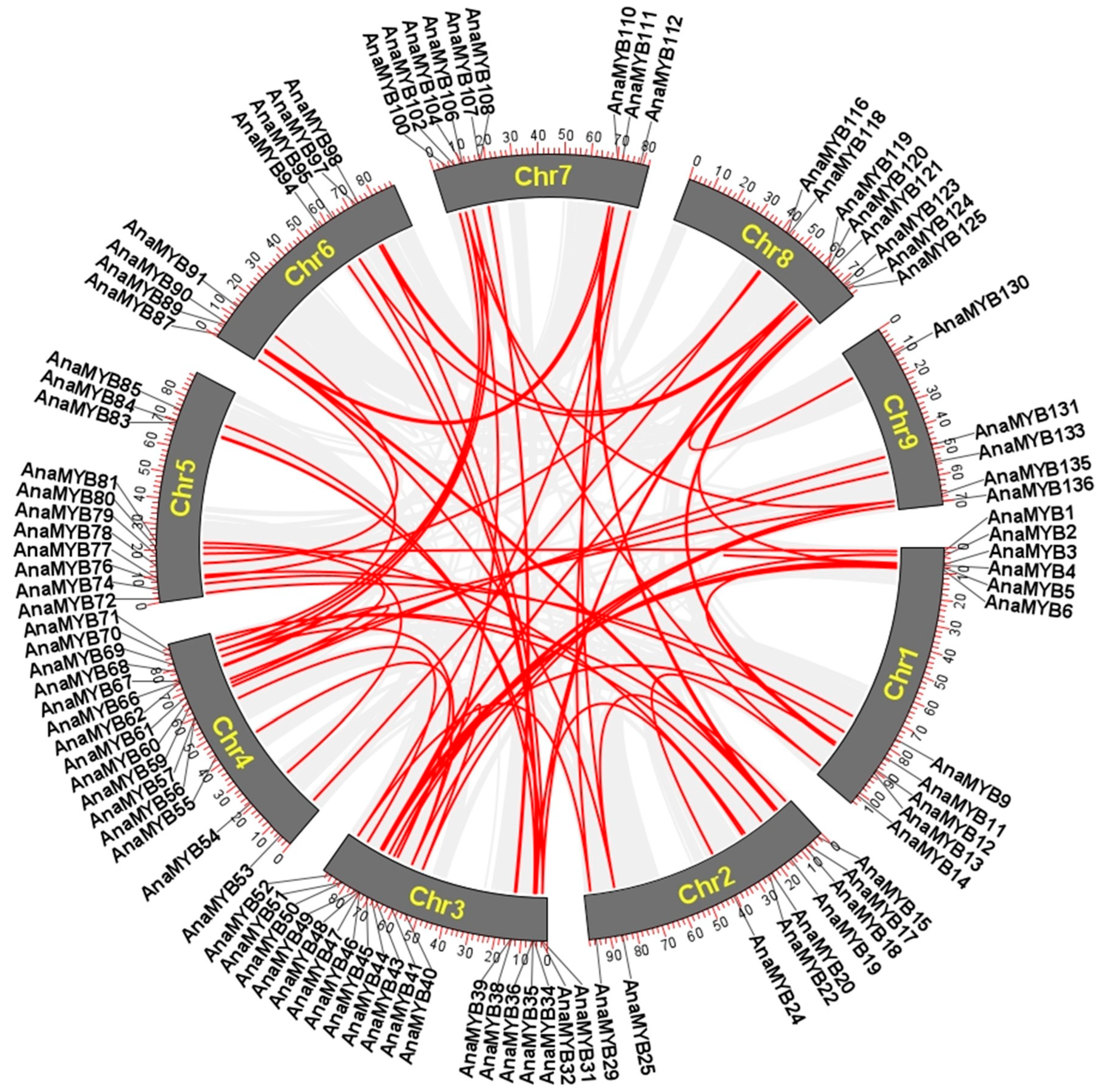
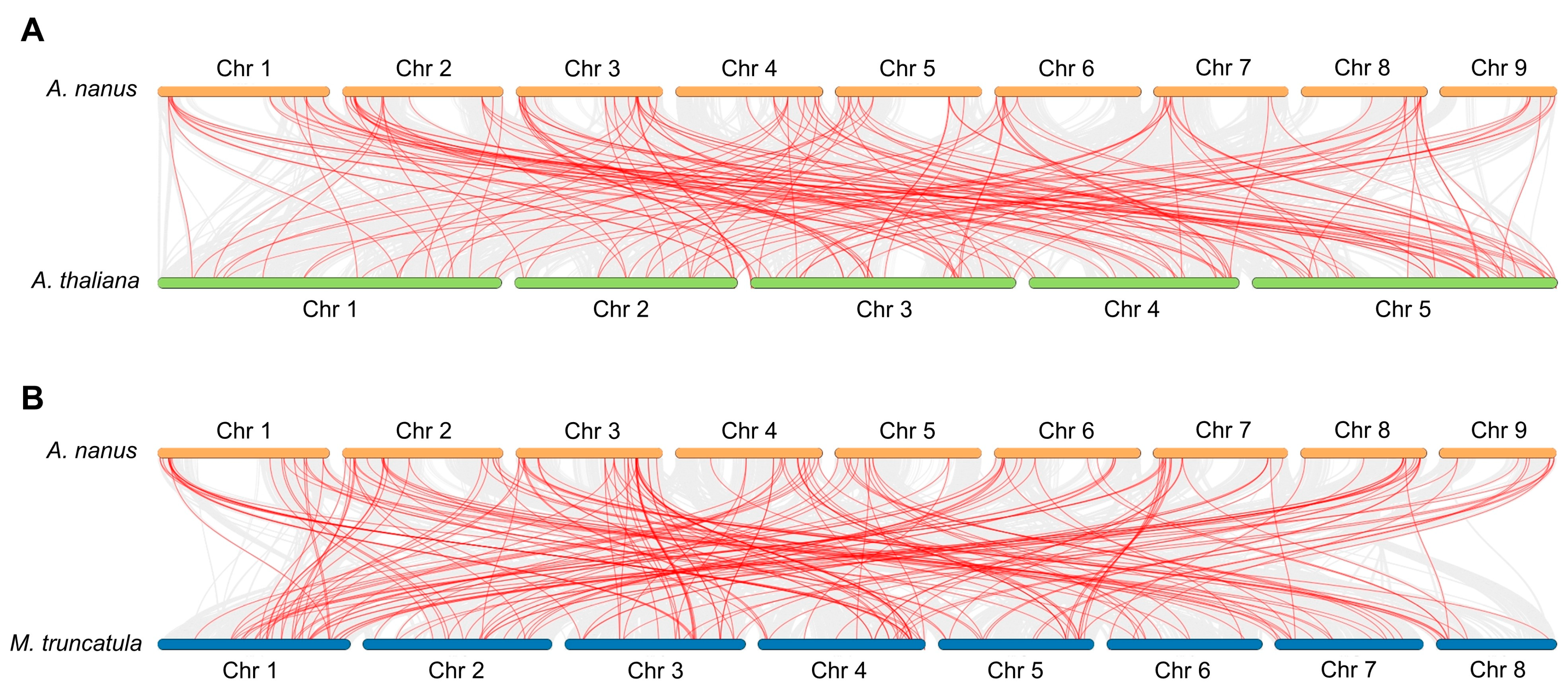
2.5. Gene Duplication Events Analysis
2.6. Identification of Cis-Acting Elements
2.7. Determination of MiRNA-Targeted R2R3-MYB Genes
2.8. Expression Analysis of R2R3-MYB Genes
2.9. qRT-PCR Analysis
2.10. Transactivation Activity Assay
2.11. Subcellular Localization Analysis
2.12. Transient Expression of AnaMYB87 in Apples
2.13. Dual-Luciferase Reporter Assay in Arabidopsis
2.14. Luciferase Reporter Assay in Tobacco Leaves
2.15. Determination of Anthocyanin Content
2.16. Statistical Methods
3. Results
3.1. Genome-Wide Identification of R2R3-MYB TFs
3.2. Phylogenetic Analysis of R2R3-MYB Family in A. nanus
3.3. Structural Feature of A. nanus R2R3-MYB TFs
3.4. Gene Duplication of R2R3-MYB TFs in A. nanus
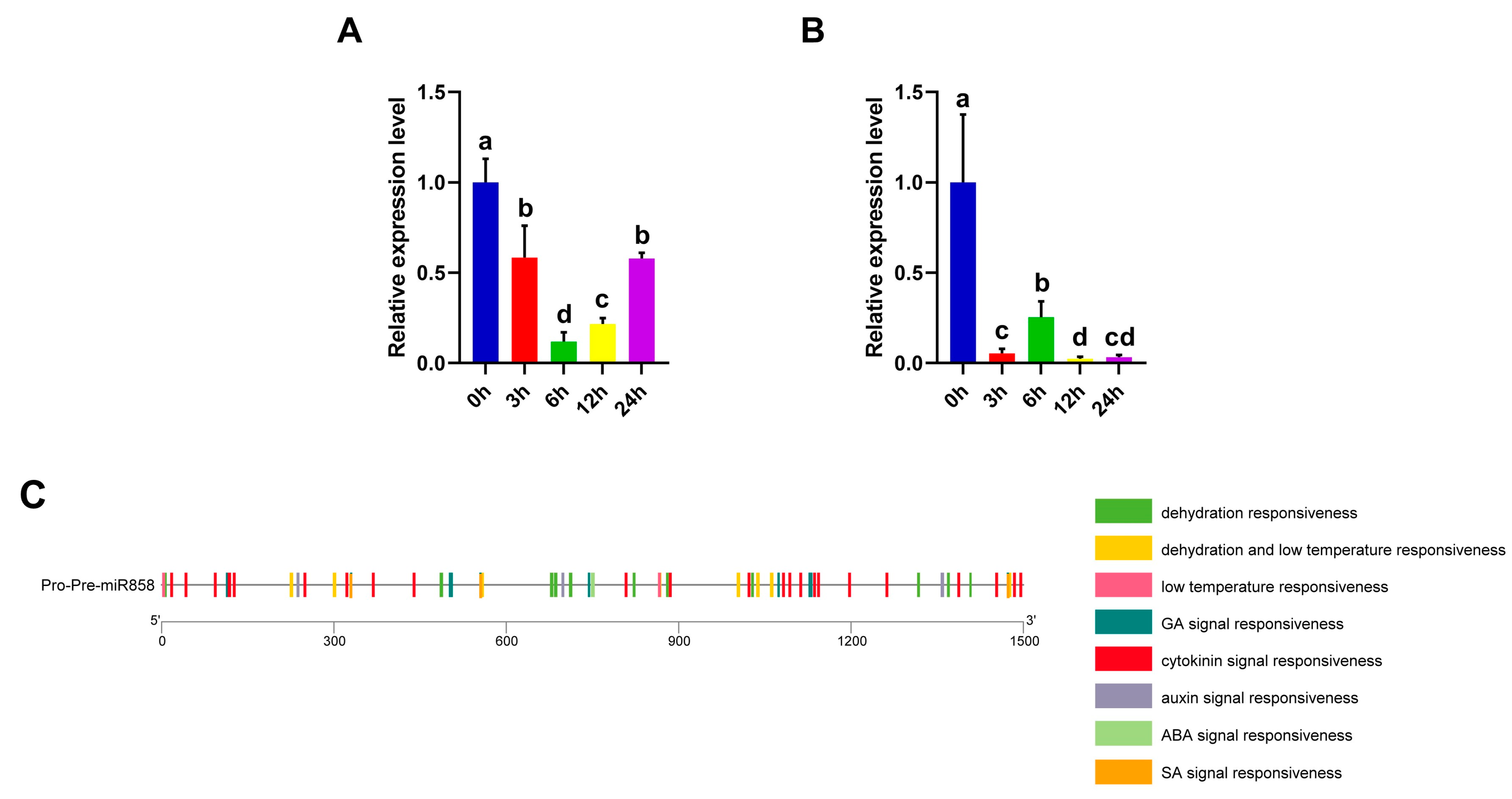
3.5. Cis-Acting Element Analysis of the Promoters of A. nanus R2R3-MYB Genes
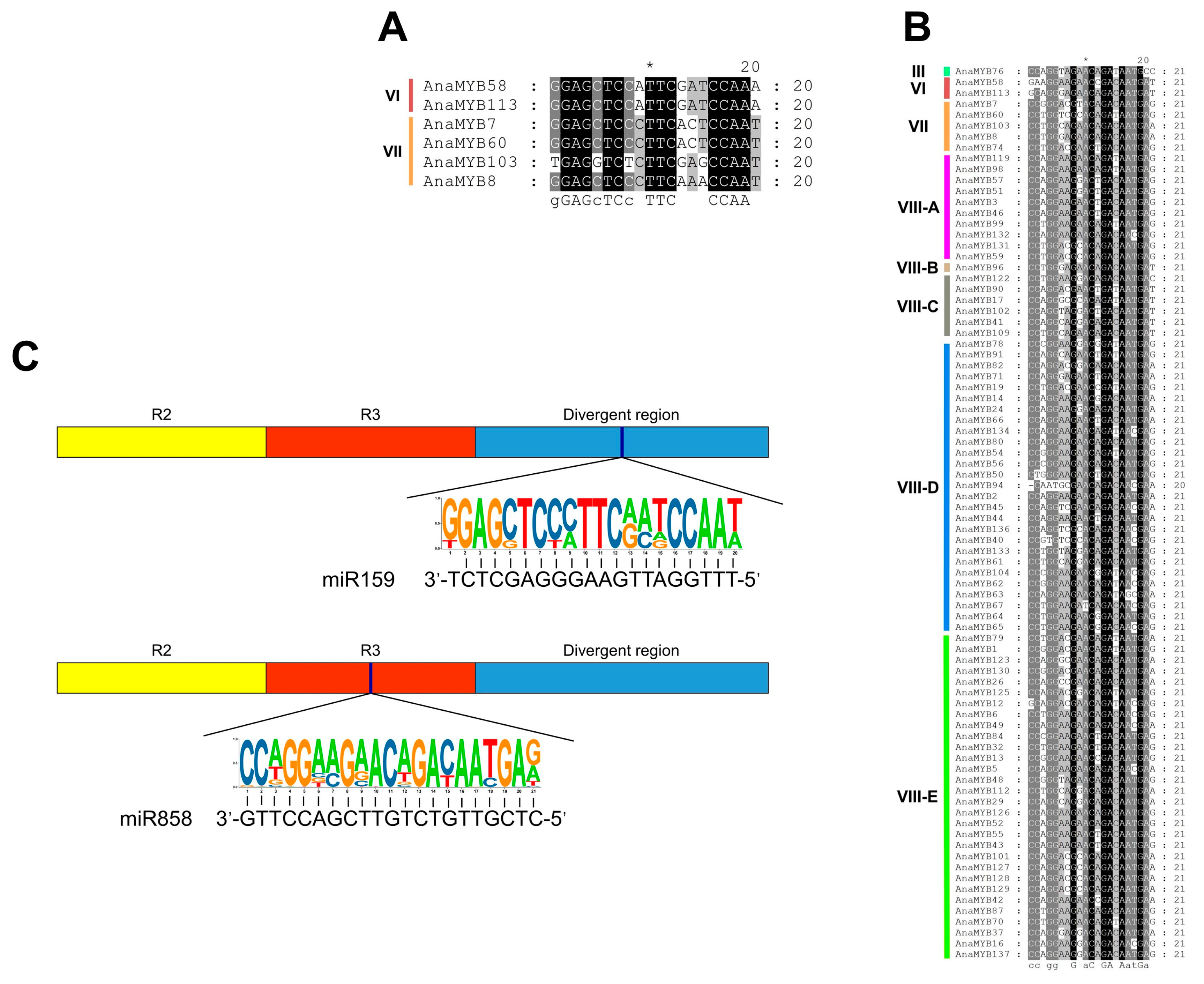
3.6. Identification of A. nanus R2R3-MYB Genes Targeted by MiRNAs
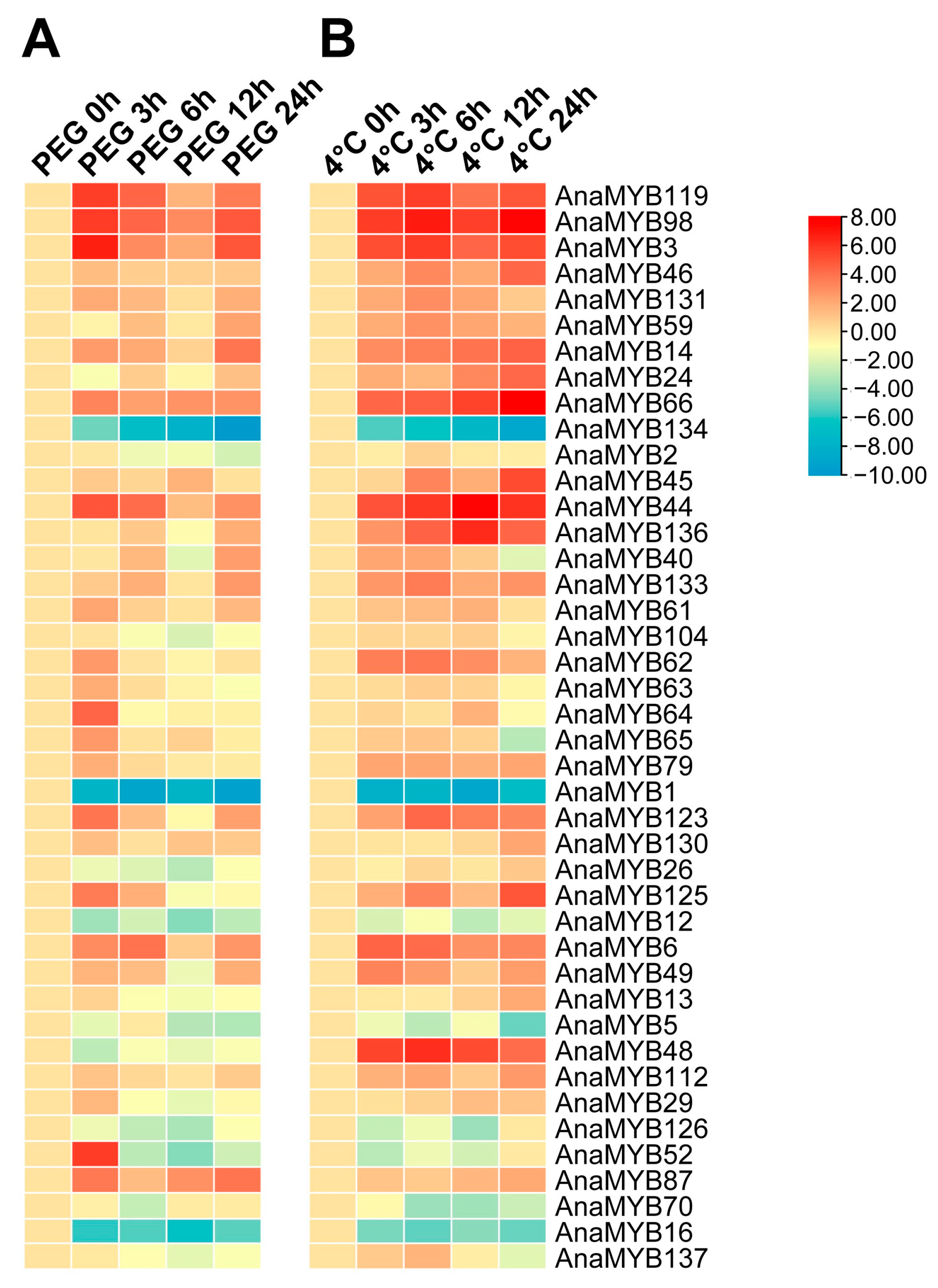
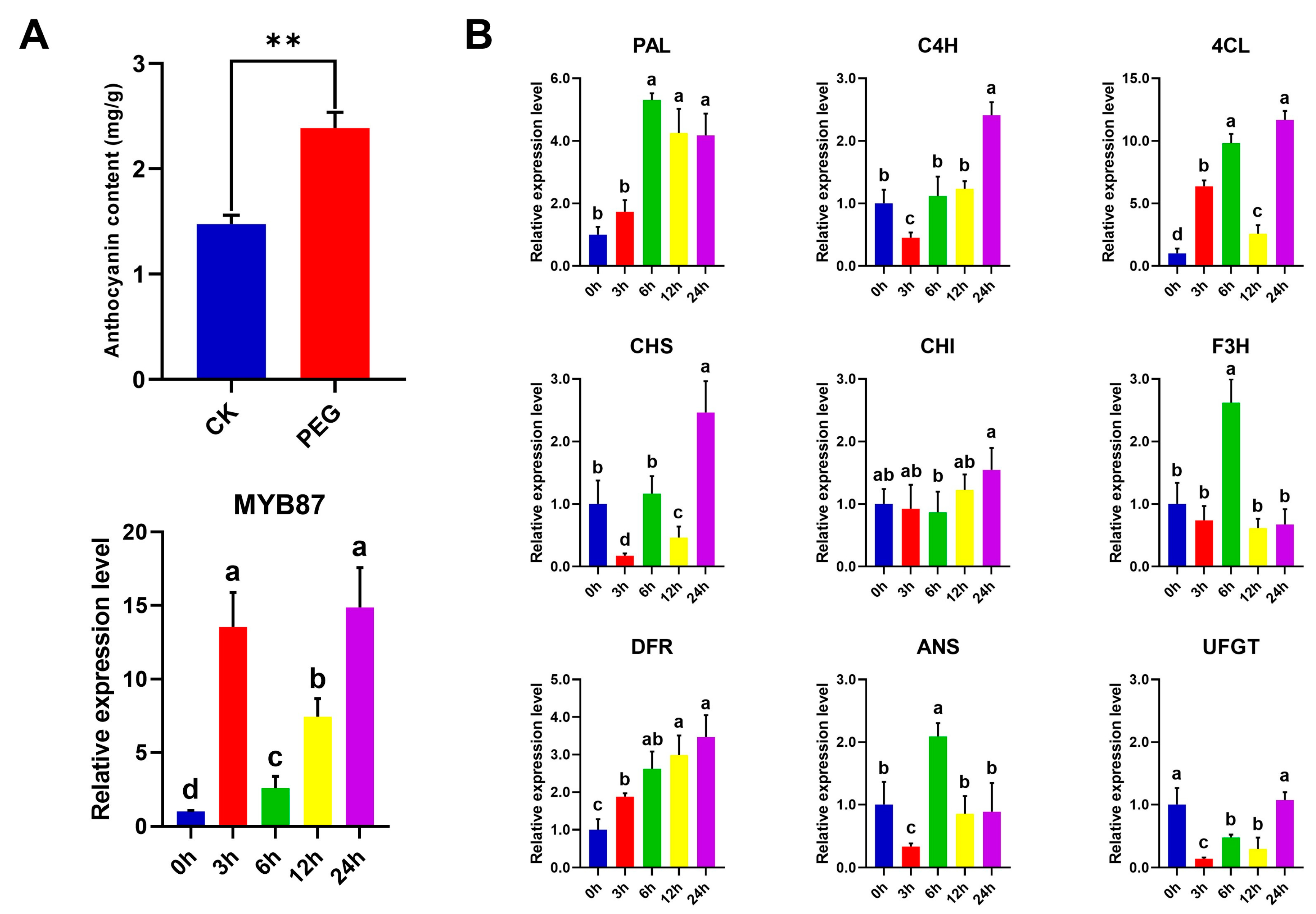
3.7. Expression of R2R3-MYB Genes in A. nanus under Cold and Osmotic Stress
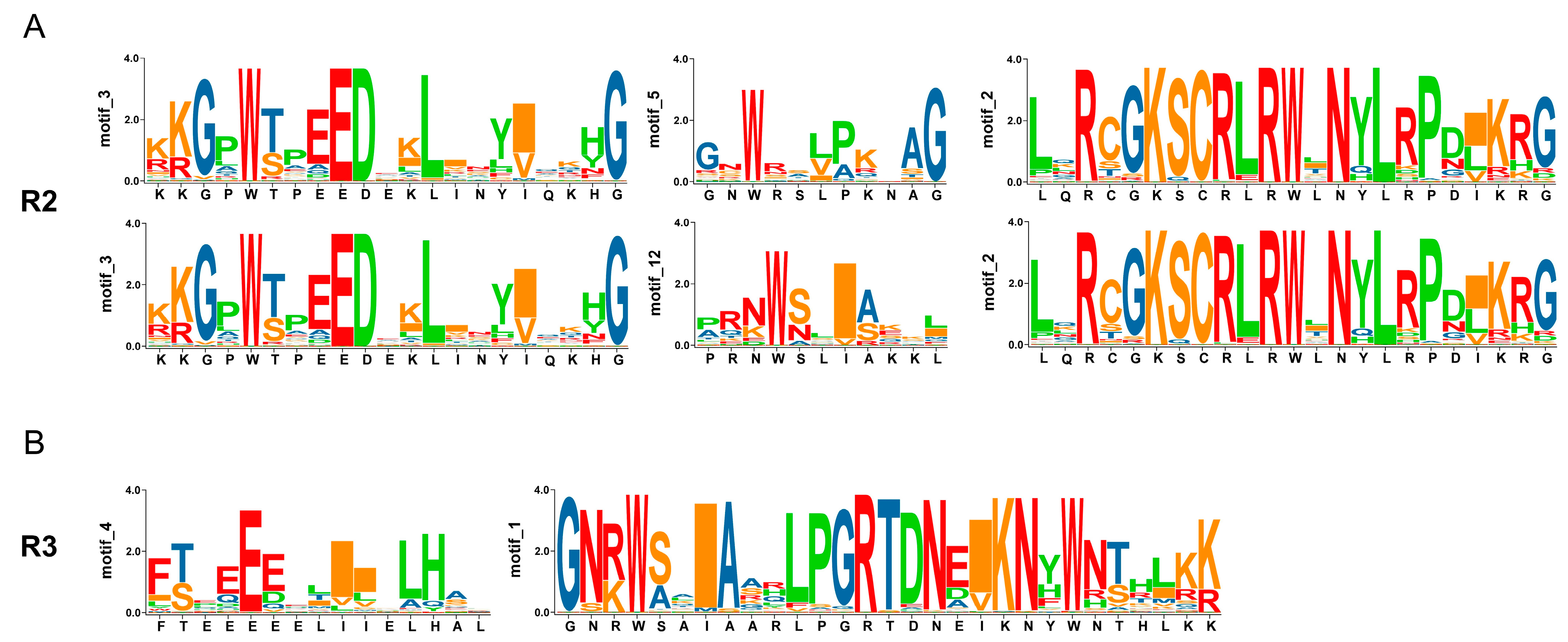
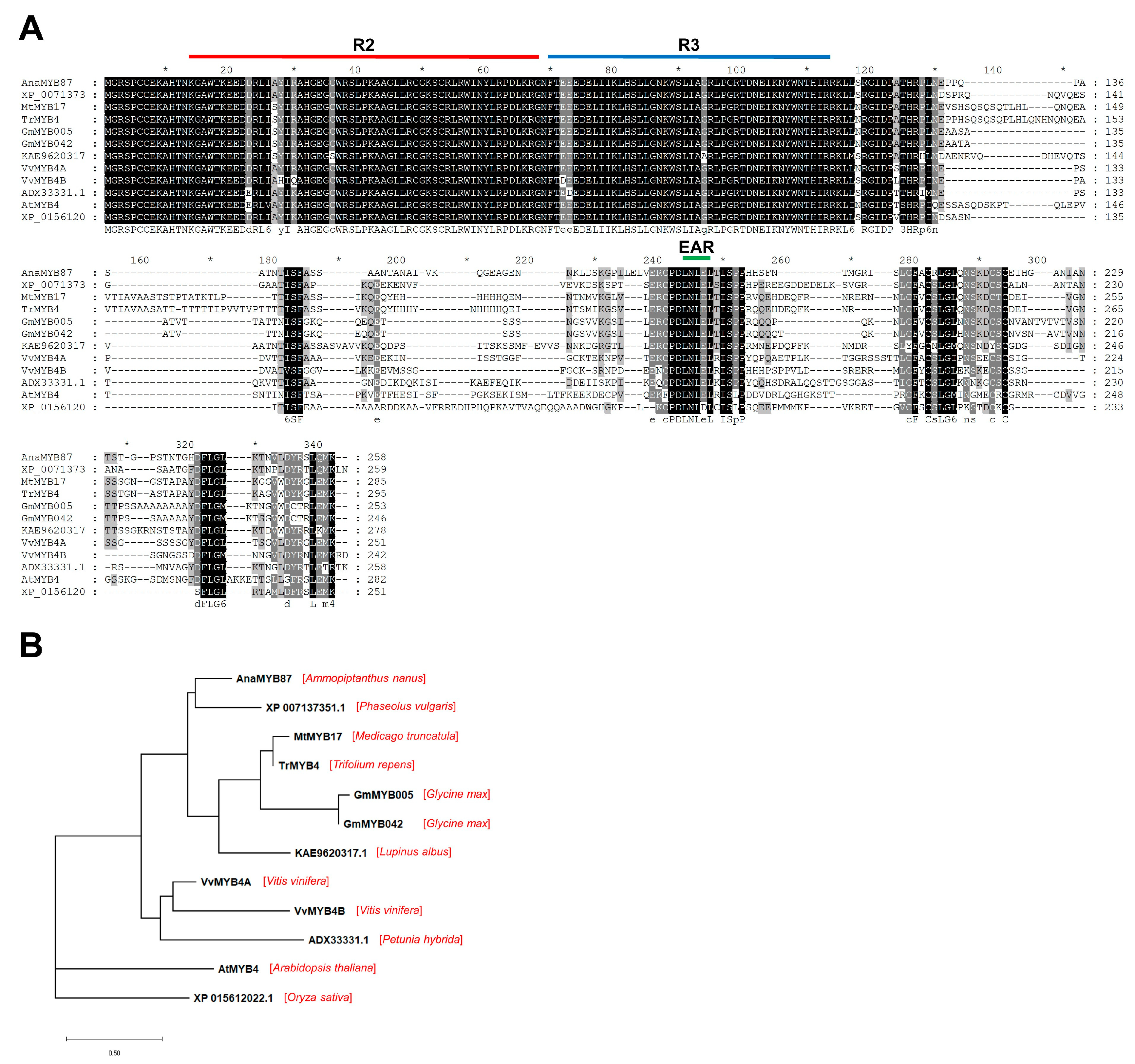
3.8. Sequence Analysis of AnaMYB87
3.9. Transactivation Activity and Subcellular Localization of AnaMYB87
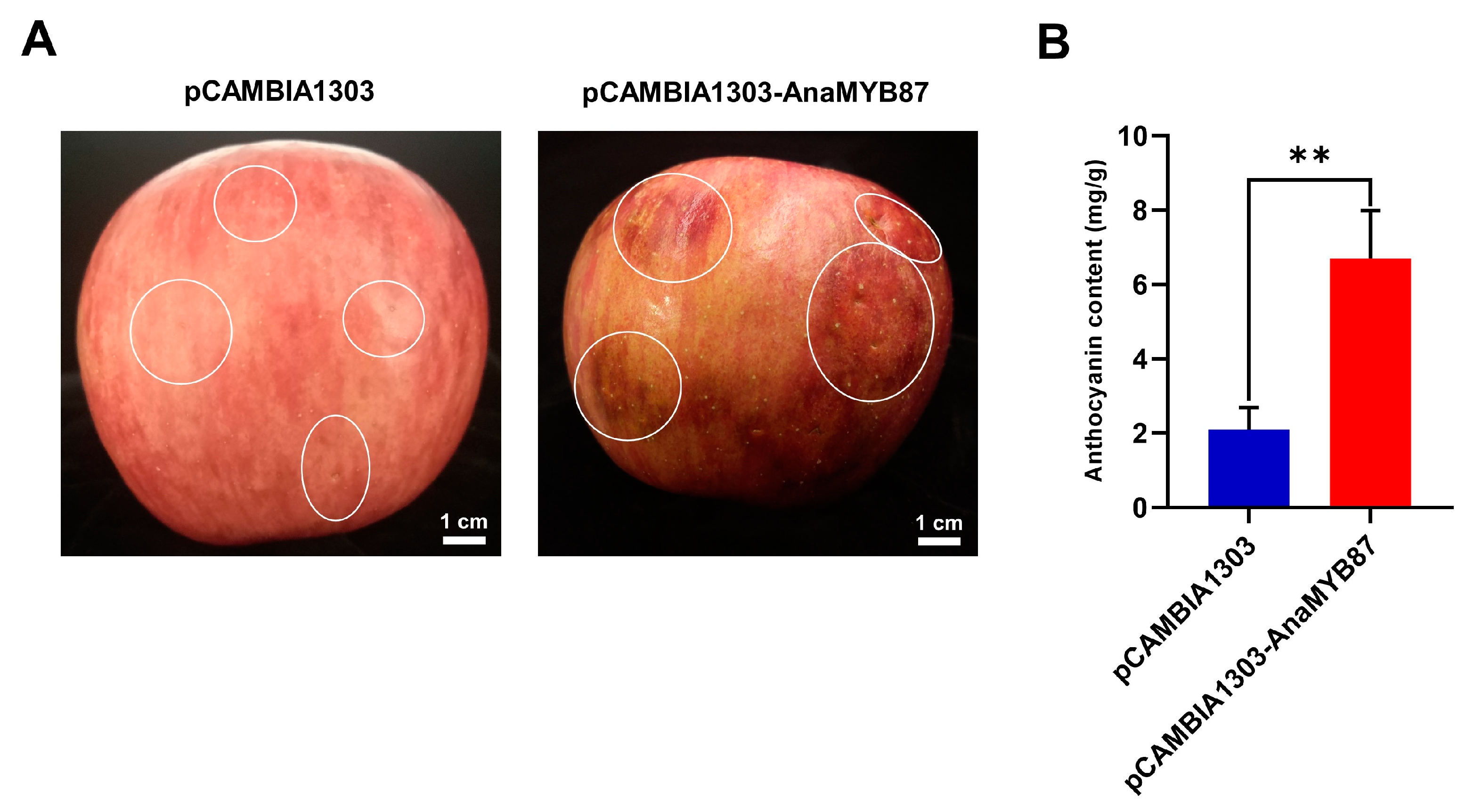
3.10. Transient Ectopic Expression of AnaMYB87 Promoted Anthocyanin Accumulation in Apples
3.11. Osmotic-Stress-Induced Anthocyanin Accumulation in A. nanus Leaves
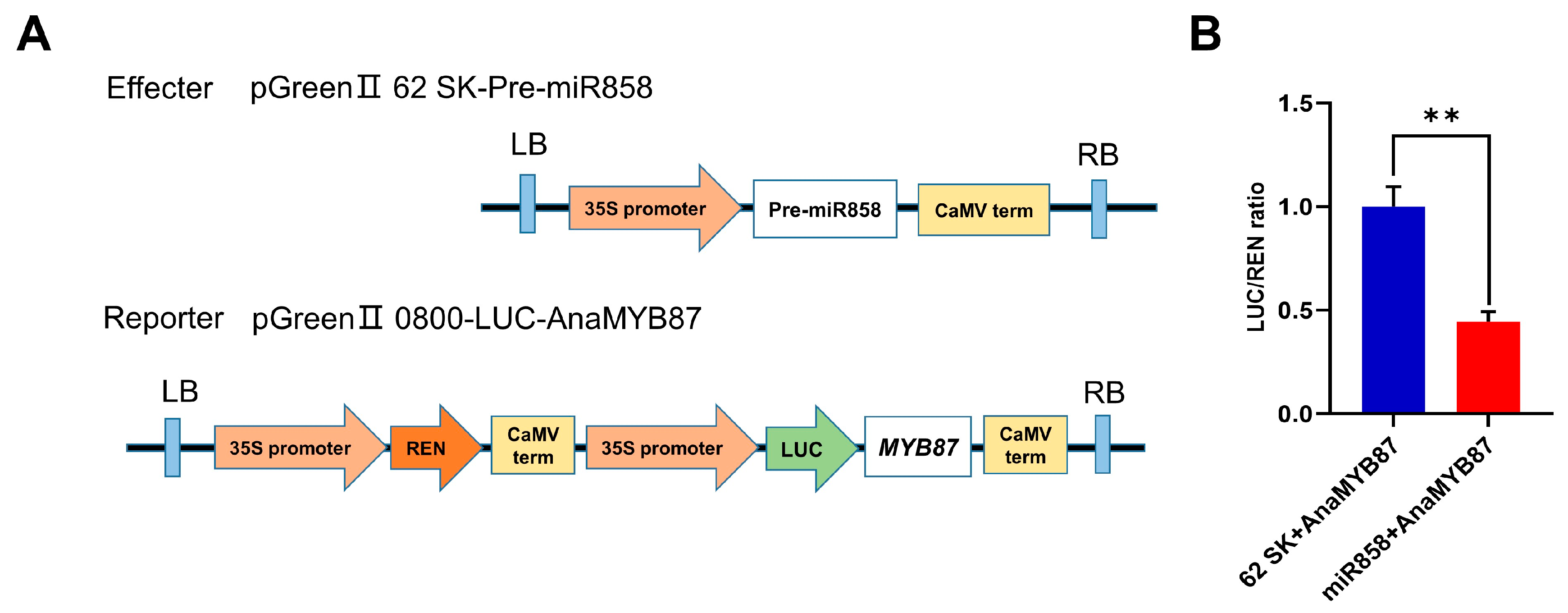
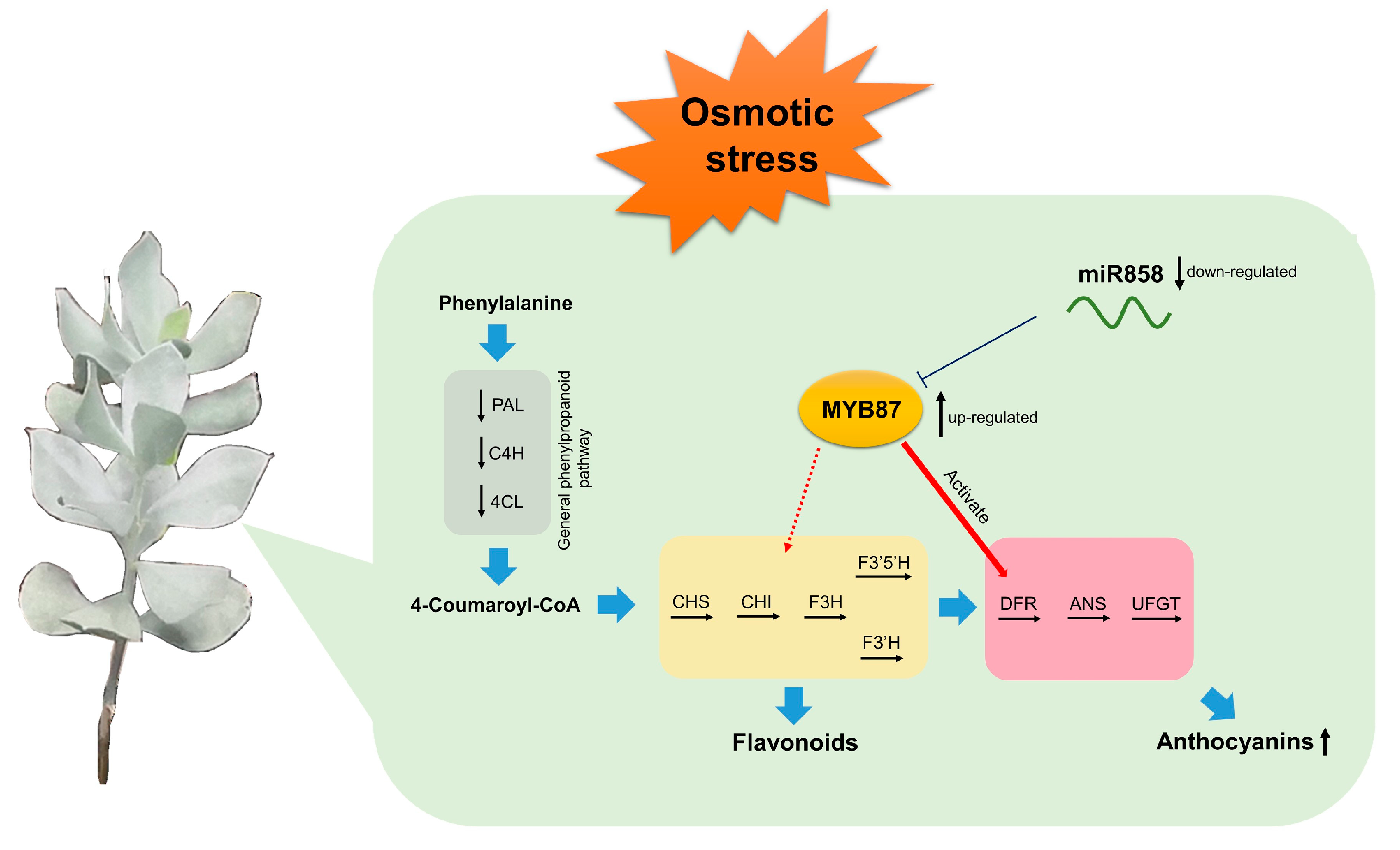
3.12. AnaMYB87 Was Targeted by MiR858
3.13. Ana-miR858 Was Down-Regulated upon Osmotic Treatment
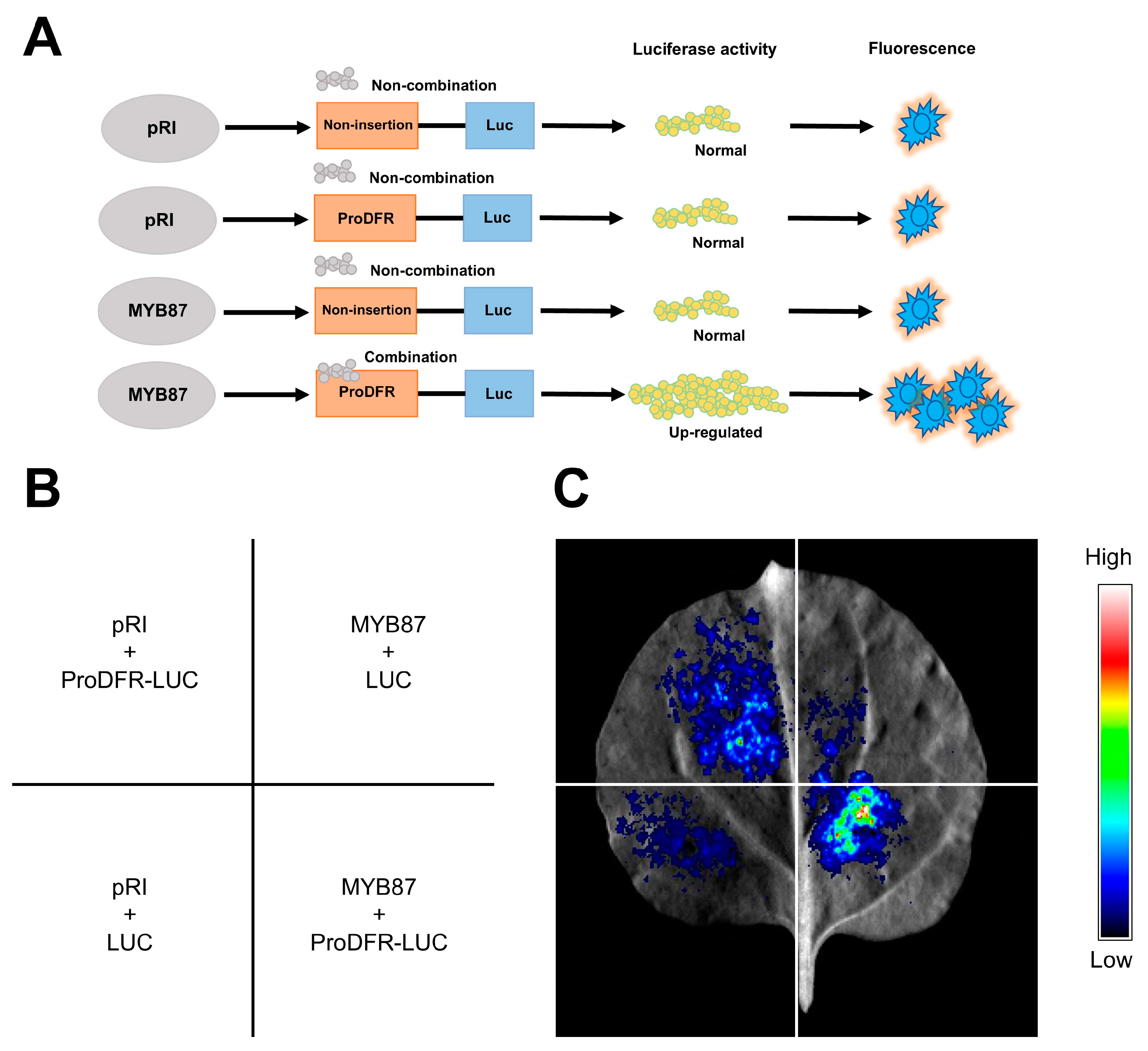
3.14. AnaMYB87 Activated the Transcription of AnaDFR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Takahashi, F.; Kuromori, T.; Urano, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Drought stress responses and resistance in plants: From cellular responses to long-distance intercellular communication. Front. Plant Sci. 2020, 11, 556972. [Google Scholar] [CrossRef] [PubMed]
- Baldoni, E.; Genga, A.; Cominelli, E. Plant MYB transcription factors: Their role in drought response mechanisms. Int. J. Mol. Sci. 2015, 16, 15811–15851. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Lin, X.; Chen, A.; Peterson, T.; Ma, K.; Bertzky, M.; Ciais, P.; Kapos, V.; Peng, C.; Poulter, B. Global priority conservation areas in the face of 21st century climate change. PLoS ONE 2013, 8, e54839. [Google Scholar] [CrossRef]
- Eshel, G.; Araus, V.; Undurraga, S.; Soto, D.C.; Moraga, C.; Montecinos, A.; Moyano, T.; Maldonado, J.; Díaz, F.P.; Varala, K.; et al. Plant ecological genomics at the limits of life in the Atacama Desert. Proc. Natl. Acad. Sci. USA 2021, 118, e2101177118. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Liu, P.L.; Duan, L.; Pan, B.R.; Su, Z.H. Evolutionary response to the Qinghai-Tibetan Plateau uplift: Phylogeny and biogeography of Ammopiptanthus and tribe Thermopsideae (Fabaceae). PeerJ 2017, 5, e3607. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.M.; Chen, S.Z.; Tang, L.; Tu, P.F. Three new isoflavonoids from the aerial parts of Ammopiptanthus mongolicus. Helv. Chim. Acta 2008, 91, 1015–1022. [Google Scholar] [CrossRef]
- Chen, G.; Sun, Y.; Yushan, D.; Shaerbayi, N.; Zhang, H.; He, H.; Jin, Y.; Chen, L. Identification and characterization of chemical constituents from Ammopiptanthus nanus stem and their metabolites in rats by UHPLC-Q-TOF-MS/MS. Planta Med. 2023. [Google Scholar] [CrossRef]
- Lu, C.; Yin, L.; Li, K. Proteome expression patterns in the stress tolerant evergreen Ammopiptanthus nanus under conditions of extreme cold. Plant Growth Regul. 2010, 62, 65–70. [Google Scholar] [CrossRef]
- Wu, Y.; Wei, W.; Pang, X.; Wang, X.; Zhang, H.; Dong, B.; Xing, Y.; Li, X.; Wang, M. Comparative transcriptome profiling of a desert evergreen shrub, Ammopiptanthus mongolicus, in response to drought and cold stresses. BMC Genomics 2014, 5, 671. [Google Scholar] [CrossRef]
- Gao, F.; Wang, J.; Wei, S.; Li, Z.; Wang, N.; Li, H.; Feng, J.; Li, H.; Zhou, Y.; Zhang, F. Transcriptomic analysis of drought stress responses in Ammopiptanthus mongolicus leaves using the RNA-Seq technique. PLoS ONE 2015, 10, e0124382. [Google Scholar] [CrossRef]
- Gao, F.; Wang, N.; Li, H.; Liu, J.; Fu, C.; Xiao, Z.; Wei, C.; Lu, X.; Feng, J.; Zhou, Y. Identification of drought-responsive microRNAs and their targets in Ammopiptanthus mongolicus by using high-throughput sequencing. Sci. Rep. 2016, 6, 34601. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Wang, X.; Li, X.; Xu, M.; Li, H.; Abla, M.; Sun, H.; Wei, S.; Feng, J.; Zhou, Y. Long-read sequencing and de novo genome assembly of Ammopiptanthus nanus, a desert shrub. Gigascience 2018, 7, giy074. [Google Scholar] [CrossRef] [PubMed]
- Klempnauer, K.H.; Gonda, T.J.; Bishop, J.M. Nucleotide sequence of the retroviral leukemia gene V-MYB and its cellular progenitor C-MYB: The architecture of a transduced oncogene. Cell 1982, 31 Pt 1, 453–463. [Google Scholar] [CrossRef] [PubMed]
- Ogata, K.; Morikawa, S.; Nakamura, H.; Hojo, H.; Yoshimura, S.; Zhang, R.; Aimoto, S.; Ametani, Y.; Hirata, Z.; Sarai, A.; et al. Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb. Nat. Struct. Biol. 1995, 2, 309–320. [Google Scholar] [CrossRef] [PubMed]
- de Mendoza, A.; Sebé-Pedrós, A.; Šestak, M.S.; Matejcic, M.; Torruella, G.; Domazet-Loso, T.; Ruiz-Trillo, I. Transcription factor evolution in eukaryotes and the assembly of the regulatory toolkit in multicellular lineages. Proc. Natl. Acad. Sci. USA 2013, 110, E4858–E4866. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Liang, Z.; Zhao, S.; Nan, M.G.; Tran, L.S.; Lu, K.; Huang, Y.B.; Li, J.N. The evolutionary history of R2R3-MYB proteins across 50 eukaryotes: New insights into subfamily classification and expansion. Sci. Rep. 2015, 5, 11037. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Pratyusha, D.S.; Sarada, D.V.L. MYB transcription factors-master regulators of phenylpropanoid biosynthesis and diverse developmental and stress responses. Plant Cell Rep. 2022, 41, 2245–2260. [Google Scholar] [CrossRef]
- Yang, J.; Chen, Y.; Xiao, Z.; Shen, H.; Li, Y.; Wang, Y. Multilevel regulation of anthocyanin-promoting R2R3-MYB transcription factors in plants. Front. Plant Sci. 2022, 13, 1008829. [Google Scholar] [CrossRef]
- Naing, A.H.; Kim, C.K. Roles of R2R3-MYB transcription factors in transcriptional regulation of anthocyanin biosynthesis in horticultural plants. Plant Mol. Biol. 2018, 98, 1–18. [Google Scholar] [CrossRef]
- Li, S.; Zhang, Y.; Shi, L.; Cao, S.; Chen, W.; Yang, Z. Involvement of a MYB transcription factor in anthocyanin biosynthesis during Chinese Bayberry (Morella rubra) fruit ripening. Biology 2023, 12, 894. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Zhao, C.; Wang, S.; Gao, N.; Wang, X.X.; Na, X.; Wang, X.M.; Bi, Y. PIF4-PAP1 interaction affects MYB-bHLH-WD40 complex formation and anthocyanin accumulation in Arabidopsis. J. Plant Physiol. 2022, 268, 153558. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Plunkert, M.; Teng, Z.; Mackenzie, K.; Guo, L.; Luo, Y.; Hytönen, T.; Liu, Z. Two MYB activators of anthocyanin biosynthesis exhibit specialized activities in petiole and fruit of diploid strawberry. J. Exp. Bot. 2023, 74, 1517–1531. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.J.; Lee, Z.; Kim, S.; Jeong, E.; Shim, J.S. Modulation of lignin biosynthesis for drought tolerance in plants. Front. Plant Sci. 2023, 14, 1116426. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, R.; Dalton, S.; Roberts, L.A.; Moron-Garcia, O.M.; Iacono, R.; Kosik, O.; Gallagher, J.A.; Bosch, M. Modified expression of ZmMYB167 in Brachypodium distachyon and Zea mays leads to increased cell wall lignin and phenolic content. Sci. Rep. 2019, 9, 8800. [Google Scholar] [CrossRef] [PubMed]
- Chezem, W.R.; Memon, A.; Li, F.S.; Weng, J.K.; Clay, N.K. SG2-type R2R3-MYB transcription factor MYB15 controls defense-induced lignification and basal immunity in Arabidopsis. Plant Cell 2017, 29, 1907–1926. [Google Scholar] [CrossRef]
- Song, Q.; Kong, L.; Yang, X.; Jiao, B.; Hu, J.; Zhang, Z.; Xu, C.; Luo, K. PtoMYB142, a poplar R2R3-MYB transcription factor, contributes to drought tolerance by regulating wax biosyn-thesis. Tree Physio 2022, 42, 2133–2147. [Google Scholar]
- Shim, J.S.; Jung, C.; Lee, S.; Min, K.; Lee, Y.W.; Choi, Y.; Lee, J.S.; Song, J.T.; Kim, J.K.; Choi, Y.D. AtMYB44 regulates WRKY70 expression and modulates antagonistic interaction between salicylic acid and jasmonic acid signaling. Plant J. 2013, 73, 483–495. [Google Scholar] [CrossRef]
- Zhang, L.; Song, J.; Lin, R.; Tang, M.; Shao, S.; Yu, J.; Zhou, Y. Tomato SlMYB15 transcription factor targeted by sly-miR156e-3p positively regulates ABA-mediated cold tolerance. J. Exp. Bot. 2022, 73, 7538–7551. [Google Scholar] [CrossRef]
- Cao, Z.H.; Zhang, S.Z.; Wang, R.K.; Zhang, R.F.; Ha, Y.J. Genome wide analysis of the apple MYB transcription factor family allows the identification of MdoMYB121 gene confering abiotic stress tolerance in plants. PLoS ONE 2013, 8, e69955. [Google Scholar] [CrossRef]
- Nakabayashi, R.; Yonekura-Sakakibara, K.; Urano, K.; Suzuki, M.; Yamada, Y.; Nishizawa, T.; Matsuda, F.; Kojima, M.; Sakakibara, H.; Shinozaki, K.; et al. Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 2014, 77, 367–379. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wu, Y.; Yu, Z.; Gao, Z.; Ding, Q.; Shah, S.H.A.; Lin, W.; Li, Y.; Hou, X. BcMYB111 responds to BcCBF2 and induces flavonol biosynthesis to enhance tolerance under cold stress in non-heading Chinese cabbage. Int. J. Mol. Sci. 2023, 24, 8670. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Guo, D.-D.; Min, D.-H.; Cao, T.; Ning, L.; Jiang, Q.-Y.; Sun, X.-J.; Zhang, H.; Tang, W.-S.; Gao, S.-Q.; et al. Foxtail millet MYB-like transcription factor SiMYB16 confers salt tolerance in transgenic rice by regulating phenylpropane pathway. Plant Physiol. Biochem. 2023, 195, 310–321. [Google Scholar] [CrossRef]
- Zhan, J.; Meyers, B.C. Plant small RNAs: Their biogenesis, regulatory roles, and functions. Annu. Rev. Plant Biol. 2023, 74, 21–51. [Google Scholar] [CrossRef] [PubMed]
- Millar, A.A.; Lohe, A.; Wong, G. Biology and function of miR159 in plants. Plants 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Xu, S.; Mei, Y.; Li, J.; Gu, Y.; Zhang, W.; Wang, J. MicroRNAs in medicinal plants. Int. J. Mol. Sci. 2022, 23, 10477. [Google Scholar] [CrossRef] [PubMed]
- Kavas, M.; Abdulla, M.F.; Mostafa, K.; Seçgin, Z.; Yerlikaya, B.A.; Otur, Ç.; Gökdemir, G.; Kurt, K.A.; Al-Khayri, J.M.; Jain, S.M. Investigation and expression analysis of R2R3-MYBs and anthocyanin biosynthesis-related genes during seed color development of common bean (Phaseolus vulgaris). Plants 2022, 11, 3386. [Google Scholar] [CrossRef]
- Solofoharivelo, M.C.; van der Walt, A.P.; Stephan, D.; Burger, J.T.; Murray, S.L. MicroRNAs in fruit trees: Discovery, diversity and future research directions. Plant Biol. 2013, 6, 856–865. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, L.; Meng, S.; Liu, Y.; Zhao, X.; Pang, C.; Zhang, H.; Xu, T.; He, Y.; Qi, M.; et al. Expression of galactinol synthase from Ammopiptanthus nanus in tomato improves tolerance to cold stress. J. Exp. Bot. 2020, 71, 435–449. [Google Scholar] [CrossRef]
- Yu, H.; Zheng, H.; Liu, Y.; Yang, Q.; Li, W.; Zhang, Y.; Fu, F. Antifreeze protein from Ammopiptanthus nanus functions in temperature-stress through domain A. Sci. Rep. 2021, 11, 8458. [Google Scholar] [CrossRef]
- Pang, X.; Xue, M.; Ren, M.; Nan, D.; Wu, Y.; Guo, H. Ammopiptanthus mongolicus stress-responsive NAC gene enhances the tolerance of transgenic Arabidopsis thaliana to drought and cold stresses. Genet. Mol. Biol. 2019, 42, 624–634. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.S.; Bai, L.P.; Zhang, L.; Chen, G.; Fan, J.G.; Xu, S.; Guo, Z.F. Identification and characterization of AnICE1 and AnCBFs involved in cold tolerance from Ammopiptanthus nanus. Plant Physiol. Biochem. 2021, 168, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.R. Accelerated profile HMM searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef] [PubMed]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, w202–w208. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wu, N.; Song, W.; Yin, G.; Qin, Y.; Yan, Y.; Hu, Y. Soybean (Glycine max) expansin gene superfamily origins: Segmental and tandem duplication events followed by divergent selection among subfamilies. BMC Plant Biol. 2014, 14, 93. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef] [PubMed]
- Higo, K.; Ugawa, Y.; Iwamoto, M.; Korenaga, T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999, 27, 297–300. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Wang., X.; Zhou, Y.; Tan, J.; Zhou, Y.; Gao, F. Small RNA sequencing revealed that miR4415, a legume-specific miRNA, was involved in the cold acclimation of Ammopiptanthus nanus by targeting an L-Ascorbate Oxidase gene and regulating the redox state of apoplast. Front. Genet. 2022, 13, 870446. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Zhuang, Z.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server. Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Zhu, M.; Liu, Q.; Liu, F.; Zheng, L.; Bing, J.; Zhou, Y.; Gao, F. Gene profiling of the Ascorbate Oxidase family genes under osmotic and cold stress reveals the role of AnAO5 in cold adaptation in Ammopiptanthus nanus. Plants 2023, 12, 677. [Google Scholar] [CrossRef]
- Shi, J.; Liu, M.; Shi, J.; Zheng, G.; Wang, Y.; Wang, J.; Chen, Y.; Lu, C.; Yin, W. Reference gene selection for qPCR in Ammopiptanthus mongolicus under abiotic stresses and expression analysis of seven ROS-scavenging enzyme genes. Plant Cell Rep. 2012, 31, 1245–1254. [Google Scholar] [CrossRef]
- Clément, T.; Salone, V.; Rederstorff, M. Dual luciferase gene reporter assays to study miRNA function. Methods Mol. Biol. 2015, 1296, 187–198. [Google Scholar] [PubMed]
- Jing, Y.; Guo, Q.; Lin, R. The chromatin-remodeling factor PICKLE antagonizes polycomb repression of FT to promote flowering. Plant Physiol. 2019, 181, 656–668. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Jiang, Z.; Wang, H.; Lin, R. The central circadian clock proteins CCA1 and LHY regulate iron homeostasis in Arabidopsis. J. Integr. Plant Biol. 2019, 61, 168–181. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.C.; Enderle, B.; Kathare, P.K.; Islam, R.; Hiltbrunner, A.; Huq, E. PCH1 and PCHL directly interact with PIF1, promote its degradation, and inhibit its transcriptional function during photomorphogenesis. Mol. Plant 2020, 13, 499–514. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.H.; Kim, D.H.; Kim, J.K.; Lee, J.Y.; Ha, S.H. A radish basic helix-loop-helix transcription factor, RsTT8 acts a positive regulator for anthocyanin biosynthesis. Front. Plant Sci. 2017, 8, 1917. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.K.; Rao, G.Y. Insights into the diversification and evolution of R2R3-MYB transcription factors in plants. Plant Physiol. 2020, 183, 637–655. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef]
- Weston, K. Myb proteins in life, death and differentiation. Curr. Opin. Genet. Dev. 1998, 8, 76–81. [Google Scholar] [CrossRef]
- Li, W.; Liu, Y.; Zhao, J.; Zhen, X.; Guo, C.; Shu, Y. Genome-wide identification and characterization of R2R3-MYB genes in Medicago truncatula. Genet. Mol. Biol. 2019, 42, 611–623. [Google Scholar] [CrossRef]
- Lee, E.; Liu, X.; Eglit, Y.; Sack, F. FOUR LIPS and MYB88 conditionally restrict the G1/S transition during stomatal formation. J. Exp. Bot. 2013, 64, 5207–5219. [Google Scholar] [CrossRef]
- Wang, H.-Z.; Yang, K.-Z.; Zou, J.-J.; Zhu, L.-L.; Xie, Z.D.; Morita, M.T.; Tasaka, M.; Friml, J.; Grotewold, E.; Beeckman, T.; et al. Transcriptional regulation of PIN genes by FOUR LIPS and MYB88 during Arabidopsis root gravitropism. Nat. Commun. 2015, 6, 8822. [Google Scholar] [CrossRef] [PubMed]
- Xiao, R.; Zhang, C.; Guo, X.; Li, H.; Lu, H. MYB transcription factors and its regulation in secondary cell wall formation and lignin biosynthesis during xylem development. Int. J. Mol. Sci. 2021, 22, 3560. [Google Scholar] [CrossRef] [PubMed]
- Oshima, Y.; Shikata, M.; Koyama, T.; Ohtsubo, N.; Mitsuda, N.; Ohme-Takagi, M. MIXTA-like transcription factors and WAX INDUCER1/SHINE1 coordinately regulate cuticle development in Arabidopsis and Torenia fournieri. Plant Cell. 2013, 25, 1609–1624. [Google Scholar] [CrossRef] [PubMed]
- Loreti, E.; Perata, P. Mobile plant microRNAs allow communication within and between organisms. New Phytol. 2022, 235, 2176–2182. [Google Scholar] [CrossRef]
- Yang, K.; Han, H.; Li, Y.; Ye, J.; Xu, F. Significance of miRNA in enhancement of flavonoid biosynthesis. Plant Biol. 2022, 24, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Badola, P.K.; Bhatia, C.; Sharma, D.; Trivedi, P.K. Primary transcript of miR858 encodes regulatory peptide and controls flavonoid biosynthesis and development in Arabidopsis. Nat. Plants 2020, 6, 1262–1274. [Google Scholar] [CrossRef]
- Maloney, G.S.; DiNapoli, K.T.; Muday, G.K. The anthocyanin reduced tomato mutant demonstrates the role of flavonols in tomato lateral root and root hair development. Plant Physiol. 2014, 166, 614–631. [Google Scholar] [CrossRef]
- Li, P.; Li, Y.J.; Zhang, F.J.; Zhang, G.Z.; Jiang, X.Y.; Yu, H.M.; Hou, B.K. The Arabidopsis UDP-glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. Plant J. 2017, 89, 85–103. [Google Scholar] [CrossRef]
- Tamagnone, L.; Merida, A.; Parr, A.; Mackay, S.; Culianez-Macia, F.A.; Roberts, K.; Martin, C. The AmMYB308 and AmMYB330 transcription factors from antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell 1998, 10, 135–154. [Google Scholar] [CrossRef]
- Wang, X.C.; Wu, J.; Guan, M.L.; Zhao, C.H.; Geng, P.; Zhao, Q. Arabidopsis MYB4 plays dual roles in flavonoid biosynthesis. Plant J. 2020, 101, 637–652. [Google Scholar] [CrossRef]
- Cavallini, E.; Matus, J.T.; Finezzo, L.; Zenoni, S.; Loyola, R.; Guzzo, F.; Schlechter, R.; Ageorges, A.; Arce-Johnson, P.; Tornielli, G.B. The phenylpropanoid pathway is controlled at different branches by a set of R2R3-MYB C2 repressors in grapevine. Plant Physiol. 2015, 167, 1448–1470. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Reichelt, M.; Yoshida, K.; Gershenzon, J.; Constabel, C.P. Two R2R3-MYB proteins are broad repressors of flavonoid and phenylpropanoid metabolism in poplar. Plant J. 2018, 96, 949–965. [Google Scholar] [CrossRef] [PubMed]




| ID | Core Sequence | Cis-Acting Element | Putative Function | FLP | II | III | IV | V | ARP | VI | VII | VIII-A | VIII-B | VIII-C | VIII-D | VIII-E | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S000175 | CTAACCA | MYBATRD22 | Responsive to dehydration | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 3 | 2 | 2 | 6 | 11 | 28 |
| S000176 | CNGTTR | MYBCORE | Responsive to dehydration | 2 | 5 | 2 | 6 | 5 | 2 | 2 | 5 | 15 | 5 | 9 | 36 | 41 | 135 |
| S000177 | TAACTG | MYB2AT | Responsive to dehydration | 0 | 1 | 1 | 6 | 2 | 2 | 2 | 3 | 10 | 4 | 1 | 12 | 17 | 61 |
| S000180 | GGATA | MYBST1 | Responsive to dehydration | 2 | 5 | 2 | 6 | 5 | 2 | 2 | 5 | 14 | 5 | 10 | 36 | 39 | 133 |
| S000408 | WAACCA | MYB1AT | Responsive to dehydration | 2 | 5 | 2 | 6 | 5 | 2 | 2 | 5 | 14 | 5 | 9 | 36 | 39 | 132 |
| S000409 | YAACKG | MYB2CONSENSUSAT | Responsive to dehydration | 2 | 4 | 2 | 6 | 4 | 2 | 2 | 4 | 15 | 5 | 4 | 29 | 32 | 111 |
| S000413 | CATGTG | MYCATERD1 | Responsive to dehydration | 2 | 5 | 1 | 3 | 5 | 2 | 2 | 5 | 11 | 4 | 7 | 23 | 31 | 101 |
| S000414 | ACGTG | ABRELATERD1 | Responsive to dehydration | 1 | 3 | 2 | 5 | 4 | 2 | 2 | 2 | 15 | 4 | 8 | 30 | 32 | 110 |
| S000415 | ACGT | ACGTATERD1 | Responsive to dehydration | 2 | 5 | 2 | 6 | 5 | 2 | 2 | 3 | 15 | 5 | 9 | 35 | 37 | 128 |
| S000497 | RYCGAC | CBFHV | Responsive to dehydration | 0 | 2 | 1 | 2 | 3 | 0 | 2 | 2 | 6 | 2 | 6 | 14 | 18 | 58 |
| S000418 | RCCGAC | DRECRTCOREAT | Responsive to dehydration and low temperature | 0 | 2 | 1 | 2 | 1 | 0 | 0 | 2 | 4 | 1 | 4 | 8 | 17 | 42 |
| S000407 | CANNTG | MYCCONSENSUSAT | Responsive to dehydration and low temperature | 2 | 5 | 2 | 6 | 5 | 2 | 2 | 5 | 15 | 5 | 10 | 37 | 41 | 137 |
| S000411 | GTCGAC | CRTDREHVCBF2 | Responsive to low temperature | 0 | 0 | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 1 | 0 | 2 | 2 | 8 |
| S000153 | CCGAC | LTRECOREATCOR15 | Responsive to low temperature | 0 | 3 | 2 | 2 | 2 | 0 | 0 | 3 | 10 | 2 | 5 | 19 | 24 | 72 |
| S000250 | CCGAAA | LTRE-1 | Responsive to low temperature | 0 | 2 | 2 | 3 | 2 | 0 | 0 | 3 | 3 | 0 | 1 | 13 | 12 | 41 |
| S000174 | CACATG | MYCATRD22 | Responsive to ABA | 2 | 5 | 1 | 3 | 5 | 2 | 2 | 4 | 11 | 4 | 7 | 23 | 29 | 98 |
| S000394 | ACGTGKC | ACGTABREMOTIFA2OSEM | Responsive to ABA | 0 | 2 | 2 | 1 | 1 | 0 | 2 | 2 | 0 | 4 | 1 | 9 | 9 | 33 |
| S000403 | TATCCA | TATCCAOSAMY | Regulation of GA and other hormones | 1 | 3 | 2 | 4 | 2 | 2 | 0 | 2 | 12 | 2 | 9 | 26 | 26 | 91 |
| S000298 | TTTTTTCC | Pyrimidine box | Necessity of GA induction | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 3 | 7 | 2 | 2 | 4 | 12 | 35 |
| S000181 | TAACAAA | MYBGAHV | Response to GA signal | 2 | 2 | 2 | 4 | 5 | 1 | 2 | 1 | 7 | 1 | 4 | 20 | 25 | 76 |
| S000259 | CCTTTT | PYRIMIDINEBOXOSRAMY1A | Response to GA signal | 2 | 5 | 2 | 4 | 5 | 2 | 1 | 4 | 14 | 3 | 9 | 37 | 35 | 123 |
| S000416 | TATCCAC | TATCCAC box | Response to GA signal | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 5 | 0 | 5 | 3 | 2 | 17 |
| S000419 | TAACAGA | GARE | Response to GA signal | 0 | 2 | 1 | 5 | 2 | 1 | 1 | 1 | 7 | 3 | 2 | 9 | 15 | 49 |
| S000420 | TAACGTA | GARE2OSREP1 | Response to GA signal | 1 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 1 | 1 | 8 | 4 | 18 |
| S000438 | ACGTGTC | GADOWNAT | Response to GA signal | 0 | 2 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 4 | 5 | 17 |
| S000439 | TAACAAR | GAREAT | Response to GA signal | 2 | 4 | 2 | 4 | 5 | 2 | 2 | 3 | 10 | 2 | 7 | 28 | 32 | 103 |
| S000447 | TGAC | WRKY71OS | Response to GA signal, WRKY binding site | 2 | 5 | 2 | 6 | 5 | 2 | 2 | 5 | 15 | 5 | 10 | 37 | 41 | 137 |
| S000454 | NGATT | ARR1AT | Response to cytokinin signaling | 2 | 5 | 2 | 6 | 5 | 2 | 2 | 5 | 15 | 5 | 10 | 37 | 41 | 137 |
| S000491 | TATTAG | CPBCSPOR | Response to cytokinin signaling | 2 | 5 | 0 | 5 | 4 | 1 | 2 | 5 | 13 | 4 | 10 | 34 | 32 | 117 |
| S000370 | CATATG | CATATGGMSAUR | Response to auxin signals | 1 | 0 | 1 | 1 | 2 | 1 | 1 | 2 | 12 | 2 | 6 | 20 | 21 | 70 |
| S000270 | TGTCTC | ARFAT | Response to auxin signals | 2 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 11 | 1 | 6 | 23 | 23 | 78 |
| S000499 | GAGAC | SURE | Response to auxin signals | 2 | 4 | 1 | 4 | 4 | 2 | 2 | 4 | 15 | 4 | 10 | 35 | 35 | 122 |
| S000024 | TGACG | ASF-1 binding site | Response to auxin and SA signals | 1 | 4 | 2 | 4 | 3 | 2 | 2 | 4 | 8 | 2 | 4 | 26 | 24 | 86 |
| S000037 | AWTTCAAA | ERE | Response to ethylene signal | 1 | 1 | 1 | 5 | 5 | 1 | 1 | 2 | 11 | 3 | 5 | 24 | 18 | 78 |
| S000458 | AACGTG | T/G-box | Response to JA signal | 0 | 0 | 0 | 2 | 2 | 2 | 1 | 2 | 8 | 3 | 4 | 14 | 12 | 50 |
| S000292 | ACACNNG | DPBFCOREDCDC3 | Induced by abscisic acid | 2 | 5 | 2 | 4 | 3 | 2 | 1 | 5 | 14 | 5 | 9 | 33 | 39 | 124 |
| S000390 | TTGAC | WBOXATNPR1 | Response to SA and other stress signals | 2 | 5 | 2 | 6 | 5 | 2 | 2 | 5 | 15 | 5 | 10 | 37 | 40 | 136 |
| S000391 | YTGTCWC | SEBF | Response to pathogenic signals | 1 | 3 | 0 | 4 | 2 | 2 | 2 | 2 | 10 | 1 | 9 | 23 | 24 | 83 |
| S000453 | GAAAAA | GT-1 motif | Response to pathogens and high salt stress | 1 | 5 | 2 | 5 | 5 | 2 | 2 | 5 | 15 | 5 | 10 | 36 | 36 | 129 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sumbur, B.; Gao, F.; Liu, Q.; Feng, D.; Bing, J.; Dorjee, T.; Li, X.; Sun, H.; Zhou, Y. The Characterization of R2R3-MYB Genes in Ammopiptanthus nanus Uncovers That the miR858-AnaMYB87 Module Mediates the Accumulation of Anthocyanin under Osmotic Stress. Biomolecules 2023, 13, 1721. https://doi.org/10.3390/biom13121721
Sumbur B, Gao F, Liu Q, Feng D, Bing J, Dorjee T, Li X, Sun H, Zhou Y. The Characterization of R2R3-MYB Genes in Ammopiptanthus nanus Uncovers That the miR858-AnaMYB87 Module Mediates the Accumulation of Anthocyanin under Osmotic Stress. Biomolecules. 2023; 13(12):1721. https://doi.org/10.3390/biom13121721
Chicago/Turabian StyleSumbur, Batu, Fei Gao, Qi Liu, Dandan Feng, Jie Bing, Tashi Dorjee, Xuting Li, Huigai Sun, and Yijun Zhou. 2023. "The Characterization of R2R3-MYB Genes in Ammopiptanthus nanus Uncovers That the miR858-AnaMYB87 Module Mediates the Accumulation of Anthocyanin under Osmotic Stress" Biomolecules 13, no. 12: 1721. https://doi.org/10.3390/biom13121721
APA StyleSumbur, B., Gao, F., Liu, Q., Feng, D., Bing, J., Dorjee, T., Li, X., Sun, H., & Zhou, Y. (2023). The Characterization of R2R3-MYB Genes in Ammopiptanthus nanus Uncovers That the miR858-AnaMYB87 Module Mediates the Accumulation of Anthocyanin under Osmotic Stress. Biomolecules, 13(12), 1721. https://doi.org/10.3390/biom13121721






