Non-Coding RNAs as Potential Targets for Diagnosis and Treatment of Oral Lichen Planus: A Narrative Review
Abstract
1. Introduction
2. OLP Pathogenesis
2.1. Etiology and Initiation of OLP
2.2. Progression of OLP
2.3. Molecular Features of OLP Pathogenesis
2.3.1. Foxp3 (Forkhead Box Protein 3)
2.3.2. mTOR (Mammalian Target of Rapamycin)
2.3.3. HIF-1α (Hypoxia-Inducible Factor-1 Alpha)
2.3.4. TRIM21 (Tripartite Motif-Containing Protein 21)
3. Implications of miRNA for OLP Pathogenesis
3.1. Molecular Functions of miRNA
3.2. Roles and Biological Relevance of miRNAs in OLP
3.2.1. hsa-miR-155
3.2.2. hsa-miR-21
3.2.3. hsa-miR-146a
4. Implications of lncRNA for OLP Pathogenesis
4.1. Molecular Functions of lncRNA
4.2. Roles and Biological Relevance of lncRNAs in OLP
4.2.1. SPRR2C (Small Proline-Rich Protein 2C)
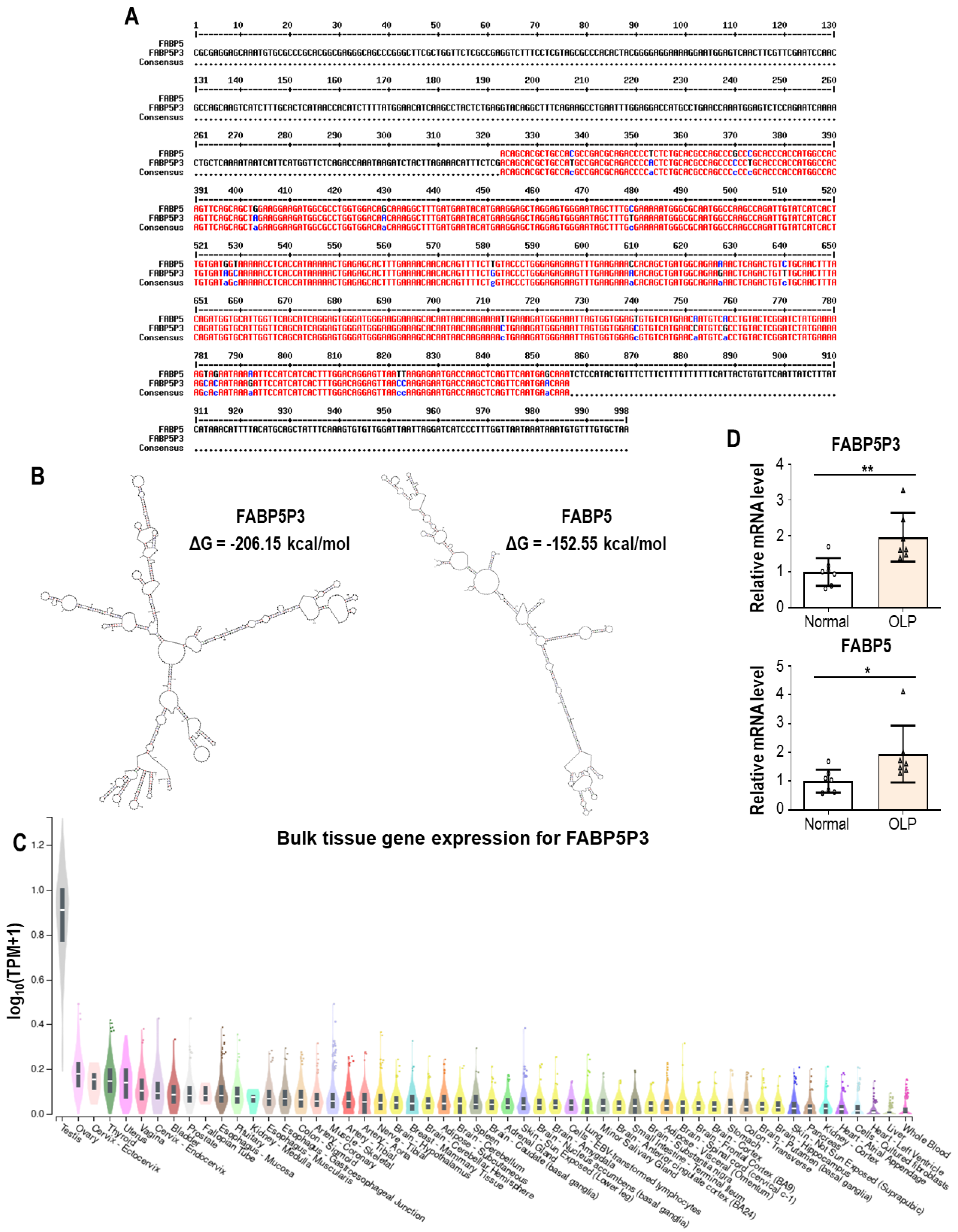
4.2.2. FABP5P3 (Fatty Acid Binding Protein 5 Pseudogene 3)
4.2.3. PRINS (Psoriasis Susceptibility-Related RNA Gene Induced by Stress)
4.2.4. DQ786243
5. Implications of circRNA for OLP Pathogenesis
5.1. Molecular Functions of circRNA
5.2. Roles and Biological Relevance of circRNAs in OLP
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gonzalez-Moles, M.A.; Warnakulasuriya, S.; Gonzalez-Ruiz, I.; Gonzalez-Ruiz, L.; Ayen, A.; Lenouvel, D.; Ruiz-Avila, I.; Ramos-Garcia, P. Worldwide prevalence of oral lichen planus: A systematic review and meta-analysis. Oral Dis. 2021, 27, 813–828. [Google Scholar] [CrossRef]
- Gonzalez-Moles, M.A.; Ruiz-Avila, I.; Gonzalez-Ruiz, L.; Ayen, A.; Gil-Montoya, J.A.; Ramos-Garcia, P. Malignant transformation risk of oral lichen planus: A systematic review and comprehensive meta-analysis. Oral Oncol. 2019, 96, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Tang, X.; Zheng, X.; Ge, S.; Wen, H.; Lin, X.; Chen, Z.; Lu, L. Global Prevalence and Incidence Estimates of Oral Lichen Planus: A Systematic Review and Meta-analysis. JAMA Dermatol. 2020, 156, 172–181. [Google Scholar] [CrossRef]
- Keller, L.M.; Lombardi, T. Gingival lichen planus: A clinical and pathological study. J. Stomatol. Oral Maxillofac. Surg. 2023, 124, 101354. [Google Scholar] [CrossRef] [PubMed]
- Lucchese, A.; Dolci, A.; Minervini, G.; Salerno, C.; Di Stasio, D.; Minervini, G.; Laino, L.; Silvestre, F.; Serpico, R. Vulvovaginal gingival lichen planus: Report of two cases and review of literature. Oral Implantol. 2016, 9, 54–60. [Google Scholar] [CrossRef]
- Warnakulasuriya, S.; Johnson, N.W.; van der Waal, I. Nomenclature and classification of potentially malignant disorders of the oral mucosa. J. Oral Pathol. Med. 2007, 36, 575–580. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Jawanda, M.K. Oral Lichen Planus: An Update on Etiology, Pathogenesis, Clinical Presentation, Diagnosis and Management. Indian. J. Dermatol. 2015, 60, 222–229. [Google Scholar] [CrossRef]
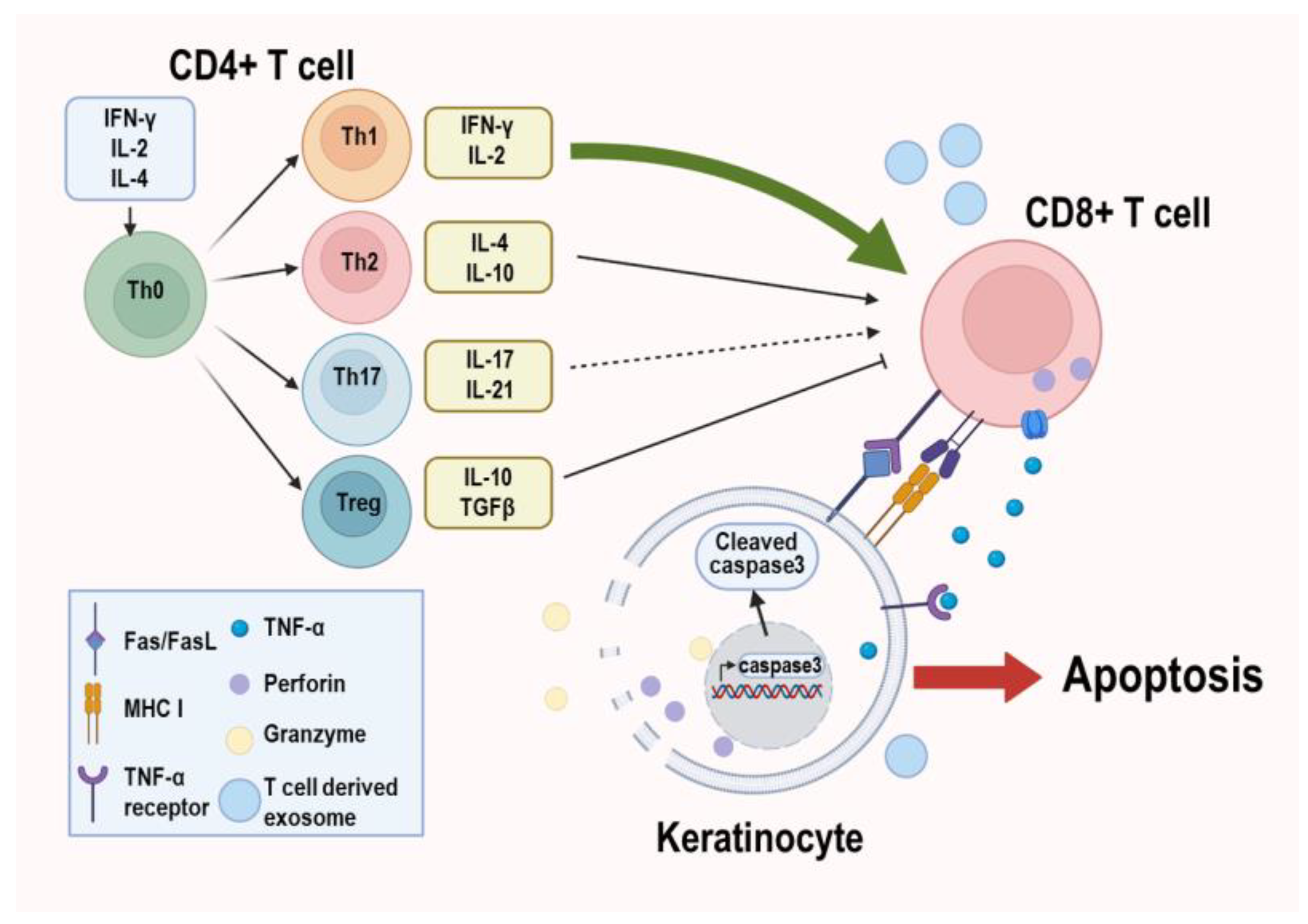
- Yang, J.Y.; Tan, Y.Q.; Zhou, G. T cell-derived exosomes containing cytokines induced keratinocytes apoptosis in oral lichen planus. Oral Dis. 2022, 28, 682–690. [Google Scholar] [CrossRef]
- Sun, Y.M.; Chen, Y.Q. Principles and innovative technologies for decrypting noncoding RNAs: From discovery and functional prediction to clinical application. J. Hematol. Oncol. 2020, 13, 109. [Google Scholar] [CrossRef] [PubMed]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Balatti, V.; Nigita, G.; Veneziano, D.; Drusco, A.; Stein, G.S.; Messier, T.L.; Farina, N.H.; Lian, J.B.; Tomasello, L.; Liu, C.G.; et al. tsRNA signatures in cancer. Proc. Natl. Acad. Sci. USA 2017, 114, 8071–8076. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Tan, Y.Q.; Zhang, J.; Zhou, G. Familial oral lichen planus in a 3-year-old boy: A case report with eight years of follow-up. BMC Oral Health 2020, 20, 341. [Google Scholar] [CrossRef]
- Jung, W.; Jang, S. Oral Microbiome Research on Oral Lichen Planus: Current Findings and Perspectives. Biology 2022, 11, 723. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yao, H.; Cui, B.; Ning, G.; Tang, G.Y. Genetic linkage analysis of oral lichen planus in a Chinese family. Genet. Mol. Res. 2011, 10, 1427–1433. [Google Scholar] [CrossRef]
- Farthing, P.M.; Cruchley, A.T. Expression of MHC class II antigens (HLA DR, DP and DQ) by keratinocytes in oral lichen planus. J. Oral Pathol. Med. 1989, 18, 305–309. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, M.; Abbaszadeh, H.; Mohtasham, N.; Salehiniya, H.; Shafaie, E. The association between high-risk human papillomavirus and oral lichen planus. Clin. Exp. Dent. Res. 2023, 9, 93–99. [Google Scholar] [CrossRef]
- Vijayan, A.K.; Muthukrishnan, A.; Velayudhannair, V.; Varun, J.; Vidyadharan, M.; James, J. Expression of human papillomavirus 16 and 18 DNA in oral lichen planus using polymerase chain reaction. J. Oral Maxillofac. Pathol. 2022, 26, 495–500. [Google Scholar] [CrossRef]
- Vijayan, A.K.; Muthukrishnan, A.; Vidyadharan, M.; Nair, A.M. Role of Human Papilloma Virus in Malignant Transformation of Oral Lichen Planus: A Systematic Review. J. Pharm. Bioallied Sci. 2021, 13, S62–S67. [Google Scholar] [CrossRef]
- Baek, K.; Lee, J.; Lee, A.; Lee, J.; Yoon, H.J.; Park, H.K.; Chun, J.; Choi, Y. Characterization of intratissue bacterial communities and isolation of Escherichia coli from oral lichen planus lesions. Sci. Rep. 2020, 10, 3495. [Google Scholar] [CrossRef]
- Wang, J.; Yang, J.; Xia, W.; Zhang, M.; Tang, H.; Wang, K.; Zhou, C.; Qian, L.; Fan, Y. Escherichia coli enhances Th17/Treg imbalance via TLR4/NF-kappaB signaling pathway in oral lichen planus. Int. Immunopharmacol. 2023, 119, 110175. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, S. Psychosocial stressors in oral lichen planus. Aust. Dent. J. 2004, 49, 192–195. [Google Scholar] [CrossRef] [PubMed]
- McCartan, B.E. Psychological factors associated with oral lichen planus. J. Oral Pathol. Med. 1995, 24, 273–275. [Google Scholar] [CrossRef] [PubMed]
- Eisen, D. The clinical features, malignant potential, and systemic associations of oral lichen planus: A study of 723 patients. J. Am. Acad. Dermatol. 2002, 46, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Cerqueira, J.D.M.; Moura, J.R.; Arsati, F.; Lima-Arsati, Y.B.O.; Bittencourt, R.A.; Freitas, V.S. Psychological disorders and oral lichen planus: A systematic review. J. Investig. Clin. Dent. 2018, 9, e12363. [Google Scholar] [CrossRef]
- Gheorghe, C.; Mihai, L.; Parlatescu, I.; Tovaru, S. Association of oral lichen planus with chronic C hepatitis. Review of the data in literature. Maedica 2014, 9, 98–103. [Google Scholar]
- Petrou-Amerikanou, C.; Markopoulos, A.K.; Belazi, M.; Karamitsos, D.; Papanayotou, P. Prevalence of oral lichen planus in diabetes mellitus according to the type of diabetes. Oral Dis. 1998, 4, 37–40. [Google Scholar] [CrossRef]
- Kaomongkolgit, R. Oral lichenoid drug reaction associated with antihypertensive and hypoglycemic drugs. J. Drugs Dermatol. 2010, 9, 73–75. [Google Scholar]
- Li, D.; Li, J.; Li, C.; Chen, Q.; Hua, H. The Association of Thyroid Disease and Oral Lichen Planus: A Literature Review and Meta-analysis. Front. Endocrinol. 2017, 8, 310. [Google Scholar] [CrossRef]
- Dreiher, J.; Shapiro, J.; Cohen, A.D. Lichen planus and dyslipidaemia: A case-control study. Br. J. Dermatol. 2009, 161, 626–629. [Google Scholar] [CrossRef]
- Hasan, S.; Ahmed, S.; Kiran, R.; Panigrahi, R.; Thachil, J.M.; Saeed, S. Oral lichen planus and associated comorbidities: An approach to holistic health. J. Family Med. Prim. Care 2019, 8, 3504–3517. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Gao, R.; Wang, Y.; Nguyen, T.; Yang, F.; Shi, Y.; Liu, T.; Liao, W.; Li, R.; Zhang, F.; et al. MicroRNA-26a/b have protective roles in oral lichen planus. Cell Death Dis. 2020, 11, 15. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, Y.; Du, G.; Zhang, W.; Cao, T.; Shi, L.; Wang, Y.; Mi, J.; Tang, G. Down-regulation of miRNA-27b-3p suppresses keratinocytes apoptosis in oral lichen planus. J. Cell Mol. Med. 2019, 23, 4326–4337. [Google Scholar] [CrossRef]
- Ge, X.; Yuan, L.; Wei, J.; Nguyen, T.; Tang, C.; Liao, W.; Li, R.; Yang, F.; Zhang, F.; Zhao, B.; et al. Vitamin D/VDR signaling induces miR-27a/b expression in oral lichen planus. Sci. Rep. 2020, 10, 301. [Google Scholar] [CrossRef]
- Dewhurst, G.; Wood, D.A.; Walker, F.; Lampe, F.C.; Jeffreys, M.; Cooper, M.; Williams, J.D. A population survey of cardiovascular disease in elderly people: Design, methods and prevalence results. Age Ageing 1991, 20, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Sachdeva, S.; Sachdeva, S.; Kapoor, P. Wickham striae: Etiopathogenensis and clinical significance. Indian. J. Dermatol. 2011, 56, 442–443. [Google Scholar] [CrossRef]
- Aragane, Y.; Riemann, H.; Bhardwaj, R.S.; Schwarz, A.; Sawada, Y.; Yamada, H.; Luger, T.A.; Kubin, M.; Trinchieri, G.; Schwarz, T. IL-12 is expressed and released by human keratinocytes and epidermoid carcinoma cell lines. J. Immunol. 1994, 153, 5366–5372. [Google Scholar] [CrossRef]
- Yamamoto, T.; Osaki, T. Characteristic cytokines generated by keratinocytes and mononuclear infiltrates in oral lichen planus. J. Investig. Dermatol. 1995, 104, 784–788. [Google Scholar] [CrossRef]
- Jana, A.; Thomas, J.; Ghosh, P. Erosive oral lichen planus inflicts higher cellular stress than reticular type. J. Oral Maxillofac. Pathol. 2021, 25, 279–285. [Google Scholar] [CrossRef]
- Alves, M.G.; Balducci, I.; Carvalho, Y.R.; Nunes, F.D.; Almeida, J.D. Oral lichen planus: A histopathological study. Histopathology 2015, 66, 463–464. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhou, J.; Fu, S.; Wang, C.; Zhou, B. A Study of Association Between Oral Lichen Planus and Immune Balance of Th1/Th2 Cells. Inflammation 2015, 38, 1874–1879. [Google Scholar] [CrossRef] [PubMed]
- Sugerman, P.B.; Savage, N.W.; Walsh, L.J.; Seymour, G.J. Disease mechanisms in oral lichen planus. A possible role for autoimmunity. Australas. J. Dermatol. 1993, 34, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, D.; Han, Q.; Zhao, X.; Zeng, X.; Xu, Y.; Sun, Z.; Chen, Q. Role of distinct CD4+ T helper subset in pathogenesis of oral lichen planus. J. Oral Pathol. Med. 2016, 45, 385–393. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, Y.; Zhao, Z.; Li, P.; Chen, D.; Wang, W.; Han, Y.; Zou, S.; Jin, X.; Zhao, J.; et al. Paeoniflorin drives the immunomodulatory effects of mesenchymal stem cells by regulating Th1/Th2 cytokines in oral lichen planus. Sci. Rep. 2022, 12, 18678. [Google Scholar] [CrossRef]
- Xie, S.; Ding, L.; Xiong, Z.; Zhu, S. Implications of Th1 and Th17 cells in pathogenesis of oral lichen planus. J. Huazhong Univ. Sci. Technolog. Med. Sci. 2012, 32, 451–457. [Google Scholar] [CrossRef]
- Zhang, D.; Wang, J.; Li, Z.; Zhou, M.; Chen, Q.; Zeng, X.; Chen, Y. The Activation of NF-kappaB in Infiltrated Mononuclear Cells Negatively Correlates with Treg Cell Frequency in Oral Lichen Planus. Inflammation 2015, 38, 1683–1689. [Google Scholar] [CrossRef]
- Wang, J.; Zhai, X.; Guo, J.; Li, Y.; Yang, Y.; Wang, L.; Yang, L.; Liu, F. Long non-coding RNA DQ786243 modulates the induction and function of CD4+ Treg cells through Foxp3-miR-146a-NF-kappaB axis: Implications for alleviating oral lichen planus. Int. Immunopharmacol. 2019, 75, 105761. [Google Scholar] [CrossRef]
- Tao, X.A.; Xia, J.; Chen, X.B.; Wang, H.; Dai, Y.H.; Rhodus, N.L.; Cheng, B. FOXP3 T regulatory cells in lesions of oral lichen planus correlated with disease activity. Oral Dis. 2010, 16, 76–82. [Google Scholar] [CrossRef]
- Zhou, X.J.; Sugerman, P.B.; Savage, N.W.; Walsh, L.J.; Seymour, G.J. Intra-epithelial CD8+ T cells and basement membrane disruption in oral lichen planus. J. Oral Pathol. Med. 2002, 31, 23–27. [Google Scholar] [CrossRef]
- Sugerman, P.B.; Satterwhite, K.; Bigby, M. Autocytotoxic T-cell clones in lichen planus. Br. J. Dermatol. 2000, 142, 449–456. [Google Scholar] [CrossRef]
- Sklavounou-Andrikopoulou, A.; Chrysomali, E.; Iakovou, M.; Garinis, G.A.; Karameris, A. Elevated serum levels of the apoptosis related molecules TNF-alpha, Fas/Apo-1 and Bcl-2 in oral lichen planus. J. Oral Pathol. Med. 2004, 33, 386–390. [Google Scholar] [CrossRef]
- Santoro, A.; Majorana, A.; Bardellini, E.; Gentili, F.; Festa, S.; Sapelli, P.; Facchetti, F. Cytotoxic molecule expression and epithelial cell apoptosis in oral and cutaneous lichen planus. Am. J. Clin. Pathol. 2004, 121, 758–764. [Google Scholar] [CrossRef]
- Santoro, A.; Majorana, A.; Bardellini, E.; Festa, S.; Sapelli, P.; Facchetti, F. NF-kappaB expression in oral and cutaneous lichen planus. J. Pathol. 2003, 201, 466–472. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Zhang, J.; Lichtenheld, M.G.; Meadows, G.G. A role for NF-kappa B activation in perforin expression of NK cells upon IL-2 receptor signaling. J. Immunol. 2002, 169, 1319–1325. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.Z.; Sugerman, P.B.; Zhou, X.J.; Walsh, L.J.; Savage, N.W. Mast cell degranulation and the role of T cell RANTES in oral lichen planus. Oral Dis. 2001, 7, 246–251. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wang, F.; Shi, Y.; Zhong, L.; Duan, S.; Kuang, W.; Liu, N.; Luo, E.; Zhou, Y.; Jiang, L.; et al. Single-cell immune profiling reveals immune responses in oral lichen planus. Front. Immunol. 2023, 14, 1182732. [Google Scholar] [CrossRef] [PubMed]
- Lei, L.; Zhan, L.; Tan, W.; Chen, S.; Li, Y.; Reynolds, M. Foxp3 gene expression in oral lichen planus: A clinicopathological study. Mol. Med. Rep. 2014, 9, 928–934. [Google Scholar] [CrossRef][Green Version]
- Wang, F.; Zhang, J.; Zhou, G. The mTOR-glycolytic pathway promotes T-cell immunobiology in oral lichen planus. Immunobiology 2020, 225, 151933. [Google Scholar] [CrossRef]
- Ma, R.J.; Tan, Y.Q.; Zhou, G. Aberrant IGF1-PI3K/AKT/MTOR signaling pathway regulates the local immunity of oral lichen planus. Immunobiology 2019, 224, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Zhang, J.; Zhou, G. HIF1alpha/PLD2 axis linked to glycolysis induces T-cell immunity in oral lichen planus. Biochim. Biophys. Acta Gen. Subj. 2020, 1864, 129602. [Google Scholar] [CrossRef]
- Ding, M.; Xu, J.Y.; Fan, Y. Altered expression of mRNA for HIF-1alpha and its target genes RTP801 and VEGF in patients with oral lichen planus. Oral Dis. 2010, 16, 299–304. [Google Scholar] [CrossRef]
- de Carvalho Fraga, C.A.; Alves, L.R.; Marques-Silva, L.; de Sousa, A.A.; Jorge, A.S.; de Jesus, S.F.; Vilela, D.N.; Pinheiro, U.B.; Jones, K.M.; de Paula, A.M.; et al. High HIF-1alpha expression genotypes in oral lichen planus. Clin. Oral Investig. 2013, 17, 2011–2015. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Wang, Y.; Sun, Q.; Jiang, C.; Zhu, M.; Song, C.; Li, C.; Du, G.; Deng, Y.; Nie, H.; et al. Enhanced T-cell proliferation and IL-6 secretion mediated by overexpression of TRIM21 in oral lesions of patients with oral lichen planus. J. Oral Pathol. Med. 2020, 49, 350–356. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Wei, W.; Wang, Y.; Song, C.; Pan, L.; Sun, K.; Du, G.; Deng, Y.; Tang, G. TRIM21 causes abnormal expression of IL-6 in oral lichen planus via the TRIB2-MAPK signal axis. Am. J. Transl. Res. 2020, 12, 4648–4658. [Google Scholar] [PubMed]
- Yao, Y.; Pan, L.; Wei, Y.; Feng, M.; Li, X.; Sun, L.; Tang, G.; Wang, Y. TRIM21 promotes inflammation by ubiquitylating NF-kappaB in T cells of oral lichen planus. J. Oral Pathol. Med. 2023, 52, 448–455. [Google Scholar] [CrossRef] [PubMed]
- Scotto, G.; Fazio, V.; Massa, S.; Lo Muzio, L.; Spirito, F. COVID-19 and Oral Lichen Planus: Between an “Intriguing Plot” and the “Fata Morgana Effect”. J. Clin. Med. 2023, 12, 4829. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Vasudevan, S.; Tong, Y.; Steitz, J.A. Switching from repression to activation: microRNAs can up-regulate translation. Science 2007, 318, 1931–1934. [Google Scholar] [CrossRef]
- Lin, C.C.; Liu, L.Z.; Addison, J.B.; Wonderlin, W.F.; Ivanov, A.V.; Ruppert, J.M. A KLF4-miRNA-206 autoregulatory feedback loop can promote or inhibit protein translation depending upon cell context. Mol. Cell Biol. 2011, 31, 2513–2527. [Google Scholar] [CrossRef]
- Orom, U.A.; Nielsen, F.C.; Lund, A.H. MicroRNA-10a binds the 5’UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell 2008, 30, 460–471. [Google Scholar] [CrossRef]
- Dharap, A.; Pokrzywa, C.; Murali, S.; Pandi, G.; Vemuganti, R. MicroRNA miR-324-3p induces promoter-mediated expression of RelA gene. PLoS ONE 2013, 8, e79467. [Google Scholar] [CrossRef] [PubMed]
- Miao, L.; Yao, H.; Li, C.; Pu, M.; Yao, X.; Yang, H.; Qi, X.; Ren, J.; Wang, Y. A dual inhibition: microRNA-552 suppresses both transcription and translation of cytochrome P450 2E1. Biochim. Biophys. Acta 2016, 1859, 650–662. [Google Scholar] [CrossRef] [PubMed]
- Xiao, M.; Li, J.; Li, W.; Wang, Y.; Wu, F.; Xi, Y.; Zhang, L.; Ding, C.; Luo, H.; Li, Y.; et al. MicroRNAs activate gene transcription epigenetically as an enhancer trigger. RNA Biol. 2017, 14, 1326–1334. [Google Scholar] [CrossRef] [PubMed]
- Benhamed, M.; Herbig, U.; Ye, T.; Dejean, A.; Bischof, O. Senescence is an endogenous trigger for microRNA-directed transcriptional gene silencing in human cells. Nat. Cell Biol. 2012, 14, 266–275. [Google Scholar] [CrossRef] [PubMed]
- Park, C.K.; Xu, Z.Z.; Berta, T.; Han, Q.; Chen, G.; Liu, X.J.; Ji, R.R. Extracellular microRNAs activate nociceptor neurons to elicit pain via TLR7 and TRPA1. Neuron 2014, 82, 47–54. [Google Scholar] [CrossRef]
- Lehmann, S.M.; Kruger, C.; Park, B.; Derkow, K.; Rosenberger, K.; Baumgart, J.; Trimbuch, T.; Eom, G.; Hinz, M.; Kaul, D.; et al. An unconventional role for miRNA: Let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat. Neurosci. 2012, 15, 827–835. [Google Scholar] [CrossRef]
- Han, Q.; Liu, D.; Convertino, M.; Wang, Z.; Jiang, C.; Kim, Y.H.; Luo, X.; Zhang, X.; Nackley, A.; Dokholyan, N.V.; et al. miRNA-711 Binds and Activates TRPA1 Extracellularly to Evoke Acute and Chronic Pruritus. Neuron 2018, 99, 449–463.e6. [Google Scholar] [CrossRef]
- Ma, H.; Wu, Y.; Yang, H.; Liu, J.; Dan, H.; Zeng, X.; Zhou, Y.; Jiang, L.; Chen, Q. MicroRNAs in oral lichen planus and potential miRNA-mRNA pathogenesis with essential cytokines: A review. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2016, 122, 164–173. [Google Scholar] [CrossRef]
- Sharma, S.; Opyrchal, M.; Lu, X. Harnessing tumorous flaws for immune supremacy: Is miRNA-155 the weak link in breast cancer progression? J. Clin. Investig. 2022, 132, e163010. [Google Scholar] [CrossRef]
- Yin, Q.; Wang, X.; McBride, J.; Fewell, C.; Flemington, E. B-cell receptor activation induces BIC/miR-155 expression through a conserved AP-1 element. J. Biol. Chem. 2008, 283, 2654–2662. [Google Scholar] [CrossRef]
- Bruning, U.; Cerone, L.; Neufeld, Z.; Fitzpatrick, S.F.; Cheong, A.; Scholz, C.C.; Simpson, D.A.; Leonard, M.O.; Tambuwala, M.M.; Cummins, E.P.; et al. MicroRNA-155 promotes resolution of hypoxia-inducible factor 1alpha activity during prolonged hypoxia. Mol. Cell Biol. 2011, 31, 4087–4096. [Google Scholar] [CrossRef] [PubMed]
- Kluiver, J.; van den Berg, A.; de Jong, D.; Blokzijl, T.; Harms, G.; Bouwman, E.; Jacobs, S.; Poppema, S.; Kroesen, B.J. Regulation of pri-microRNA BIC transcription and processing in Burkitt lymphoma. Oncogene 2007, 26, 3769–3776. [Google Scholar] [CrossRef] [PubMed]
- Vigorito, E.; Kohlhaas, S.; Lu, D.; Leyland, R. miR-155: An ancient regulator of the immune system. Immunol. Rev. 2013, 253, 146–157. [Google Scholar] [CrossRef] [PubMed]
- Monnot, G.C.; Martinez-Usatorre, A.; Lanitis, E.; Lopes, S.F.; Cheng, W.C.; Ho, P.C.; Irving, M.; Coukos, G.; Donda, A.; Romero, P. miR-155 Overexpression in OT-1 CD8+ T Cells Improves Anti-Tumor Activity against Low-Affinity Tumor Antigen. Mol. Ther. Oncolytics 2020, 16, 111–123. [Google Scholar] [CrossRef] [PubMed]
- Dudda, J.C.; Salaun, B.; Ji, Y.; Palmer, D.C.; Monnot, G.C.; Merck, E.; Boudousquie, C.; Utzschneider, D.T.; Escobar, T.M.; Perret, R.; et al. MicroRNA-155 is required for effector CD8+ T cell responses to virus infection and cancer. Immunity 2013, 38, 742–753. [Google Scholar] [CrossRef]
- Gracias, D.T.; Stelekati, E.; Hope, J.L.; Boesteanu, A.C.; Doering, T.A.; Norton, J.; Mueller, Y.M.; Fraietta, J.A.; Wherry, E.J.; Turner, M.; et al. The microRNA miR-155 controls CD8+ T cell responses by regulating interferon signaling. Nat. Immunol. 2013, 14, 593–602. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Wrzesinski, C.; Yu, Z.; Hu, J.; Gautam, S.; Hawk, N.V.; Telford, W.G.; Palmer, D.C.; Franco, Z.; Sukumar, M.; et al. miR-155 augments CD8+ T-cell antitumor activity in lymphoreplete hosts by enhancing responsiveness to homeostatic gammac cytokines. Proc. Natl. Acad. Sci. USA 2015, 112, 476–481. [Google Scholar] [CrossRef]
- Thai, T.H.; Calado, D.P.; Casola, S.; Ansel, K.M.; Xiao, C.; Xue, Y.; Murphy, A.; Frendewey, D.; Valenzuela, D.; Kutok, J.L.; et al. Regulation of the germinal center response by microRNA-155. Science 2007, 316, 604–608. [Google Scholar] [CrossRef] [PubMed]
- Arbore, G.; Henley, T.; Biggins, L.; Andrews, S.; Vigorito, E.; Turner, M.; Leyland, R. MicroRNA-155 is essential for the optimal proliferation and survival of plasmablast B cells. Life Sci. Alliance 2019, 2, e201800244. [Google Scholar] [CrossRef] [PubMed]
- Teng, G.; Hakimpour, P.; Landgraf, P.; Rice, A.; Tuschl, T.; Casellas, R.; Papavasiliou, F.N. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity 2008, 28, 621–629. [Google Scholar] [CrossRef]
- Taganov, K.D.; Boldin, M.P.; Chang, K.J.; Baltimore, D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA 2006, 103, 12481–12486. [Google Scholar] [CrossRef]
- O’Connell, R.M.; Taganov, K.D.; Boldin, M.P.; Cheng, G.; Baltimore, D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. USA 2007, 104, 1604–1609. [Google Scholar] [CrossRef] [PubMed]
- Tili, E.; Michaille, J.J.; Cimino, A.; Costinean, S.; Dumitru, C.D.; Adair, B.; Fabbri, M.; Alder, H.; Liu, C.G.; Calin, G.A.; et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J. Immunol. 2007, 179, 5082–5089. [Google Scholar] [CrossRef] [PubMed]
- Kurowska-Stolarska, M.; Alivernini, S.; Ballantine, L.E.; Asquith, D.L.; Millar, N.L.; Gilchrist, D.S.; Reilly, J.; Ierna, M.; Fraser, A.R.; Stolarski, B.; et al. MicroRNA-155 as a proinflammatory regulator in clinical and experimental arthritis. Proc. Natl. Acad. Sci. USA 2011, 108, 11193–11198. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Zheng, L.Y.; Zhang, P.; Yu, C.Q. miR-146a and miR-155 expression in PBMCs from patients with Sjogren’s syndrome. J. Oral Pathol. Med. 2014, 43, 792–797. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.Q.; Zilahi, E.; Papp, G.; Sipka, S.; Zeher, M. Simultaneously increased expression of microRNA-155 and suppressor of cytokine signaling 1 (SOCS1) gene in the peripheral blood mononuclear cells of patients with primary Sjogren’s syndrome. Int. J. Rheum. Dis. 2017, 20, 609–613. [Google Scholar] [CrossRef]
- Alevizos, I.; Alexander, S.; Turner, R.J.; Illei, G.G. MicroRNA expression profiles as biomarkers of minor salivary gland inflammation and dysfunction in Sjogren’s syndrome. Arthritis Rheum. 2011, 63, 535–544. [Google Scholar] [CrossRef]
- Zhang, J.; Zhu, L.; Shi, H.; Zheng, H. Protective effects of miR-155-5p silencing on IFN-gamma-induced apoptosis and inflammation in salivary gland epithelial cells. Exp. Ther. Med. 2021, 22, 882. [Google Scholar] [CrossRef]
- Hu, J.Y.; Zhang, J.; Ma, J.Z.; Liang, X.Y.; Chen, G.Y.; Lu, R.; Du, G.F.; Zhou, G. MicroRNA-155-IFN-gamma Feedback Loop in CD4+T Cells of Erosive type Oral Lichen Planus. Sci. Rep. 2015, 5, 16935. [Google Scholar] [CrossRef]
- Wang, L.; Wu, W.; Chen, J.; Li, Y.; Xu, M.; Cai, Y. MicroRNA Microarray-Based Identification of Involvement of miR-155 and miR-19a in Development of Oral Lichen Planus (OLP) by Modulating Th1/Th2 Balance via Targeting eNOS and Toll-Like Receptor 2 (TLR2). Med. Sci. Monit. 2018, 24, 3591–3603. [Google Scholar] [CrossRef]
- Cheng, J.; Zhang, Y.; Yang, J.; Wang, Y.; Xu, J.; Fan, Y. MiR-155-5p modulates inflammatory phenotype of activated oral lichen-planus-associated-fibroblasts by targeting SOCS1. Mol. Biol. Rep. 2022, 49, 7783–7792. [Google Scholar] [CrossRef]
- Danielsson, K.; Wahlin, Y.B.; Gu, X.; Boldrup, L.; Nylander, K. Altered expression of miR-21, miR-125b, and miR-203 indicates a role for these microRNAs in oral lichen planus. J. Oral Pathol. Med. 2012, 41, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Kumarswamy, R.; Volkmann, I.; Thum, T. Regulation and function of miRNA-21 in health and disease. RNA Biol. 2011, 8, 706–713. [Google Scholar] [CrossRef]
- Fujita, S.; Ito, T.; Mizutani, T.; Minoguchi, S.; Yamamichi, N.; Sakurai, K.; Iba, H. miR-21 Gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. J. Mol. Biol. 2008, 378, 492–504. [Google Scholar] [CrossRef]
- Loffler, D.; Brocke-Heidrich, K.; Pfeifer, G.; Stocsits, C.; Hackermuller, J.; Kretzschmar, A.K.; Burger, R.; Gramatzki, M.; Blumert, C.; Bauer, K.; et al. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood 2007, 110, 1330–1333. [Google Scholar] [CrossRef] [PubMed]
- McClure, C.; McPeak, M.B.; Youssef, D.; Yao, Z.Q.; McCall, C.E.; El Gazzar, M. Stat3 and C/EBPbeta synergize to induce miR-21 and miR-181b expression during sepsis. Immunol. Cell Biol. 2017, 95, 42–55. [Google Scholar] [CrossRef]
- Ribas, J.; Ni, X.; Castanares, M.; Liu, M.M.; Esopi, D.; Yegnasubramanian, S.; Rodriguez, R.; Mendell, J.T.; Lupold, S.E. A novel source for miR-21 expression through the alternative polyadenylation of VMP1 gene transcripts. Nucleic Acids Res. 2012, 40, 6821–6833. [Google Scholar] [CrossRef] [PubMed]
- Hashimi, S.T.; Fulcher, J.A.; Chang, M.H.; Gov, L.; Wang, S.; Lee, B. MicroRNA profiling identifies miR-34a and miR-21 and their target genes JAG1 and WNT1 in the coordinate regulation of dendritic cell differentiation. Blood 2009, 114, 404–414. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.X.; Munitz, A.; Rothenberg, M.E. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J. Immunol. 2009, 182, 4994–5002. [Google Scholar] [CrossRef] [PubMed]
- Xi, J.; Huang, Q.; Wang, L.; Ma, X.; Deng, Q.; Kumar, M.; Zhou, Z.; Li, L.; Zeng, Z.; Young, K.H.; et al. miR-21 depletion in macrophages promotes tumoricidal polarization and enhances PD-1 immunotherapy. Oncogene 2018, 37, 3151–3165. [Google Scholar] [CrossRef] [PubMed]
- Canfran-Duque, A.; Rotllan, N.; Zhang, X.; Fernandez-Fuertes, M.; Ramirez-Hidalgo, C.; Araldi, E.; Daimiel, L.; Busto, R.; Fernandez-Hernando, C.; Suarez, Y. Macrophage deficiency of miR-21 promotes apoptosis, plaque necrosis, and vascular inflammation during atherogenesis. EMBO Mol. Med. 2017, 9, 1244–1262. [Google Scholar] [CrossRef] [PubMed]
- Carissimi, C.; Carucci, N.; Colombo, T.; Piconese, S.; Azzalin, G.; Cipolletta, E.; Citarella, F.; Barnaba, V.; Macino, G.; Fulci, V. miR-21 is a negative modulator of T-cell activation. Biochimie 2014, 107 Pt B, 319–326. [Google Scholar] [CrossRef]
- Wang, L.; He, L.; Zhang, R.; Liu, X.; Ren, Y.; Liu, Z.; Zhang, X.; Cheng, W.; Hua, Z.C. Regulation of T lymphocyte activation by microRNA-21. Mol. Immunol. 2014, 59, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Q.; Wang, P.; Wang, T.; Qi, J.; Wei, M.; Wang, S.; Fan, T.; Johnson, D.; Wan, X.; Shi, W.; et al. MicroRNA-21 regulates T-cell apoptosis by directly targeting the tumor suppressor gene Tipe2. Cell Death Dis. 2014, 5, e1095. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.X.; Hartner, J.; Lim, E.J.; Fabry, V.; Mingler, M.K.; Cole, E.T.; Orkin, S.H.; Aronow, B.J.; Rothenberg, M.E. MicroRNA-21 limits in vivo immune response-mediated activation of the IL-12/IFN-gamma pathway, Th1 polarization, and the severity of delayed-type hypersensitivity. J. Immunol. 2011, 187, 3362–3373. [Google Scholar] [CrossRef] [PubMed]
- Sheedy, F.J.; Palsson-McDermott, E.; Hennessy, E.J.; Martin, C.; O’Leary, J.J.; Ruan, Q.; Johnson, D.S.; Chen, Y.; O’Neill, L.A. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat. Immunol. 2010, 11, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Meisgen, F.; Xu, N.; Wei, T.; Janson, P.C.; Obad, S.; Broom, O.; Nagy, N.; Kauppinen, S.; Kemeny, L.; Stahle, M.; et al. MiR-21 is up-regulated in psoriasis and suppresses T cell apoptosis. Exp. Dermatol. 2012, 21, 312–314. [Google Scholar] [CrossRef] [PubMed]
- Guinea-Viniegra, J.; Jimenez, M.; Schonthaler, H.B.; Navarro, R.; Delgado, Y.; Concha-Garzon, M.J.; Tschachler, E.; Obad, S.; Dauden, E.; Wagner, E.F. Targeting miR-21 to treat psoriasis. Sci. Transl. Med. 2014, 6, 225re221. [Google Scholar] [CrossRef] [PubMed]
- Mehdipour, M.; Shahidi, M.; Manifar, S.; Jafari, S.; Mashhadi Abbas, F.; Barati, M.; Mortazavi, H.; Shirkhoda, M.; Farzanegan, A.; Elmi Rankohi, Z. Diagnostic and prognostic relevance of salivary microRNA-21, -125a, -31 and -200a levels in patients with oral lichen planus—A short report. Cell Oncol. 2018, 41, 329–334. [Google Scholar] [CrossRef]
- Lagos-Quintana, M.; Rauhut, R.; Yalcin, A.; Meyer, J.; Lendeckel, W.; Tuschl, T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002, 12, 735–739. [Google Scholar] [CrossRef]
- Kutty, R.K.; Nagineni, C.N.; Samuel, W.; Vijayasarathy, C.; Jaworski, C.; Duncan, T.; Cameron, J.E.; Flemington, E.K.; Hooks, J.J.; Redmond, T.M. Differential regulation of microRNA-146a and microRNA-146b-5p in human retinal pigment epithelial cells by interleukin-1beta, tumor necrosis factor-alpha, and interferon-gamma. Mol. Vis. 2013, 19, 737–750. [Google Scholar]
- Paterson, M.R.; Kriegel, A.J. MiR-146a/b: A family with shared seeds and different roots. Physiol. Genom. 2017, 49, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Xia, P.; Fang, X.; Zhang, Z.H.; Huang, Q.; Yan, K.X.; Kang, K.F.; Han, L.; Zheng, Z.Z. Dysregulation of miRNA146a versus IRAK1 induces IL-17 persistence in the psoriatic skin lesions. Immunol. Lett. 2012, 148, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Niimoto, T.; Nakasa, T.; Ishikawa, M.; Okuhara, A.; Izumi, B.; Deie, M.; Suzuki, O.; Adachi, N.; Ochi, M. MicroRNA-146a expresses in interleukin-17 producing T cells in rheumatoid arthritis patients. BMC Musculoskelet. Disord. 2010, 11, 209. [Google Scholar] [CrossRef]
- Yin, Y.; Li, F.; Shi, J.; Li, S.; Cai, J.; Jiang, Y. MiR-146a Regulates Inflammatory Infiltration by Macrophages in Polymyositis/Dermatomyositis by Targeting TRAF6 and Affecting IL-17/ICAM-1 Pathway. Cell Physiol. Biochem. 2016, 40, 486–498. [Google Scholar] [CrossRef]
- Tang, Y.; Luo, X.; Cui, H.; Ni, X.; Yuan, M.; Guo, Y.; Huang, X.; Zhou, H.; de Vries, N.; Tak, P.P.; et al. MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum. 2009, 60, 1065–1075. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.G.; Sun, Y.R.; Chen, G.Y.; Liang, X.Y.; Zhang, J.; Zhou, G. Different Expression of MicroRNA-146a in Peripheral Blood CD4+ T Cells and Lesions of Oral Lichen Planus. Inflammation 2016, 39, 860–866. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, L.; Wang, L.; Yang, Y.; Wang, Y. Forkhead box p3 controls progression of oral lichen planus by regulating microRNA-146a. J. Cell Biochem. 2018, 119, 8862–8871. [Google Scholar] [CrossRef]
- Guo, C.J.; Ma, X.K.; Xing, Y.H.; Zheng, C.C.; Xu, Y.F.; Shan, L.; Zhang, J.; Wang, S.; Wang, Y.; Carmichael, G.G.; et al. Distinct Processing of lncRNAs Contributes to Non-conserved Functions in Stem Cells. Cell 2020, 181, 621–636.e22. [Google Scholar] [CrossRef] [PubMed]
- Hezroni, H.; Koppstein, D.; Schwartz, M.G.; Avrutin, A.; Bartel, D.P.; Ulitsky, I. Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Rep. 2015, 11, 1110–1122. [Google Scholar] [CrossRef] [PubMed]
- Mele, M.; Mattioli, K.; Mallard, W.; Shechner, D.M.; Gerhardinger, C.; Rinn, J.L. Chromatin environment, transcriptional regulation, and splicing distinguish lincRNAs and mRNAs. Genome Res. 2017, 27, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Carlevaro-Fita, J.; Rahim, A.; Guigo, R.; Vardy, L.A.; Johnson, R. Cytoplasmic long noncoding RNAs are frequently bound to and degraded at ribosomes in human cells. RNA 2016, 22, 867–882. [Google Scholar] [CrossRef]
- Wang, K.C.; Yang, Y.W.; Liu, B.; Sanyal, A.; Corces-Zimmerman, R.; Chen, Y.; Lajoie, B.R.; Protacio, A.; Flynn, R.A.; Gupta, R.A.; et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 2011, 472, 120–124. [Google Scholar] [CrossRef] [PubMed]
- Kuo, C.C.; Hanzelmann, S.; Senturk Cetin, N.; Frank, S.; Zajzon, B.; Derks, J.P.; Akhade, V.S.; Ahuja, G.; Kanduri, C.; Grummt, I.; et al. Detection of RNA-DNA binding sites in long noncoding RNAs. Nucleic Acids Res. 2019, 47, e32. [Google Scholar] [CrossRef] [PubMed]
- Yap, K.; Mukhina, S.; Zhang, G.; Tan, J.S.C.; Ong, H.S.; Makeyev, E.V. A Short Tandem Repeat-Enriched RNA Assembles a Nuclear Compartment to Control Alternative Splicing and Promote Cell Survival. Mol. Cell 2018, 72, 525–540.e13. [Google Scholar] [CrossRef]
- Gong, C.; Maquat, L.E. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 2011, 470, 284–288. [Google Scholar] [CrossRef]
- Grelet, S.; Link, L.A.; Howley, B.; Obellianne, C.; Palanisamy, V.; Gangaraju, V.K.; Diehl, J.A.; Howe, P.H. A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nat. Cell Biol. 2017, 19, 1105–1115. [Google Scholar] [CrossRef] [PubMed]
- Lv, D.; Chang, Z.; Cai, Y.; Li, J.; Wang, L.; Jiang, Q.; Xu, K.; Ding, N.; Li, X.; Xu, J.; et al. TransLnc: A comprehensive resource for translatable lncRNAs extends immunopeptidome. Nucleic Acids Res. 2022, 50, D413–D420. [Google Scholar] [CrossRef] [PubMed]
- Barczak, W.; Carr, S.M.; Liu, G.; Munro, S.; Nicastri, A.; Lee, L.N.; Hutchings, C.; Ternette, N.; Klenerman, P.; Kanapin, A.; et al. Long non-coding RNA-derived peptides are immunogenic and drive a potent anti-tumour response. Nat. Commun. 2023, 14, 1078. [Google Scholar] [CrossRef] [PubMed]
- Tsoi, L.C.; Iyer, M.K.; Stuart, P.E.; Swindell, W.R.; Gudjonsson, J.E.; Tejasvi, T.; Sarkar, M.K.; Li, B.; Ding, J.; Voorhees, J.J.; et al. Analysis of long non-coding RNAs highlights tissue-specific expression patterns and epigenetic profiles in normal and psoriatic skin. Genome Biol. 2015, 16, 24. [Google Scholar] [CrossRef]
- South, A.P.; Cabral, A.; Ives, J.H.; James, C.H.; Mirza, G.; Marenholz, I.; Mischke, D.; Backendorf, C.; Ragoussis, J.; Nizetic, D. Human epidermal differentiation complex in a single 2.5 Mbp long continuum of overlapping DNA cloned in bacteria integrating physical and transcript maps. J. Investig. Dermatol. 1999, 112, 910–918. [Google Scholar] [CrossRef]
- Cabral, A.; Sayin, A.; de Winter, S.; Fischer, D.F.; Pavel, S.; Backendorf, C. SPRR4, a novel cornified envelope precursor: UV-dependent epidermal expression and selective incorporation into fragile envelopes. J. Cell Sci. 2001, 114, 3837–3843. [Google Scholar] [CrossRef]
- Kartasova, T.; van de Putte, P. Isolation, characterization, and UV-stimulated expression of two families of genes encoding polypeptides of related structure in human epidermal keratinocytes. Mol. Cell Biol. 1988, 8, 2195–2203. [Google Scholar] [CrossRef]
- Ziegler, C.; Graf, J.; Faderl, S.; Schedlbauer, J.; Strieder, N.; Forstl, B.; Spang, R.; Bruckmann, A.; Merkl, R.; Hombach, S.; et al. The long non-coding RNA LINC00941 and SPRR5 are novel regulators of human epidermal homeostasis. EMBO Rep. 2019, 20, e46612. [Google Scholar] [CrossRef] [PubMed]
- Wilflingseder, J.; Kainz, A.; Perco, P.; Korbely, R.; Mayer, B.; Oberbauer, R. Molecular predictors for anaemia after kidney transplantation. Nephrol. Dial. Transplant. 2009, 24, 1015–1023. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shefler, A.; Patrick, M.T.; Wasikowski, R.; Chen, J.; Sarkar, M.K.; Gudjonsson, J.E.; Tsoi, L.C. Skin-Expressing lncRNAs in Inflammatory Responses. Front. Genet. 2022, 13, 835740. [Google Scholar] [CrossRef]
- Yin, X.; Yang, Z.; Zhu, M.; Chen, C.; Sun, Q. Role of the long non-coding RNA, SPRR2C, based on an in vitro psoriatic keratinocyte cell model. Eur. J. Dermatol. 2022, 32, 171–180. [Google Scholar] [CrossRef]
- Luo, M.; Huang, P.; Pan, Y.; Zhu, Z.; Zhou, R.; Yang, Z.; Wang, C. Weighted gene coexpression network and experimental analyses identify lncRNA SPRR2C as a regulator of the IL-22-stimulated HaCaT cell phenotype through the miR-330/STAT1/S100A7 axis. Cell Death Dis. 2021, 12, 86. [Google Scholar] [CrossRef] [PubMed]
- Breunig, S.; Wallner, V.; Kobler, K.; Wimmer, H.; Steinbacher, P.; Streubel, M.K.; Bischof, J.; Duschl, J.; Neuhofer, C.; Gruber, W.; et al. The life in a gradient: Calcium, the lncRNA SPRR2C and mir542/mir196a meet in the epidermis to regulate the aging process. Aging 2021, 13, 19127–19144. [Google Scholar] [CrossRef] [PubMed]
- George Warren, W.; Osborn, M.; Yates, A.; Wright, K.; O’Sullivan, S.E. The emerging role of fatty acid binding protein 5 (FABP5) in cancers. Drug Discov. Today 2023, 28, 103628. [Google Scholar] [CrossRef] [PubMed]
- Smathers, R.L.; Petersen, D.R. The human fatty acid-binding protein family: Evolutionary divergences and functions. Hum. Genomics 2011, 5, 170–191. [Google Scholar] [CrossRef] [PubMed]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zeng, J.; Yin, G.; Deng, Z.; Wang, L.; Liu, J.; Yao, K.; Long, Z.; Jiang, X.; Tan, J. Long non-coding RNA FABP5P3/miR-22 axis improves TGFbeta1-induced fatty acid oxidation deregulation and fibrotic changes in proximal tubular epithelial cells of renal fibrosis. Cell Cycle 2023, 22, 433–449. [Google Scholar] [CrossRef] [PubMed]
- Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988, 16, 10881–10890. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Luo, Z.; Lu, G.; Gui, F.; Wu, J.; Li, F.; Ni, Y. LncRNA FABP5P3/miR-589-5p/ZMYND19 axis contributes to hepatocellular carcinoma cell proliferation, migration and invasion. Biochem. Biophys. Res. Commun. 2018, 498, 551–558. [Google Scholar] [CrossRef]
- Huang, S.; Zhen, Y.; Yin, X.; Yang, Z.; Li, X.; Wang, R.; Wen, H.; Zhong, H.; Yan, J.; Sun, Q. KMT2C Induced by FABP5P3 Aggravates Keratinocyte Hyperproliferation and Psoriasiform Skin Inflammation by Upregulating the Transcription of PIK3R3. J. Investig. Dermatol. 2023, 143, 37–47.e8. [Google Scholar] [CrossRef]
- Danielsson, K.; Coates, P.J.; Ebrahimi, M.; Nylander, E.; Wahlin, Y.B.; Nylander, K. Genes involved in epithelial differentiation and development are differentially expressed in oral and genital lichen planus epithelium compared to normal epithelium. Acta Derm. Venereol. 2014, 94, 526–530. [Google Scholar] [CrossRef]
- Sonkoly, E.; Bata-Csorgo, Z.; Pivarcsi, A.; Polyanka, H.; Kenderessy-Szabo, A.; Molnar, G.; Szentpali, K.; Bari, L.; Megyeri, K.; Mandi, Y.; et al. Identification and characterization of a novel, psoriasis susceptibility-related noncoding RNA gene, PRINS. J. Biol. Chem. 2005, 280, 24159–24167. [Google Scholar] [CrossRef]
- Szegedi, K.; Goblos, A.; Bacsa, S.; Antal, M.; Nemeth, I.B.; Bata-Csorgo, Z.; Kemeny, L.; Dobozy, A.; Szell, M. Expression and functional studies on the noncoding RNA, PRINS. Int. J. Mol. Sci. 2012, 14, 205–225. [Google Scholar] [CrossRef]
- Szegedi, K.; Sonkoly, E.; Nagy, N.; Nemeth, I.B.; Bata-Csorgo, Z.; Kemeny, L.; Dobozy, A.; Szell, M. The anti-apoptotic protein G1P3 is overexpressed in psoriasis and regulated by the non-coding RNA, PRINS. Exp. Dermatol. 2010, 19, 269–278. [Google Scholar] [CrossRef]
- Szell, M.; Danis, J.; Bata-Csorgo, Z.; Kemeny, L. PRINS, a primate-specific long non-coding RNA, plays a role in the keratinocyte stress response and psoriasis pathogenesis. Pflugers Arch. 2016, 468, 935–943. [Google Scholar] [CrossRef]
- Abdallah, H.Y.; Tawfik, N.Z.; Soliman, N.H.; Eldeen, L.A.T. The lncRNA PRINS-miRNA-mRNA Axis Gene Expression Profile as a Circulating Biomarker Panel in Psoriasis. Mol. Diagn. Ther. 2022, 26, 451–465. [Google Scholar] [CrossRef]
- Danis, J.; Goblos, A.; Bata-Csorgo, Z.; Kemeny, L.; Szell, M. PRINS Non-Coding RNA Regulates Nucleic Acid-Induced Innate Immune Responses of Human Keratinocytes. Front. Immunol. 2017, 8, 1053. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Zhang, L.; Huo, X.S.; Yuan, J.H.; Xu, D.; Yuan, S.X.; Zhu, N.; Zhou, W.P.; Yang, G.S.; Wang, Y.Z.; et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology 2011, 54, 1679–1689. [Google Scholar] [CrossRef]
- Qiao, Y.Q.; Huang, M.L.; Xu, A.T.; Zhao, D.; Ran, Z.H.; Shen, J. LncRNA DQ786243 affects Treg related CREB and Foxp3 expression in Crohn’s disease. J. Biomed. Sci. 2013, 20, 87. [Google Scholar] [CrossRef] [PubMed]
- Viglietta, V.; Baecher-Allan, C.; Weiner, H.L.; Hafler, D.A. Loss of functional suppression by CD4+CD25+ regulatory T cells in patients with multiple sclerosis. J. Exp. Med. 2004, 199, 971–979. [Google Scholar] [CrossRef]
- Valencia, X.; Stephens, G.; Goldbach-Mansky, R.; Wilson, M.; Shevach, E.M.; Lipsky, P.E. TNF downmodulates the function of human CD4+CD25hi T-regulatory cells. Blood 2006, 108, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Cao, T.; Wang, Y.; Yao, H.; Du, G.; Chen, G.; Niu, X.; Tang, G. Frequently Increased but Functionally Impaired CD4+CD25+ Regulatory T Cells in Patients with Oral Lichen Planus. Inflammation 2016, 39, 1205–1215. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sharpless, N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014, 32, 453–461. [Google Scholar] [CrossRef]
- Enuka, Y.; Lauriola, M.; Feldman, M.E.; Sas-Chen, A.; Ulitsky, I.; Yarden, Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 2016, 44, 1370–1383. [Google Scholar] [CrossRef] [PubMed]
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol. Cell 2017, 66, 22–37.e9. [Google Scholar] [CrossRef]
- Wesselhoeft, R.A.; Kowalski, P.S.; Anderson, D.G. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat. Commun. 2018, 9, 2629. [Google Scholar] [CrossRef] [PubMed]
- Amaya, L.; Grigoryan, L.; Li, Z.; Lee, A.; Wender, P.A.; Pulendran, B.; Chang, H.Y. Circular RNA vaccine induces potent T cell responses. Proc. Natl. Acad. Sci. USA 2023, 120, e2302191120. [Google Scholar] [CrossRef]
- Nigro, J.M.; Cho, K.R.; Fearon, E.R.; Kern, S.E.; Ruppert, J.M.; Oliner, J.D.; Kinzler, K.W.; Vogelstein, B. Scrambled exons. Cell 1991, 64, 607–613. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Xi, Y.; Shen, Y.; Wu, D.; Zhang, J.; Lin, C.; Wang, L.; Yu, C.; Yu, B.; Shen, W. CircBCAR3 accelerates esophageal cancer tumorigenesis and metastasis via sponging miR-27a-3p. Mol. Cancer 2022, 21, 145. [Google Scholar] [CrossRef]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef]
- Du, W.W.; Fang, L.; Yang, W.; Wu, N.; Awan, F.M.; Yang, Z.; Yang, B.B. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017, 24, 357–370. [Google Scholar] [CrossRef]
- Huang, Q.; Guo, H.; Wang, S.; Ma, Y.; Chen, H.; Li, H.; Li, J.; Li, X.; Yang, F.; Qiu, M.; et al. A novel circular RNA, circXPO1, promotes lung adenocarcinoma progression by interacting with IGF2BP1. Cell Death Dis. 2020, 11, 1031. [Google Scholar] [CrossRef]
- Song, Y.; Xu, S.; Shao, Y.; Ge, S.; Zhou, H. Expression profile of circular RNAs in oral lichen planus. Ann. Palliat. Med. 2021, 10, 5205–5217. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, T.-J.; Kim, Y.G.; Jung, W.; Jang, S.; Ko, H.-G.; Park, C.H.; Byun, J.-S.; Kim, D.-Y. Non-Coding RNAs as Potential Targets for Diagnosis and Treatment of Oral Lichen Planus: A Narrative Review. Biomolecules 2023, 13, 1646. https://doi.org/10.3390/biom13111646
Kim T-J, Kim YG, Jung W, Jang S, Ko H-G, Park CH, Byun J-S, Kim D-Y. Non-Coding RNAs as Potential Targets for Diagnosis and Treatment of Oral Lichen Planus: A Narrative Review. Biomolecules. 2023; 13(11):1646. https://doi.org/10.3390/biom13111646
Chicago/Turabian StyleKim, Tae-Jun, Yu Gyung Kim, Won Jung, Sungil Jang, Hyoung-Gon Ko, Chan Ho Park, Jin-Seok Byun, and Do-Yeon Kim. 2023. "Non-Coding RNAs as Potential Targets for Diagnosis and Treatment of Oral Lichen Planus: A Narrative Review" Biomolecules 13, no. 11: 1646. https://doi.org/10.3390/biom13111646
APA StyleKim, T.-J., Kim, Y. G., Jung, W., Jang, S., Ko, H.-G., Park, C. H., Byun, J.-S., & Kim, D.-Y. (2023). Non-Coding RNAs as Potential Targets for Diagnosis and Treatment of Oral Lichen Planus: A Narrative Review. Biomolecules, 13(11), 1646. https://doi.org/10.3390/biom13111646





