Nuclear Localization of G3BP6 Is Essential for the Flowering Transition in Arabidopsis
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Condition
2.2. Subcellular Localization of G3BPs and Plant Transformation
2.3. Analysis of Gene Expression with Quantitative Real-Time PCR (qRT-PCR)
2.4. Yeast Two-Hybrid and Bimolecular Fluorescence Complementation Assays
2.5. Transcriptome Analysis
3. Results
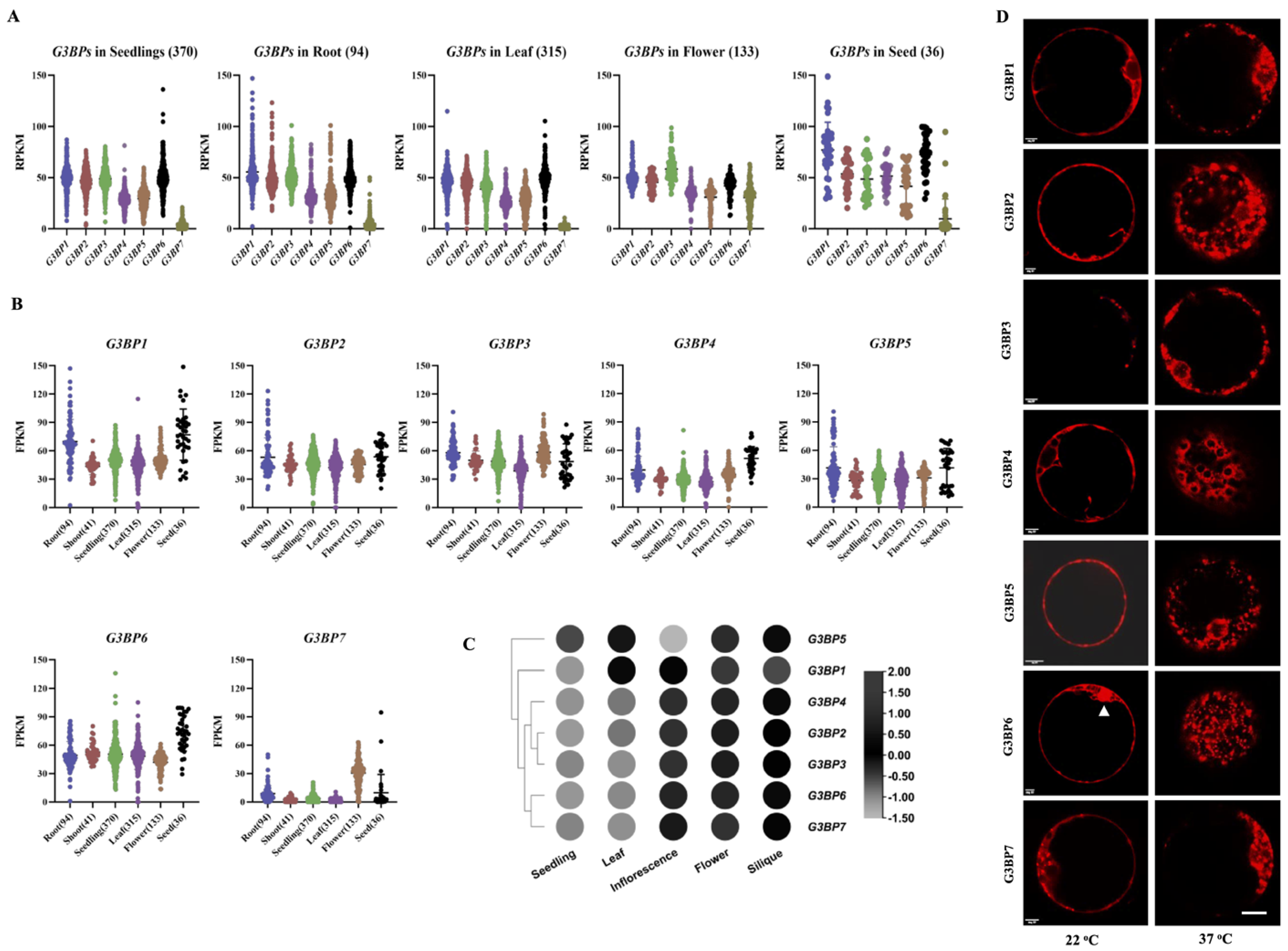
3.1. Expressional Patterns and Subcellular Localization of G3BPs in Arabidopsis
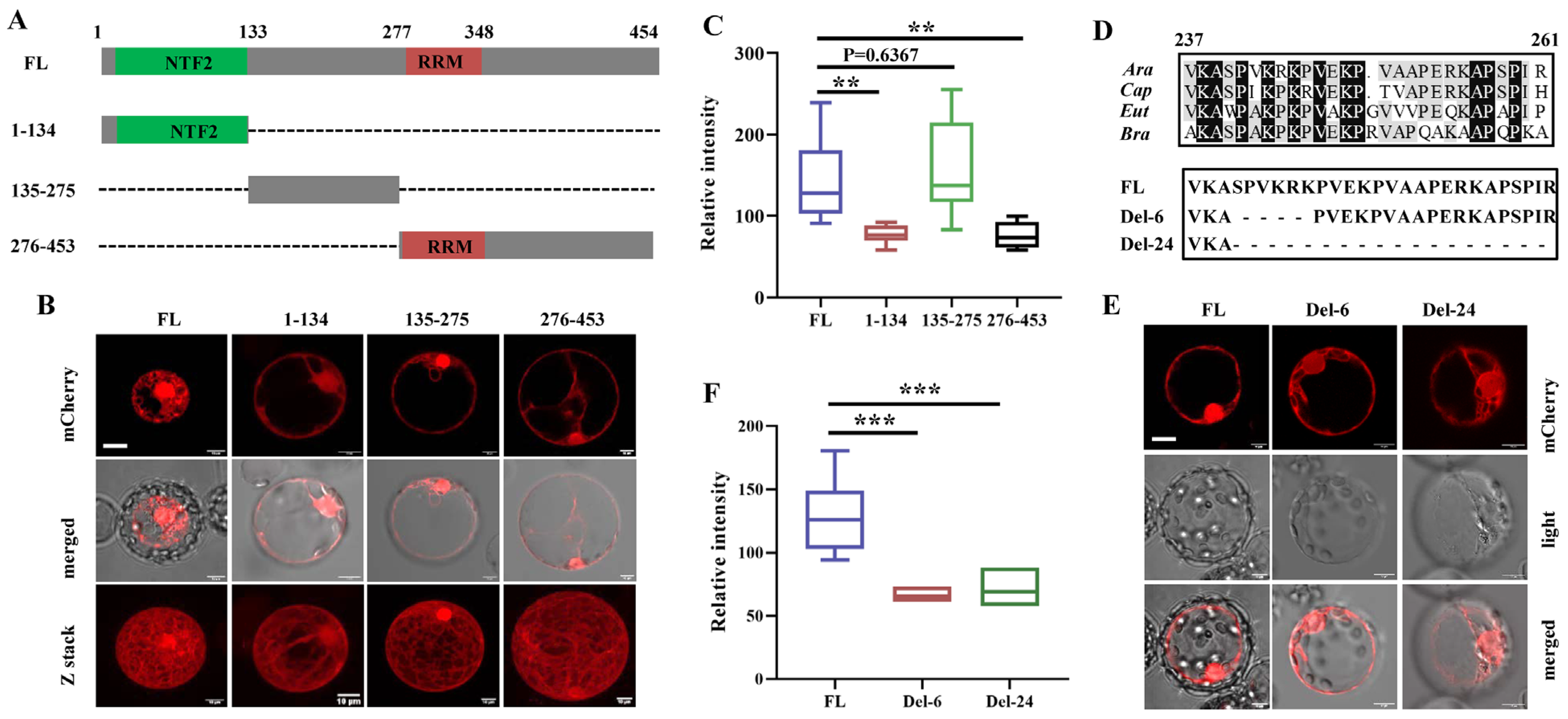
3.2. The Nuclear Localization Signal (NLS) Mainly Determines the Nuclear Localization of G3BP6
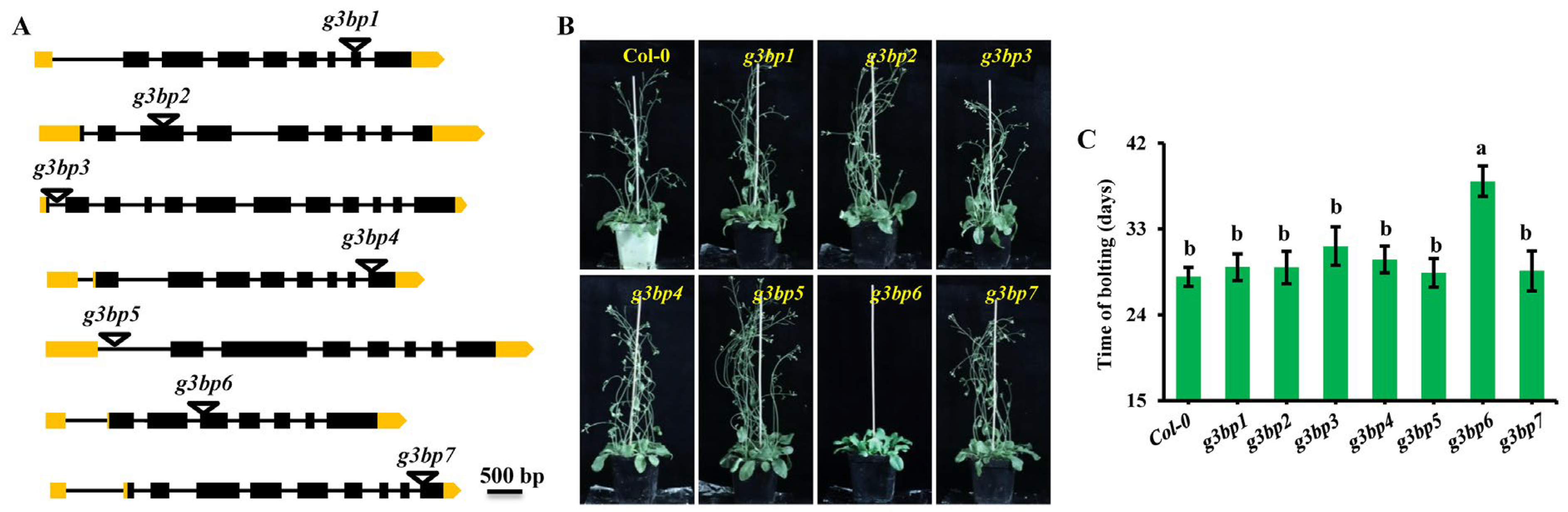
3.3. g3bp6 Mutation Significantly Delays the Flowering Time
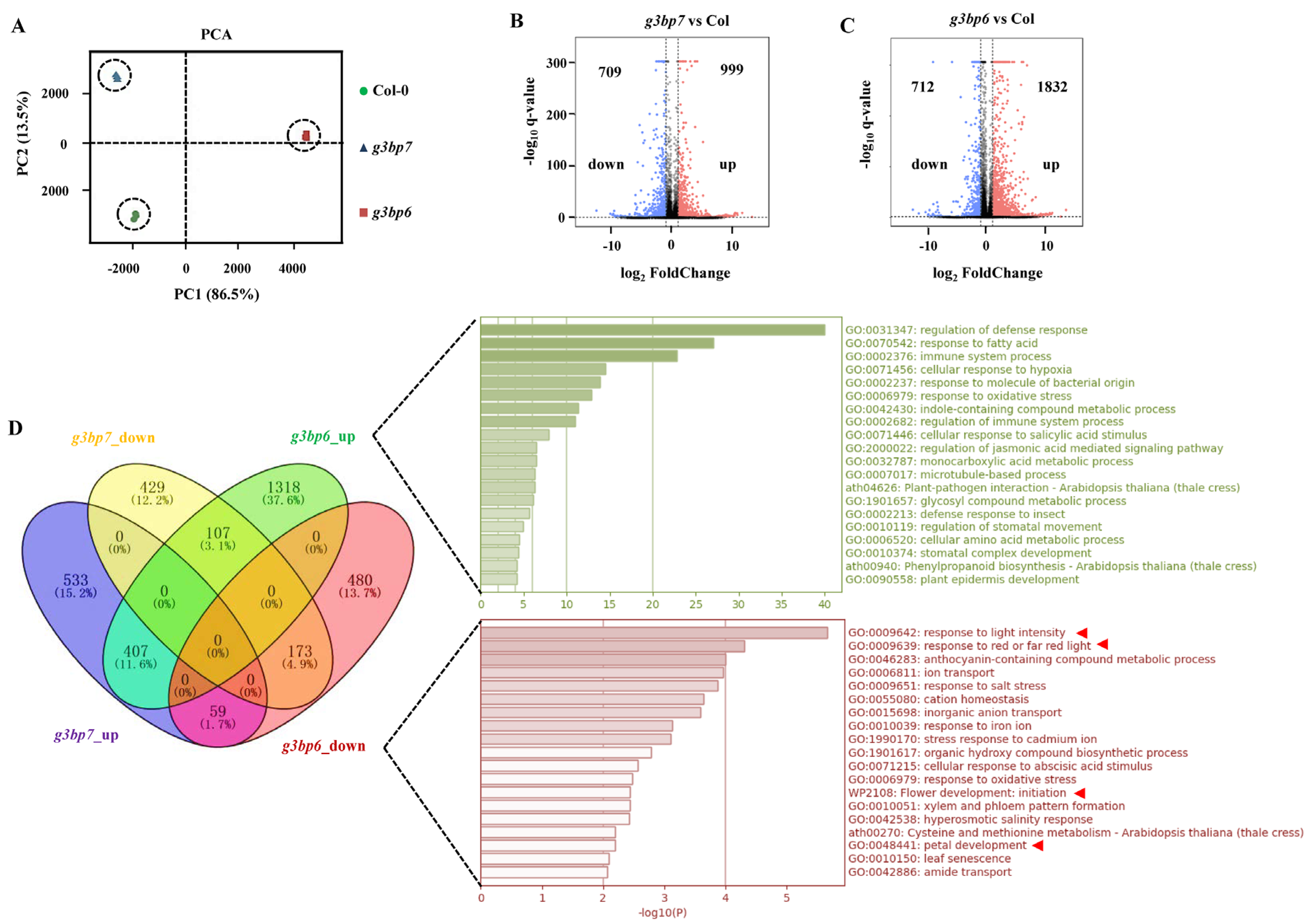
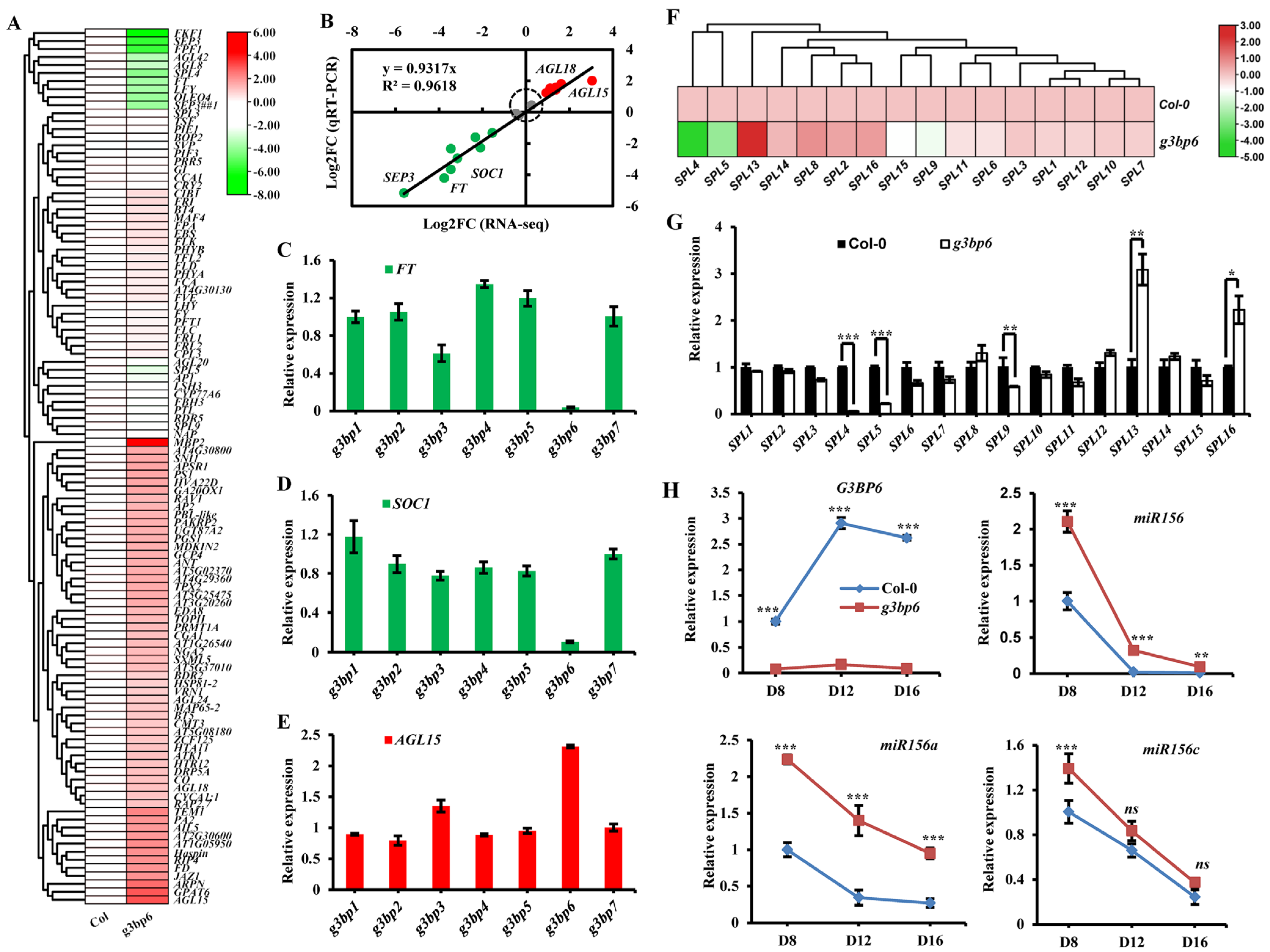
3.4. G3BP6 Controls Global Gene Expression with Specific Roles in the Flowering Transition
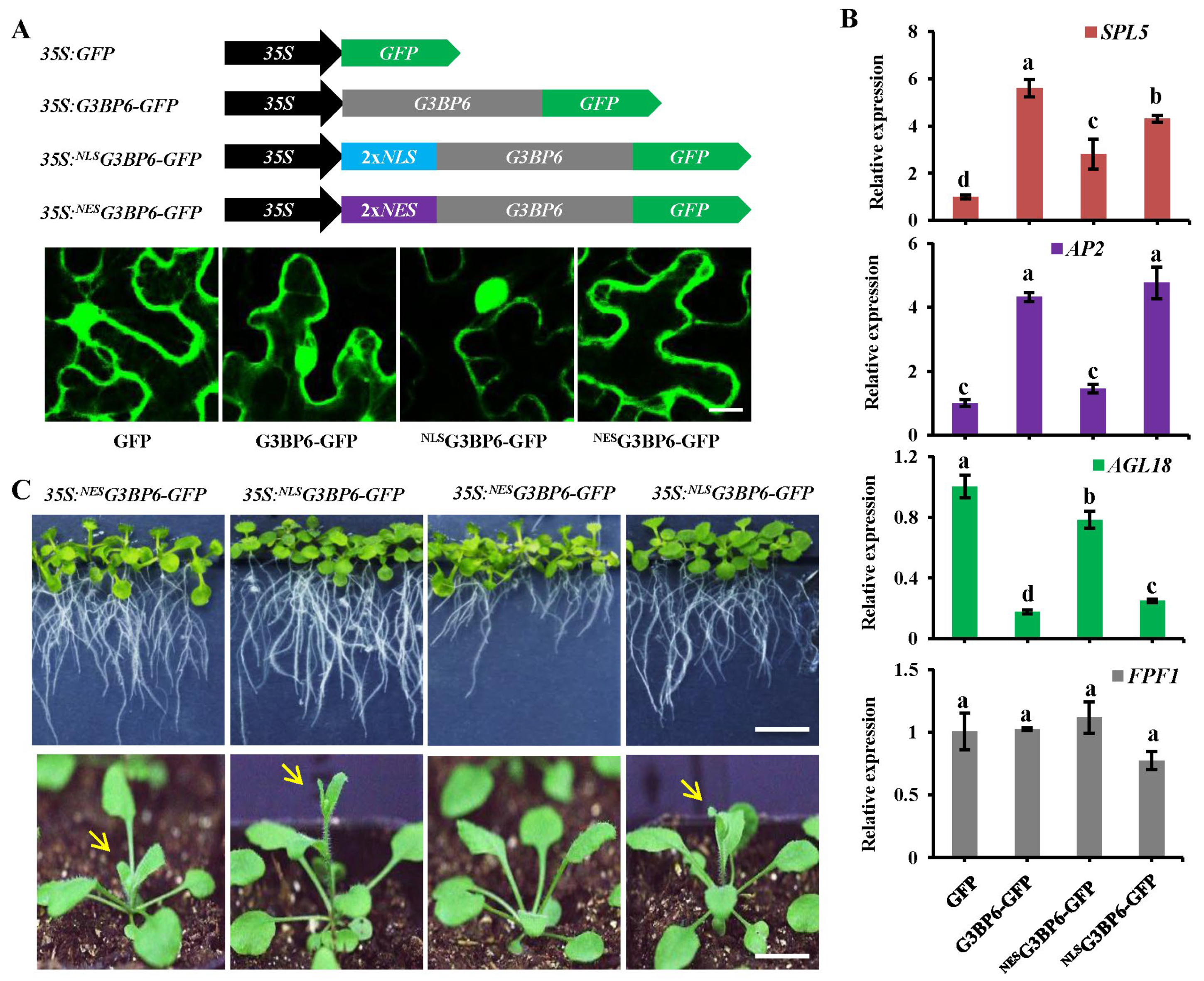
3.5. Nuclear Localization of G3BP6 Is Essential for Flowering Transition
3.6. Scaffold Protein RACK1 Contributes to the Nuclear Translocation of G3BP6
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Briggs, S.D.; Bryant, S.S.; Jove, R.; Sanderson, S.D.; Smithgall, T.E. The Ras GTPase-activating protein (GAP) is an SH3 domain-binding protein and substrate for the Src-related tyrosine kinase, Hck. J. Biol. Chem. 1995, 270, 14718–14724. [Google Scholar] [CrossRef] [PubMed]
- Alam, U.; Kennedy, D. Rasputin a decade on and more promiscuous than ever? A review of G3BPs. Biochim. Biophys. Acta Mol. Cell Res. 2019, 1866, 360–370. [Google Scholar] [CrossRef] [PubMed]
- Steggerda, S.M.; Black, B.E.; Paschal, B.M. Monoclonal antibodies to NTF2 inhibit nuclear protein import by preventing nuclear translocation of the GTPase Ran. Mol. Biol. Cell 2000, 11, 703–719. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Katahira, J.; Dimitrova, L.; Imai, Y.; Hurt, E. NTF2-like domain of Tap plays a critical role in cargo mRNA recognition and export. Nucleic Acids Res. 2015, 43, 1894–1904. [Google Scholar] [CrossRef]
- Vognsen, T.; Moller, I.R.; Kristensen, O. Crystal structures of the human G3BP1 NTF2-like domain visualize FxFG Nup repeat specificity. PLoS ONE 2013, 8, e80947. [Google Scholar] [CrossRef]
- Burd, C.G.; Dreyfuss, G. Conserved structures and diversity of functions of RNA-binding proteins. Science 1994, 265, 615–621. [Google Scholar] [CrossRef]
- Guillen-Boixet, J.; Kopach, A.; Holehouse, A.S.; Wittmann, S.; Jahnel, M.; Schlussler, R.; Kim, K.; Trussina, I.; Wang, J.; Mateju, D.; et al. RNA-induced conformational switching and clustering of G3BP drive stress granule assembly by condensation. Cell 2020, 181, 346–361.e17. [Google Scholar] [CrossRef] [PubMed]
- Isabelle, M.; Gagne, J.P.; Gallouzi, I.E.; Poirier, G.G. Quantitative proteomics and dynamic imaging reveal that G3BP-mediated stress granule assembly is poly(ADP-ribose)-dependent following exposure to MNNG-induced DNA alkylation. J. Cell Sci. 2012, 125, 4555–4566. [Google Scholar] [CrossRef]
- Kang, W.; Wang, Y.; Yang, W.; Zhang, J.; Zheng, H.; Li, D. Research progress on the structure and function of G3BP. Front. Immunol. 2021, 12, 718548. [Google Scholar] [CrossRef]
- Onomoto, K.; Yoneyama, M.; Fung, G.; Kato, H.; Fujita, T. Antiviral innate immunity and stress granule responses. Trends Immunol. 2014, 35, 420–428. [Google Scholar] [CrossRef]
- Ge, Y.; Jin, J.; Li, J.; Ye, M.; Jin, X. The roles of G3BP1 in human diseases (review). Gene 2022, 821, 146294. [Google Scholar] [CrossRef] [PubMed]
- Reuper, H.; Amari, K.; Krenz, B. Analyzing the G3BP-like gene family of Arabidopsis thaliana in early turnip mosaic virus infection. Sci. Rep. 2021, 11, 2187. [Google Scholar] [CrossRef] [PubMed]
- Abulfaraj, A.A.; Ohyanagi, H.; Goto, K.; Mineta, K.; Gojobori, T.; Hirt, H.; Rayapuram, N. Comprehensive evolutionary analysis and nomenclature of plant G3BPs. Life Sci. Alliance 2022, 5, e202101328. [Google Scholar] [CrossRef] [PubMed]
- Reuper, H.; Gotte, B.; Williams, L.; Tan, T.J.C.; McInerney, G.M.; Panas, M.D.; Krenz, B. Arabidopsis thaliana G3BP ortholog rescues mammalian stress granule phenotype across kingdoms. Int. J. Mol. Sci. 2021, 22, 6287. [Google Scholar] [CrossRef]
- Abulfaraj, A.A.; Mariappan, K.; Bigeard, J.; Manickam, P.; Blilou, I.; Guo, X.; Al-Babili, S.; Pflieger, D.; Hirt, H.; Rayapuram, N. The Arabidopsis homolog of human G3BP1 is a key regulator of stomatal and apoplastic immunity. Life Sci. Alliance 2018, 1, e201800046. [Google Scholar] [CrossRef]
- Chen, J.G.; Ullah, H.; Temple, B.; Liang, J.; Guo, J.; Alonso, J.M.; Ecker, J.R.; Jones, A.M. RACK1 mediates multiple hormone responsiveness and developmental processes in Arabidopsis. J. Exp. Bot. 2006, 57, 2697–2708. [Google Scholar] [CrossRef]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Zeng, P.; Vadnais, D.A.; Zhang, Z.; Polacco, J.C. Refined glufosinate selection in Agrobacterium-mediated transformation of soybean [Glycine max (L.) Merrill]. Plant Cell Rep. 2004, 22, 478–482. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Barnes, C.J.; Li, F.; Mandal, M.; Yang, Z.; Sahin, A.A.; Kumar, R. Heregulin induces expression, ATPase activity, and nuclear localization of G3BP, a Ras signaling component, in human breast tumors. Cancer Res. 2002, 62, 1251–1255. [Google Scholar] [PubMed]
- French, J.; Stirling, R.; Walsh, M.; Kennedy, H.D. The expression of Ras-GTPase activating protein SH3 domain-binding proteins, G3BPs, in human breast cancers. Histochem. J. 2002, 34, 223–231. [Google Scholar] [CrossRef] [PubMed]
- Prigent, M.; Barlat, I.; Langen, H.; Dargemont, C. IkappaBalpha and IkappaBalpha/NF-kappa B complexes are retained in the cytoplasm through interaction with a novel partner, RasGAP SH3-binding protein 2. J. Biol. Chem. 2000, 275, 36441–36449. [Google Scholar] [CrossRef] [PubMed]
- Nguyen Ba, A.N.; Pogoutse, A.; Provart, N.; Moses, A.M. NLStradamus: A simple Hidden Markov Model for nuclear localization signal prediction. BMC Bioinformatics 2009, 10, 202. [Google Scholar] [CrossRef]
- Corbesier, L.; Vincent, C.; Jang, S.; Fornara, F.; Fan, Q.; Searle, I.; Giakountis, A.; Farrona, S.; Gissot, L.; Turnbull, C.; et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 2007, 316, 1030–1033. [Google Scholar] [CrossRef]
- Liu, C.; Zhou, J.; Bracha-Drori, K.; Yalovsky, S.; Ito, T.; Yu, H. Specification of Arabidopsis floral meristem identity by repression of flowering time genes. Development 2007, 134, 1901–1910. [Google Scholar] [CrossRef]
- Jaeger, K.E.; Wigge, P.A. FT protein acts as a long-range signal in Arabidopsis. Curr. Biol. 2007, 17, 1050–1054. [Google Scholar] [CrossRef]
- Adamczyk, B.J.; Lehti-Shiu, M.D.; Fernandez, D.E. The MADS domain factors AGL15 and AGL18 act redundantly as repressors of the floral transition in Arabidopsis. Plant J. 2007, 50, 1007–1019. [Google Scholar] [CrossRef]
- Fernandez, D.E.; Wang, C.T.; Zheng, Y.; Adamczyk, B.J.; Singhal, R.; Hall, P.K.; Perry, S.E. The MADS-Domain factors AGAMOUS-LIKE15 and AGAMOUS-LIKE18, along with SHORT VEGETATIVE PHASE and AGAMOUS-LIKE24, are necessary to block floral gene expression during the vegetative phase. Plant Physiol. 2014, 165, 1591–1603. [Google Scholar] [CrossRef]
- Xu, M.; Hu, T.; Zhao, J.; Park, M.Y.; Earley, K.W.; Wu, G.; Yang, L.; Poethig, R.S. Developmental functions of miR156-regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes in Arabidopsis thaliana. PLoS Genet. 2016, 12, e1006263. [Google Scholar] [CrossRef] [PubMed]
- Lal, S.; Pacis, L.B.; Smith, H.M. Regulation of the SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE genes/microRNA156 module by the homeodomain proteins PENNYWISE and POUND-FOOLISH in Arabidopsis. Mol. Plant 2011, 4, 1123–1132. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.W.; Czech, B.; Weigel, D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 2009, 138, 738–749. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Hu, T.; Smith, M.R.; Poethig, R.S. Epigenetic regulation of vegetative phase change in Arabidopsis. Plant Cell 2016, 28, 28–41. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Park, M.Y.; Conway, S.R.; Wang, J.W.; Weigel, D.; Poethig, R.S. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 2009, 138, 750–759. [Google Scholar] [CrossRef]
- Chen, C.; Huang, W.; Hou, K.; Wu, W. Bolting, an important process in plant development, two types in plants. J. Plant Biol. 2019, 62, 161–169. [Google Scholar] [CrossRef]
- Adams, D.R.; Ron, D.; Kiely, P.A. RACK1, A multifaceted scaffolding protein: Structure and function. Cell Commun. Signal 2011, 9, 22. [Google Scholar] [CrossRef]
- Urano, D.; Czarnecki, O.; Wang, X.; Jones, A.M.; Chen, J.G. Arabidopsis receptor of activated C kinase1 phosphorylation by WITH NO LYSINE8 KINASE. Plant Physiol. 2015, 167, 507–516. [Google Scholar] [CrossRef]
- Reuper, H.; Krenz, B. Comparison of two Turnip mosaic virus P1 proteins in their ability to co-localize with the Arabidopsis thaliana G3BP-2 protein. Virus Genes. 2021, 57, 233–237. [Google Scholar] [CrossRef]
- Paul, P.; Joshi, S.; Tian, R.; Diogo Junior, R.; Chakrabarti, M.; Perry, S.E. The MADS-domain factor AGAMOUS-Like18 promotes somatic embryogenesis. Plant Physiol. 2022, 188, 1617–1631. [Google Scholar] [CrossRef]
- Misra, A.; Sriram, G. Network component analysis provides quantitative insights on an Arabidopsis transcription factor-gene regulatory network. BMC Syst. Biol. 2013, 7, 126. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wu, G.; Poethig, R.S. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 2006, 133, 3539–3547. [Google Scholar] [CrossRef]
- Serivichyaswat, P.; Ryu, H.S.; Kim, W.; Kim, S.; Chung, K.S.; Kim, J.J.; Ahn, J.H. Expression of the floral repressor miRNA156 is positively regulated by the AGAMOUS-like proteins AGL15 and AGL18. Mol. Cells 2015, 38, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.H.; Lee, H.J.; Ryu, J.Y.; Park, C.M. SPL3/4/5 integrate developmental aging and photoperiodic signals into the FT-FD module in Arabidopsis flowering. Mol. Plant 2016, 9, 1647–1659. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, L.; Han, J.; Qian, Z.; Zhou, B.; Xu, Y.; Wu, G. The nuclear localization signal is required for the function of squamosa promoter binding protein-like gene 9 to promote vegetative phase change in Arabidopsis. Plant Mol. Biol. 2019, 100, 571–578. [Google Scholar] [CrossRef] [PubMed]
- Teotia, S.; Tang, G. To bloom or not to bloom: Role of microRNAs in plant flowering. Mol. Plant 2015, 8, 359–377. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Yoo, S.J.; Lee, J.H.; Kim, W.; Yoo, S.K.; Fitzgerald, H.; Carrington, J.C.; Ahn, J.H. Genetic framework for flowering-time regulation by ambient temperature-responsive miRNAs in Arabidopsis. Nucleic Acids Res. 2010, 38, 3081–3093. [Google Scholar] [CrossRef]
- He, J.; Xu, M.; Willmann, M.R.; McCormick, K.; Hu, T.; Yang, L.; Starker, C.G.; Voytas, D.F.; Meyers, B.C.; Poethig, R.S. Threshold-dependent repression of SPL gene expression by miR156/miR157 controls vegetative phase change in Arabidopsis thaliana. PLoS Genet. 2018, 14, e1007337. [Google Scholar] [CrossRef]
- He, M.; Yang, Z.; Abdellatif, M.; Sayed, D. GTPase activating protein (Sh3 Domain) binding protein 1 regulates the processing of microRNA-1 during cardiac hypertrophy. PLoS ONE 2015, 10, e0145112. [Google Scholar] [CrossRef]
- Kwok, H.H.; Poon, P.Y.; Mak, K.H.; Zhang, L.Y.; Liu, P.; Zhang, H.; Mak, N.K.; Yue, P.Y.; Wong, R.N. Role of G3BP1 in glucocorticoid receptor-mediated microRNA-15b and microRNA-23a biogenesis in endothelial cells. Cell Mol. Life Sci. 2017, 74, 3613–3630. [Google Scholar] [CrossRef]
- Cheng, D.; Qian, W.; Wang, Y.; Meng, M.; Wei, L.; Li, Z.; Kang, L.; Peng, J.; Xia, Q. Nuclear import of transcription factor BR-C is mediated by its interaction with RACK1. PLoS ONE 2014, 9, e109111. [Google Scholar] [CrossRef]
- Li, Z.; Fu, Y.; Wang, Y.; Liang, J. Scaffold protein RACK1 regulates BR signaling by modulating the nuclear localization of BZR1. New Phytol. 2023, 239, 1804–1818. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Li, Z.; Liang, X.; Zhou, Y.; Liang, J. Nuclear Localization of G3BP6 Is Essential for the Flowering Transition in Arabidopsis. Biomolecules 2023, 13, 1697. https://doi.org/10.3390/biom13121697
Wang Y, Li Z, Liang X, Zhou Y, Liang J. Nuclear Localization of G3BP6 Is Essential for the Flowering Transition in Arabidopsis. Biomolecules. 2023; 13(12):1697. https://doi.org/10.3390/biom13121697
Chicago/Turabian StyleWang, Yuzhu, Zhiyong Li, Xiaoju Liang, Yeling Zhou, and Jiansheng Liang. 2023. "Nuclear Localization of G3BP6 Is Essential for the Flowering Transition in Arabidopsis" Biomolecules 13, no. 12: 1697. https://doi.org/10.3390/biom13121697
APA StyleWang, Y., Li, Z., Liang, X., Zhou, Y., & Liang, J. (2023). Nuclear Localization of G3BP6 Is Essential for the Flowering Transition in Arabidopsis. Biomolecules, 13(12), 1697. https://doi.org/10.3390/biom13121697





