Novel Insights into the Role of Chromatin Remodeler MORC2 in Cancer
Abstract
:1. Introduction
2. MORC2 Domain Organization and Structure
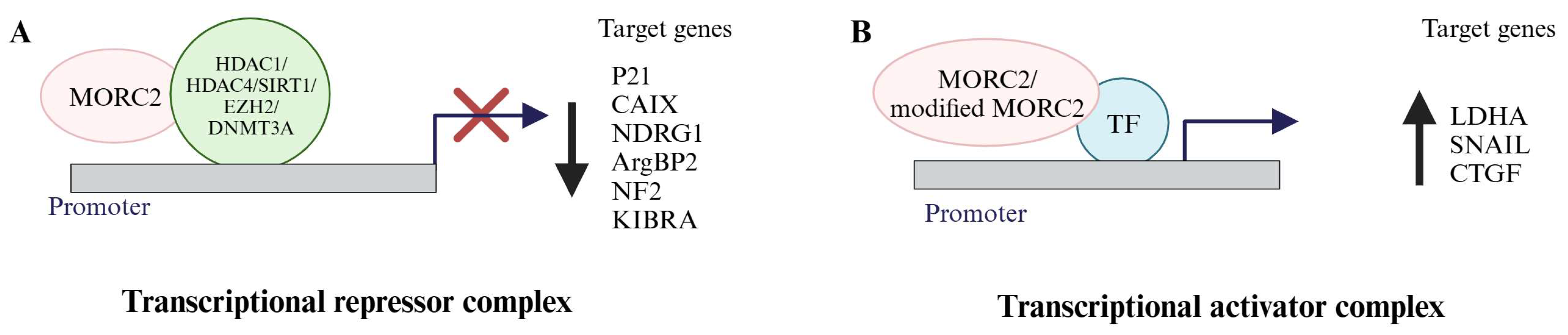
3. MORC2 Is a Transcriptional Regulator
3.1. MORC2 Acts as a Transcription Repressor
3.2. MORC2 Acts as a Transcription Activator
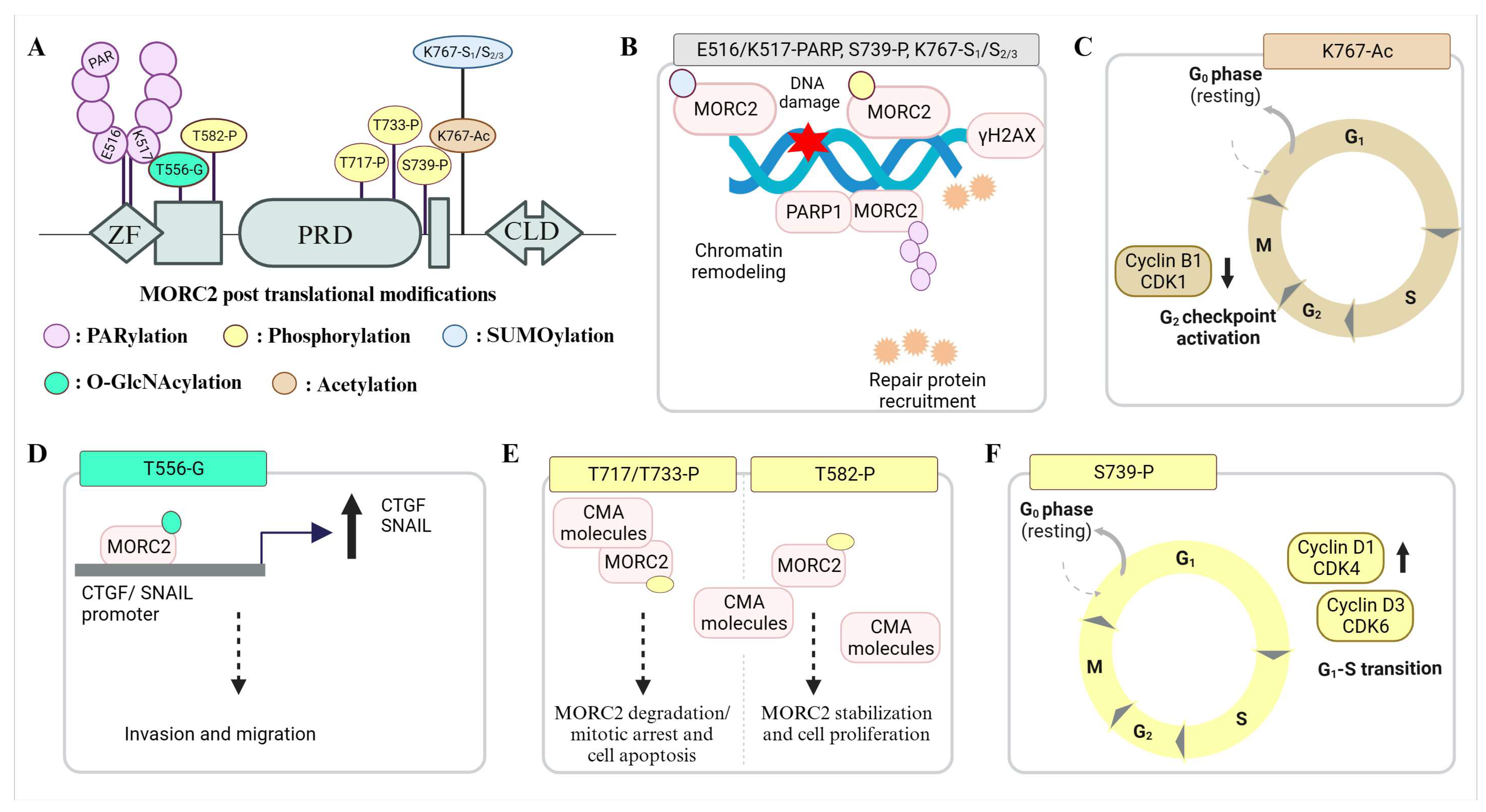
4. MORC2 Post-Translational Modifications and Their Significance
5. MORC2 Role in Cancer
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Nair, S.S.; Kumar, R. Chromatin remodeling in cancer: A gateway to regulate gene transcription. Mol. Oncol. 2012, 6, 611–619. [Google Scholar] [CrossRef]
- Reyes, A.A.; Marcum, R.D.; He, Y. Structure and Function of Chromatin Remodelers. J. Mol. Biol. 2021, 433, 166929. [Google Scholar] [CrossRef]
- Tyagi, M.; Imam, N.; Verma, K.; Patel, A.K. Chromatin remodelers: We are the drivers!! Nucleus 2016, 7, 388–404. [Google Scholar] [CrossRef]
- Zhang, P.; Torres, K.; Liu, X.; Liu, C.G.; Pollock, R.E. An Overview of Chromatin-Regulating Proteins in Cells. Curr. Protein Pept. Sci. 2016, 17, 401–410. [Google Scholar] [CrossRef]
- Clapier, C.R.; Iwasa, J.; Cairns, B.R.; Peterson, C.L. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 2017, 18, 407–422. [Google Scholar] [CrossRef]
- Magana-Acosta, M.; Valadez-Graham, V. Chromatin Remodelers in the 3D Nuclear Compartment. Front. Genet. 2020, 11, 600615. [Google Scholar] [CrossRef]
- Cairns, B.R. Emerging roles for chromatin remodeling in cancer biology. Trends Cell Biol. 2001, 11, S15–S21. [Google Scholar] [CrossRef]
- Cairns, B.R. Chromatin remodeling: Insights and intrigue from single-molecule studies. Nat. Struct. Mol. Biol. 2007, 14, 989–996. [Google Scholar] [CrossRef]
- Di Croce, L. Chromatin modifying activity of leukaemia associated fusion proteins. Hum. Mol. Genet. 2005, 14, R77–R84. [Google Scholar] [CrossRef]
- Ellis, L.; Atadja, P.W.; Johnstone, R.W. Epigenetics in cancer: Targeting chromatin modifications. Mol. Cancer Ther. 2009, 8, 1409–1420. [Google Scholar] [CrossRef]
- Mitelman, F.; Johansson, B.; Mertens, F. The impact of translocations and gene fusions on cancer causation. Nat. Rev. Cancer 2007, 7, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Vaicekauskaite, I.; Sabaliauskaite, R.; Lazutka, J.R.; Jarmalaite, S. The Emerging Role of Chromatin Remodeling Complexes in Ovarian Cancer. Int. J. Mol. Sci. 2022, 23, 13670–13690. [Google Scholar] [CrossRef]
- Chutani, N.; Singh, A.K.; Kadumuri, R.V.; Pakala, S.B.; Chavali, S. Structural and Functional Attributes of Microrchidia Family of Chromatin Remodelers. J. Mol. Biol. 2022, 434, 167664. [Google Scholar] [CrossRef]
- Watson, M.L.; Zinn, A.R.; Inoue, N.; Hess, K.D.; Cobb, J.; Handel, M.A.; Halaban, R.; Duchene, C.C.; Albright, G.M.; Moreadith, R.W. Identification of morc (microrchidia), a mutation that results in arrest of spermatogenesis at an early meiotic stage in the mouse. Proc. Natl. Acad. Sci. USA 1998, 95, 14361–14366. [Google Scholar] [CrossRef] [PubMed]
- Guddeti, R.K.; Chutani, N.; Pakala, S.B. MORC2 Interactome: Its Involvement in Metabolism and Cancer. Biophys. Rev. 2021, 13, 507–514. [Google Scholar] [CrossRef]
- Perry, J.; Zhao, Y. The CW domain, a structural module shared amongst vertebrates, vertebrate-infecting parasites and higher plants. Trends Biochem. Sci. 2003, 28, 576–580. [Google Scholar] [CrossRef] [PubMed]
- Hong, G.; Qiu, H.; Wang, C.; Jadhav, G.; Wang, H.; Tickner, J.; He, W.; Xu, J. The Emerging Role of MORC Family Proteins in Cancer Development and Bone Homeostasis. J. Cell Physiol. 2017, 232, 928–934. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, L.; Luo, Q.; Liu, J.; Wang, G. MORC protein family-related signature within human disease and cancer. Cell Death Dis. 2021, 12, 1112. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Li, Y.; Zhang, J.; Liu, D.; Liu, F.; Zhao, Y.; Shen, T.; Li, F. Involvement of histone deacetylation in MORC2-mediated down-regulation of carbonic anhydrase IX. Nucleic Acids Res. 2010, 38, 2813–2824. [Google Scholar] [CrossRef] [PubMed]
- Li, D.Q.; Nair, S.S.; Ohshiro, K.; Kumar, A.; Nair, V.S.; Pakala, S.B.; Reddy, S.D.; Gajula, R.P.; Eswaran, J.; Aravind, L.; et al. MORC2 signaling integrates phosphorylation-dependent, ATPase-coupled chromatin remodeling during the DNA damage response. Cell Rep. 2012, 2, 1657–1669. [Google Scholar] [CrossRef]
- Zhang, L.; Li, D.Q. MORC2 regulates DNA damage response through a PARP1-dependent pathway. Nucleic Acids Res. 2019, 47, 8502–8520. [Google Scholar] [CrossRef]
- Liu, H.Y.; Liu, Y.Y.; Yang, F.; Zhang, L.; Zhang, F.L.; Hu, X.; Shao, Z.M.; Li, D.Q. Acetylation of MORC2 by NAT10 regulates cell-cycle checkpoint control and resistance to DNA-damaging chemotherapy and radiotherapy in breast cancer. Nucleic Acids Res. 2020, 48, 3638–3656. [Google Scholar] [CrossRef]
- Guddeti, R.K.; Thomas, L.; Kannan, A.; Karyala, P.; Pakala, S.B. The chromatin modifier MORC2 affects glucose metabolism by regulating the expression of lactate dehydrogenase A through a feed forward loop with c-Myc. FEBS Lett. 2021, 595, 1289–1302. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Liu, H.Y.; Yu, T.J.; Lu, Q.; Zhang, F.L.; Liu, G.Y.; Shao, Z.M.; Li, D.Q. O-GlcNAcylation of MORC2 at threonine 556 by OGT couples TGF-beta signaling to breast cancer progression. Cell Death Differ. 2022, 29, 861–873. [Google Scholar] [CrossRef]
- Zhang, F.L.; Yang, S.Y.; Liao, L.; Zhang, T.M.; Zhang, Y.L.; Hu, S.Y.; Deng, L.; Huang, M.Y.; Andriani, L.; Ma, X.Y.; et al. Dynamic SUMOylation of MORC2 orchestrates chromatin remodelling and DNA repair in response to DNA damage and drives chemoresistance in breast cancer. Theranostics 2023, 13, 973–990. [Google Scholar] [CrossRef]
- Tchasovnikarova, I.A.; Timms, R.T.; Douse, C.H.; Roberts, R.C.; Dougan, G.; Kingston, R.E.; Modis, Y.; Lehner, P.J. Hyperactivation of HUSH complex function by Charcot-Marie-Tooth disease mutation in MORC2. Nat. Genet. 2017, 49, 1035–1044. [Google Scholar] [CrossRef]
- Ciquier, G.; Azzi, M.; Hebert, C.; Watkins-Martin, K.; Drapeau, M. Assessing the quality of seven clinical practice guidelines from four professional regulatory bodies in Quebec: What’s the verdict? J. Eval. Clin. Pract. 2021, 27, 25–33. [Google Scholar] [CrossRef]
- Liu, Y.; Tempel, W.; Zhang, Q.; Liang, X.; Loppnau, P.; Qin, S.; Min, J. Family-wide Characterization of Histone Binding Abilities of Human CW Domain-containing Proteins. J. Biol. Chem. 2016, 291, 9000–9013. [Google Scholar] [CrossRef]
- Yoshinaka, T.; Kosako, H.; Yoshizumi, T.; Furukawa, R.; Hirano, Y.; Kuge, O.; Tamada, T.; Koshiba, T. Structural Basis of Mitochondrial Scaffolds by Prohibitin Complexes: Insight into a Role of the Coiled-Coil Region. iScience 2019, 19, 1065–1078. [Google Scholar] [CrossRef]
- Xie, H.Y.; Zhang, T.M.; Hu, S.Y.; Shao, Z.M.; Li, D.Q. Dimerization of MORC2 through its C-terminal coiled-coil domain enhances chromatin dynamics and promotes DNA repair. Cell Commun. Signal. 2019, 17, 160. [Google Scholar] [CrossRef]
- Cristie-David, A.S.; Chen, J.; Nowak, D.B.; Bondy, A.L.; Sun, K.; Park, S.I.; Banaszak Holl, M.M.; Su, M.; Marsh, E.N.G. Coiled-Coil-Mediated Assembly of an Icosahedral Protein Cage with Extremely High Thermal and Chemical Stability. J. Am. Chem. Soc. 2019, 141, 9207–9216. [Google Scholar] [CrossRef]
- Matityahu, A.; Onn, I. A new twist in the coil: Functions of the coiled-coil domain of structural maintenance of chromosome (SMC) proteins. Curr. Genet. 2018, 64, 109–116. [Google Scholar] [CrossRef]
- Terawaki, S.; Yoshikane, A.; Higuchi, Y.; Wakamatsu, K. Structural basis for cargo binding and autoinhibition of Bicaudal-D1 by a parallel coiled-coil with homotypic registry. Biochem. Biophys. Res. Commun. 2015, 460, 451–456. [Google Scholar] [CrossRef]
- Douse, C.H.; Bloor, S.; Liu, Y.; Shamin, M.; Tchasovnikarova, I.A.; Timms, R.T.; Lehner, P.J.; Modis, Y. Neuropathic MORC2 mutations perturb GHKL ATPase dimerization dynamics and epigenetic silencing by multiple structural mechanisms. Nat. Commun. 2018, 9, 651. [Google Scholar] [CrossRef]
- Liao, X.H.; Zhang, Y.; Dong, W.J.; Shao, Z.M.; Li, D.Q. Chromatin remodeling protein MORC2 promotes breast cancer invasion and metastasis through a PRD domain-mediated interaction with CTNND1. Oncotarget 2017, 8, 97941–97954. [Google Scholar] [CrossRef]
- Wang, G.L.; Wang, C.Y.; Cai, X.Z.; Chen, W.; Wang, X.H.; Li, F. Identification and expression analysis of a novel CW-type zinc finger protein MORC2 in cancer cells. Anat. Rec. 2010, 293, 1002–1009. [Google Scholar] [CrossRef]
- Tafreshi, N.K.; Lloyd, M.C.; Bui, M.M.; Gillies, R.J.; Morse, D.L. Carbonic anhydrase IX as an imaging and therapeutic target for tumors and metastases. Subcell. Biochem. 2014, 75, 221–254. [Google Scholar] [CrossRef]
- Chen, J.; Rocken, C.; Hoffmann, J.; Kruger, S.; Lendeckel, U.; Rocco, A.; Pastorekova, S.; Malfertheiner, P.; Ebert, M.P. Expression of carbonic anhydrase 9 at the invasion front of gastric cancers. Gut 2005, 54, 920–927. [Google Scholar] [CrossRef]
- Driessen, A.; Landuyt, W.; Pastorekova, S.; Moons, J.; Goethals, L.; Haustermans, K.; Nafteux, P.; Penninckx, F.; Geboes, K.; Lerut, T.; et al. Expression of carbonic anhydrase IX (CA IX), a hypoxia-related protein, rather than vascular-endothelial growth factor (VEGF), a pro-angiogenic factor, correlates with an extremely poor prognosis in esophageal and gastric adenocarcinomas. Ann. Surg. 2006, 243, 334–340. [Google Scholar] [CrossRef]
- Besson, A.; Dowdy, S.F.; Roberts, J.M. CDK inhibitors: Cell cycle regulators and beyond. Dev. Cell 2008, 14, 159–169. [Google Scholar] [CrossRef]
- Zhang, Q.; Song, Y.; Chen, W.; Wang, X.; Miao, Z.; Cao, L.; Li, F.; Wang, G. By recruiting HDAC1, MORC2 suppresses p21 Waf1/Cip1 in gastric cancer. Oncotarget 2015, 6, 16461–16470. [Google Scholar] [CrossRef] [PubMed]
- Ou, K.; Li, Y.; Long, Y.; Luo, Y.; Tang, D.; Chen, Z. Inhibition of MORC2 Mediates HDAC4 to Promote Cellular Senescence through p53/p21 Signaling Axis. Molecules 2022, 27, 6247–6259. [Google Scholar] [CrossRef]
- Roignot, J.; Soubeyran, P. ArgBP2 and the SoHo family of adapter proteins in oncogenic diseases. Cell Adhes. Migr. 2009, 3, 167–170. [Google Scholar] [CrossRef]
- Tong, Y.; Li, Y.; Gu, H.; Wang, C.; Liu, F.; Shao, Y.; Li, J.; Cao, L.; Li, F. Microchidia protein 2, MORC2, downregulates the cytoskeleton adapter protein, ArgBP2, via histone methylation in gastric cancer cells. Biochem. Biophys. Res. Commun. 2015, 467, 821–827. [Google Scholar] [CrossRef]
- Tong, Y.; Li, Y.; Gu, H.; Wang, C.; Liu, F.; Shao, Y.; Li, F. HSF1, in association with MORC2, downregulates ArgBP2 via the PRC2 family in gastric cancer cells. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 1104–1114. [Google Scholar] [CrossRef]
- Liu, J.; Shao, Y.; He, Y.; Ning, K.; Cui, X.; Liu, F.; Wang, Z.; Li, F. MORC2 promotes development of an aggressive colorectal cancer phenotype through inhibition of NDRG1. Cancer Sci. 2019, 110, 135–146. [Google Scholar] [CrossRef]
- Wang, T.; Qin, Z.Y.; Wen, L.Z.; Guo, Y.; Liu, Q.; Lei, Z.J.; Pan, W.; Liu, K.J.; Wang, X.W.; Lai, S.J.; et al. Epigenetic restriction of Hippo signaling by MORC2 underlies stemness of hepatocellular carcinoma cells. Cell Death Differ. 2018, 25, 2086–2100. [Google Scholar] [CrossRef]
- Peinado, H.; Olmeda, D.; Cano, A. Snail, Zeb and bHLH factors in tumour progression: An alliance against the epithelial phenotype? Nat. Rev. Cancer 2007, 7, 415–428. [Google Scholar] [CrossRef]
- Kang, Y.; Siegel, P.M.; Shu, W.; Drobnjak, M.; Kakonen, S.M.; Cordon-Cardo, C.; Guise, T.A.; Massague, J. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell 2003, 3, 537–549. [Google Scholar] [CrossRef]
- Tran, H.D.; Luitel, K.; Kim, M.; Zhang, K.; Longmore, G.D.; Tran, D.D. Transient SNAIL1 expression is necessary for metastatic competence in breast cancer. Cancer Res. 2014, 74, 6330–6340. [Google Scholar] [CrossRef]
- Batlle, E.; Sancho, E.; Franci, C.; Dominguez, D.; Monfar, M.; Baulida, J.; Garcia De Herreros, A. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat. Cell Biol. 2000, 2, 84–89. [Google Scholar] [CrossRef]
- Chien, W.; O’Kelly, J.; Lu, D.; Leiter, A.; Sohn, J.; Yin, D.; Karlan, B.; Vadgama, J.; Lyons, K.M.; Koeffler, H.P. Expression of connective tissue growth factor (CTGF/CCN2) in breast cancer cells is associated with increased migration and angiogenesis. Int. J. Oncol. 2011, 38, 1741–1747. [Google Scholar] [CrossRef]
- Chen, P.S.; Wang, M.Y.; Wu, S.N.; Su, J.L.; Hong, C.C.; Chuang, S.E.; Chen, M.W.; Hua, K.T.; Wu, Y.L.; Cha, S.T.; et al. CTGF enhances the motility of breast cancer cells via an integrin-alphavbeta3-ERK1/2-dependent S100A4-upregulated pathway. J. Cell Sci. 2007, 120, 2053–2065. [Google Scholar] [CrossRef]
- Liu, H.Y.; Liu, Y.Y.; Zhang, Y.L.; Ning, Y.; Zhang, F.L.; Li, D.Q. Poly(ADP-ribosyl)ation of acetyltransferase NAT10 by PARP1 is required for its nucleoplasmic translocation and function in response to DNA damage. Cell Commun. Signal. 2022, 20, 127. [Google Scholar] [CrossRef]
- Yang, F.; Xie, H.Y.; Yang, L.F.; Zhang, L.; Zhang, F.L.; Liu, H.Y.; Li, D.Q.; Shao, Z.M. Stabilization of MORC2 by estrogen and antiestrogens through GPER1- PRKACA-CMA pathway contributes to estrogen-induced proliferation and endocrine resistance of breast cancer cells. Autophagy 2020, 16, 1061–1076. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.Y.; Qian, J.X.; Yang, S.Y.; Andriani, L.; Liao, L.; Deng, L.; Huang, M.Y.; Zhang, Y.L.; Zhang, F.L.; Shao, Z.M.; et al. Destabilization of microrchidia family CW-type zinc finger 2 via the cyclin-dependent kinase 1-chaperone-mediated autophagy pathway promotes mitotic arrest and enhances cancer cellular sensitivity to microtubule-targeting agents. Clin. Transl. Med. 2023, 13, e1210. [Google Scholar] [CrossRef]
- Zhang, S.; Guo, A.; Wang, H.; Liu, J.; Dong, C.; Ren, J.; Wang, G. Oncogenic MORC2 in cancer development and beyond. Genes. Dis. 2024, 11, 861–873. [Google Scholar] [CrossRef]
- Ding, Q.S.; Zhang, L.; Wang, B.C.; Zeng, Z.; Zou, X.Q.; Cao, P.B.; Zhou, G.M.; Tang, M.; Wu, L.; Wu, L.L.; et al. Aberrant high expression level of MORC2 is a common character in multiple cancers. Hum. Pathol. 2018, 76, 58–67. [Google Scholar] [CrossRef]
- Liao, X.; Liu, C.; Ding, Z.; Wang, C.; He, J.; Wu, S. High expression of MORC2 predicts worse neoadjuvant chemotherapy efficacy in triple negative breast cancer. Medicine 2023, 102, e34164. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yang, Y.; Dong, Y.; Liu, C. Microrchidia family CW-type zinc finger 2 promotes the proliferation, invasion, migration and epithelial-mesenchymal transition of glioma by regulating PTEN/PI3K/AKT signaling via binding to N-myc downstream regulated gene 1 promoter. Int. J. Mol. Med. 2022, 49, 16–31. [Google Scholar] [CrossRef]
- Yang, F.; Sun, R.; Hou, Z.; Zhang, F.L.; Xiao, Y.; Yang, Y.S.; Yang, S.Y.; Xie, Y.F.; Liu, Y.Y.; Luo, C.; et al. HSP90 N-terminal inhibitors target oncoprotein MORC2 for autophagic degradation and suppress MORC2-driven breast cancer progression. Clin. Transl. Med. 2022, 12, e825. [Google Scholar] [CrossRef] [PubMed]
- Saroha, H.S.; Kumar Guddeti, R.; Jacob, J.P.; Kumar Pulukuri, K.; Karyala, P.; Pakala, S.B. MORC2/beta-catenin signaling axis promotes proliferation and migration of breast cancer cells. Med. Oncol. 2022, 39, 135. [Google Scholar] [CrossRef] [PubMed]
- Liao, G.; Liu, X.; Wu, D.; Duan, F.; Xie, X.; Wen, S.; Li, Y.; Li, S. MORC2 promotes cell growth and metastasis in human cholangiocarcinoma and is negatively regulated by miR-186-5p. Aging 2019, 11, 3639–3649. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.; Ding, Q.; Guo, Q.; Guo, Y.; Wu, L.; Wu, L.; Tang, M.; Yu, H.; Zhou, F. MORC2, a novel oncogene, is upregulated in liver cancer and contributes to proliferation, metastasis and chemoresistance. Int. J. Oncol. 2018, 53, 59–72. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Song, Y.; Liu, T.; Wang, C.; Zhang, Q.; Liu, F.; Cai, X.; Miao, Z.; Xu, H.; Xu, H.; et al. PAK1-mediated MORC2 phosphorylation promotes gastric tumorigenesis. Oncotarget 2015, 6, 9877–9886. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.L.; Cao, J.L.; Xie, H.Y.; Sun, R.; Yang, L.F.; Shao, Z.M.; Li, D.Q. Cancer-Associated MORC2-Mutant M276I Regulates an hnRNPM-Mediated CD44 Splicing Switch to Promote Invasion and Metastasis in Triple-Negative Breast Cancer. Cancer Res. 2018, 78, 5780–5792. [Google Scholar] [CrossRef]
- Liberti, M.V.; Locasale, J.W. The Warburg Effect: How Does it Benefit Cancer Cells? Trends Biochem. Sci. 2016, 41, 211–218. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef]
- Guddeti, R.K.; Pacharla, H.; Yellapu, N.K.; Karyala, P.; Pakala, S.B. MORC2 and MAX contributes to the expression of glycolytic enzymes, breast cancer cell proliferation and migration. Med. Oncol. 2023, 40, 102. [Google Scholar] [CrossRef]
- Sanchez-Solana, B.; Li, D.Q.; Kumar, R. Cytosolic functions of MORC2 in lipogenesis and adipogenesis. Biochim. Biophys. Acta 2014, 1843, 316–326. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Yu, T.; Wang, Y.; Huang, X.; Wei, X. Circular RNA circDNM3OS Functions as a miR-145-5p Sponge to Accelerate Cholangiocarcinoma Growth and Glutamine Metabolism by Upregulating MORC2. Onco Targets Ther. 2021, 14, 1117–1129. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chutani, N.; Ragula, S.; Syed, K.; Pakala, S.B. Novel Insights into the Role of Chromatin Remodeler MORC2 in Cancer. Biomolecules 2023, 13, 1527. https://doi.org/10.3390/biom13101527
Chutani N, Ragula S, Syed K, Pakala SB. Novel Insights into the Role of Chromatin Remodeler MORC2 in Cancer. Biomolecules. 2023; 13(10):1527. https://doi.org/10.3390/biom13101527
Chicago/Turabian StyleChutani, Namita, Sandhya Ragula, Khajamohiddin Syed, and Suresh B. Pakala. 2023. "Novel Insights into the Role of Chromatin Remodeler MORC2 in Cancer" Biomolecules 13, no. 10: 1527. https://doi.org/10.3390/biom13101527
APA StyleChutani, N., Ragula, S., Syed, K., & Pakala, S. B. (2023). Novel Insights into the Role of Chromatin Remodeler MORC2 in Cancer. Biomolecules, 13(10), 1527. https://doi.org/10.3390/biom13101527






