Identifying Suitable Reference Gene Candidates for Quantification of DNA Damage-Induced Cellular Responses in Human U2OS Cell Culture System
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culturing and Treatments
2.2. RNA Isolation and Reverse Transcription
2.3. RT-qPCR
2.4. Statistics
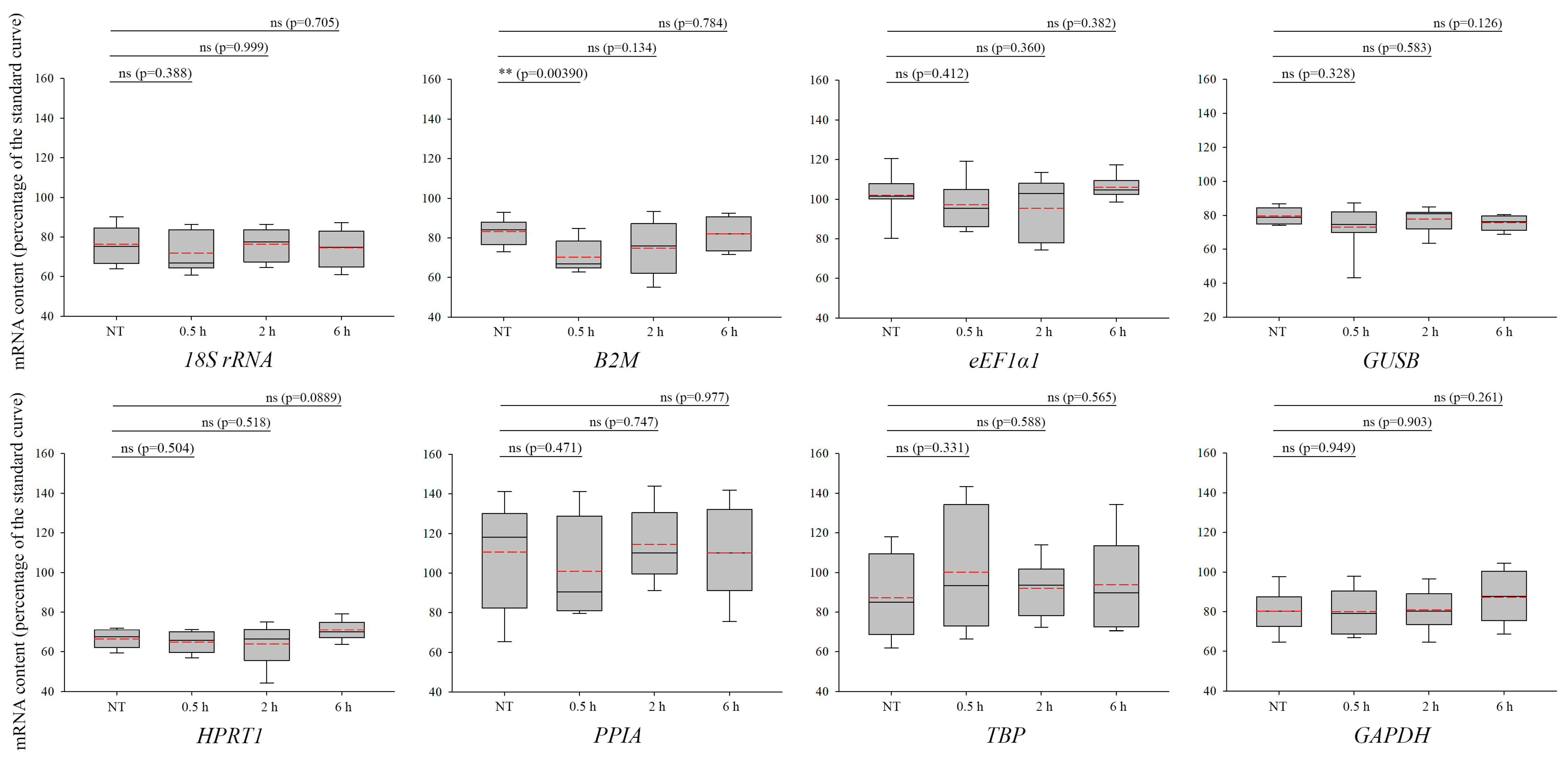
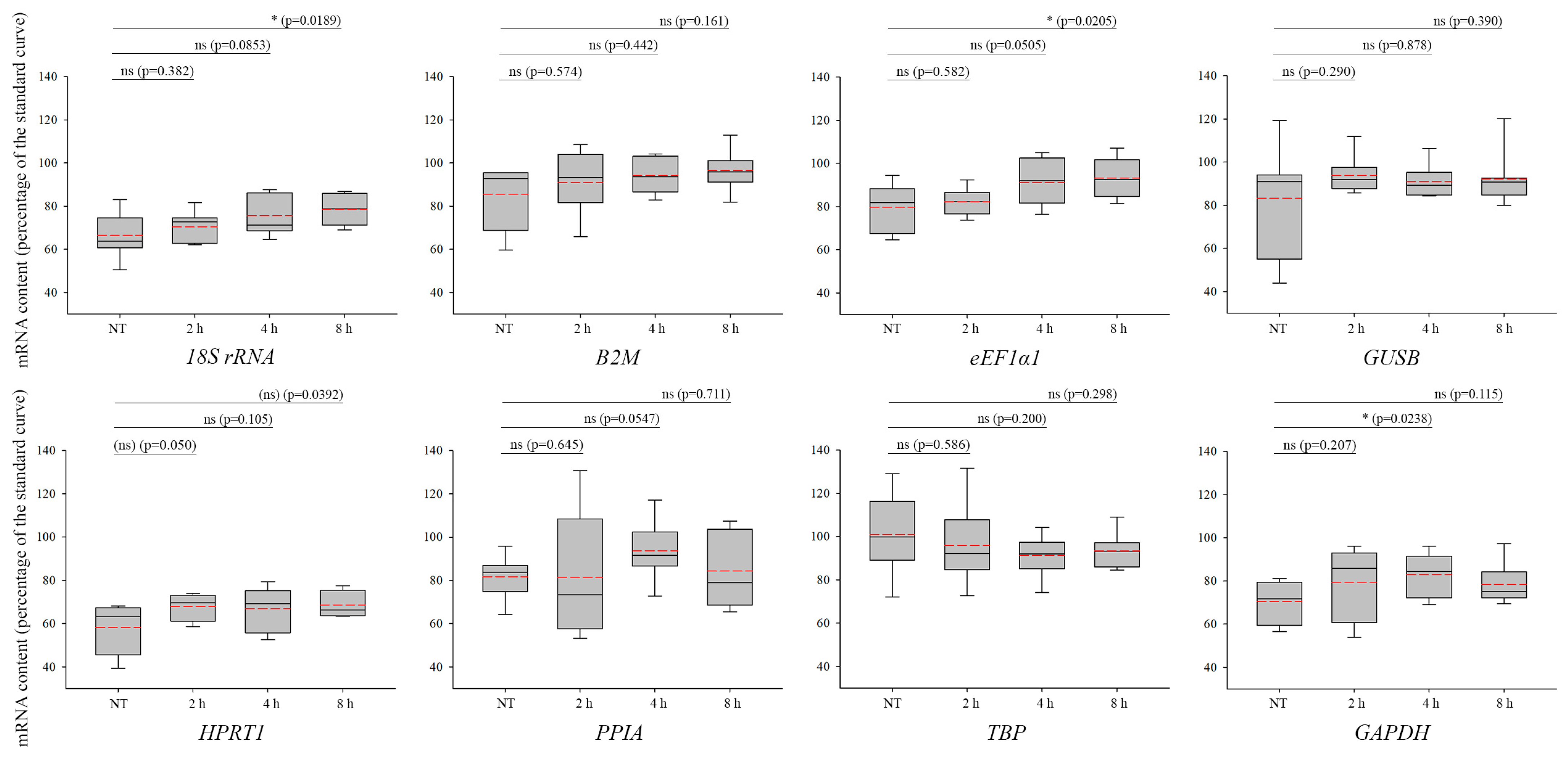
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Matsuda, S.; Murakami, M.; Ikeda, Y.; Nakagawa, Y.; Tsuji, A.; Kitagishi, Y. Role of tumor suppressor molecules in genomic perturbations and damaged DNA repair involved in the pathogenesis of cancer and neurodegeneration (Review). Biomed. Rep. 2020, 13, 10. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.H.; Wu, C.F.; Rajasekaran, N.; Shin, Y.K. Loss of Tumor Suppressor Gene Function in Human Cancer: An Overview. Cell. Physiol. Biochem. 2018, 51, 2647–2693. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.; Ding, Q.; Guo, Q.; Guo, Y.; Wu, L.; Wu, L.; Tang, M.; Yu, H.; Zhou, F. MORC2, a novel oncogene, is upregulated in liver cancer and contributes to proliferation, metastasis and chemoresistance. Int. J. Oncol. 2018, 53, 59–72. [Google Scholar] [CrossRef]
- Norbury, C.J.; Zhivotovsky, B. DNA damage-induced apoptosis. Oncogene 2004, 23, 2797–2808. [Google Scholar] [CrossRef]
- Borsos, B.N.; Huliak, I.; Majoros, H.; Ujfaludi, Z.; Gyenis, A.; Pukler, P.; Boros, I.M.; Pankotai, T. Human p53 interacts with the elongating RNAPII complex and is required for the release of actinomycin D induced transcription blockage. Sci. Rep. 2017, 7, 40960. [Google Scholar] [CrossRef] [PubMed]
- Roos, W.P.; Thomas, A.D.; Kaina, B. DNA damage and the balance between survival and death in cancer biology. Nat. Rev. Cancer 2016, 16, 20–33. [Google Scholar] [CrossRef]
- Dheda, K.; Huggett, J.F.; Bustin, S.A.; Johnson, M.A.; Rook, G.; Zumla, A. Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques 2004, 37, 112–114, 116, 118–119. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Orntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, E.; Levanon, E.Y. Human housekeeping genes, revisited. Trends Genet. 2013, 29, 569–574. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, RESEARCH0034. [Google Scholar] [CrossRef]
- Thellin, O.; Zorzi, W.; Lakaye, B.; De Borman, B.; Coumans, B.; Hennen, G.; Grisar, T.; Igout, A.; Heinen, E. Housekeeping genes as internal standards: Use and limits. J. Biotechnol. 1999, 75, 291–295. [Google Scholar] [CrossRef] [PubMed]
- de Jonge, H.J.; Fehrmann, R.S.; de Bont, E.S.; Hofstra, R.M.; Gerbens, F.; Kamps, W.A.; de Vries, E.G.; van der Zee, A.G.; te Meerman, G.J.; ter Elst, A. Evidence based selection of housekeeping genes. PLoS ONE 2007, 2, e898. [Google Scholar] [CrossRef]
- Warrington, J.A.; Nair, A.; Mahadevappa, M.; Tsyganskaya, M. Comparison of human adult and fetal expression and identification of 535 housekeeping/maintenance genes. Physiol. Genom. 2000, 2, 143–147. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Higgins, P.J.; Crawford, D.R. Control selection for RNA quantitation. Biotechniques 2000, 29, 332–337. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 2000, 25, 169–193. [Google Scholar] [CrossRef] [PubMed]
- Barber, R.D.; Harmer, D.W.; Coleman, R.A.; Clark, B.J. GAPDH as a housekeeping gene: Analysis of GAPDH mRNA expression in a panel of 72 human tissues. Physiol. Genom. 2005, 21, 389–395. [Google Scholar] [CrossRef]
- Liu, J.; Fang, H.; Chi, Z.; Wu, Z.; Wei, D.; Mo, D.; Niu, K.; Balajee, A.S.; Hei, T.K.; Nie, L.; et al. XPD localizes in mitochondria and protects the mitochondrial genome from oxidative DNA damage. Nucleic Acids Res. 2015, 43, 5476–5488. [Google Scholar] [CrossRef]
- van den Boom, J.; Wolf, M.; Weimann, L.; Schulze, N.; Li, F.; Kaschani, F.; Riemer, A.; Zierhut, C.; Kaiser, M.; Iliakis, G.; et al. VCP/p97 Extracts Sterically Trapped Ku70/80 Rings from DNA in Double-Strand Break Repair. Mol. Cell 2016, 64, 189–198. [Google Scholar] [CrossRef]
- Shah, M.Y.; Martinez-Garcia, E.; Phillip, J.M.; Chambliss, A.B.; Popovic, R.; Ezponda, T.; Small, E.C.; Will, C.; Phillip, M.P.; Neri, P.; et al. MMSET/WHSC1 enhances DNA damage repair leading to an increase in resistance to chemotherapeutic agents. Oncogene 2016, 35, 5905–5915. [Google Scholar] [CrossRef]
- Lemaitre, C.; Grabarz, A.; Tsouroula, K.; Andronov, L.; Furst, A.; Pankotai, T.; Heyer, V.; Rogier, M.; Attwood, K.M.; Kessler, P.; et al. Nuclear position dictates DNA repair pathway choice. Genes Dev. 2014, 28, 2450–2463. [Google Scholar] [CrossRef]
- Alekseev, S.; Nagy, Z.; Sandoz, J.; Weiss, A.; Egly, J.M.; Le May, N.; Coin, F. Transcription without XPB Establishes a Unified Helicase-Independent Mechanism of Promoter Opening in Eukaryotic Gene Expression. Mol. Cell 2017, 65, 504–514.e504. [Google Scholar] [CrossRef] [PubMed]
- Pankotai, T.; Bonhomme, C.; Chen, D.; Soutoglou, E. DNAPKcs-dependent arrest of RNA polymerase II transcription in the presence of DNA breaks. Nat. Struct. Mol. Biol. 2012, 19, 276–282. [Google Scholar] [CrossRef] [PubMed]
- van Cuijk, L.; van Belle, G.J.; Turkyilmaz, Y.; Poulsen, S.L.; Janssens, R.C.; Theil, A.F.; Sabatella, M.; Lans, H.; Mailand, N.; Houtsmuller, A.B.; et al. SUMO and ubiquitin-dependent XPC exchange drives nucleotide excision repair. Nat. Commun. 2015, 6, 7499. [Google Scholar] [CrossRef] [PubMed]
- Aymard, F.; Bugler, B.; Schmidt, C.K.; Guillou, E.; Caron, P.; Briois, S.; Iacovoni, J.S.; Daburon, V.; Miller, K.M.; Jackson, S.P.; et al. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat. Struct. Mol. Biol. 2014, 21, 366–374. [Google Scholar] [CrossRef]
- Shahar, O.D.; Kalousi, A.; Eini, L.; Fisher, B.; Weiss, A.; Darr, J.; Mazina, O.; Bramson, S.; Kupiec, M.; Eden, A.; et al. A high-throughput chemical screen with FDA approved drugs reveals that the antihypertensive drug Spironolactone impairs cancer cell survival by inhibiting homology directed repair. Nucleic Acids Res. 2014, 42, 5689–5701. [Google Scholar] [CrossRef]
- Dheda, K.; Huggett, J.F.; Chang, J.S.; Kim, L.U.; Bustin, S.A.; Johnson, M.A.; Rook, G.A.; Zumla, A. The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Anal. Biochem. 2005, 344, 141–143. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Bustin, S.A.; Nolan, T. Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction. J. Biomol. Tech. 2004, 15, 155–166. [Google Scholar]
- Ujfaludi, Z.; Tuzesi, A.; Majoros, H.; Rothler, B.; Pankotai, T.; Boros, I.M. Coordinated activation of a cluster of MMP genes in response to UVB radiation. Sci. Rep. 2018, 8, 2660. [Google Scholar] [CrossRef]
- Chatterjee, N.; Walker, G.C. Mechanisms of DNA damage, repair, and mutagenesis. Environ. Mol. Mutagen. 2017, 58, 235–263. [Google Scholar] [CrossRef]
- Roy, S. Impact of UV Radiation on Genome Stability and Human Health. Adv. Exp. Med. Biol. 2017, 996, 207–219. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, I.H. Free radical mechanisms in neocarzinostatin-induced DNA damage. Free Radic. Biol. Med. 1987, 3, 41–54. [Google Scholar] [CrossRef] [PubMed]
- Waring, M.J. DNA modification and cancer. Annu. Rev. Biochem. 1981, 50, 159–192. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, F.; Comez, L.; Biehl, R.; D’Amico, F.; Gessini, A.; Longo, M.; Masciovecchio, C.; Petrillo, C.; Radulescu, A.; Rossi, B.; et al. Structure of human telomere G-quadruplex in the presence of a model drug along the thermal unfolding pathway. Nucleic Acids Res. 2018, 46, 11927–11938. [Google Scholar] [CrossRef]
- Fox, J.M.; Byrne, T.D.; Woods, W.G. Actinomycin D-associated lesions mimicking DNA-DNA interstrand crosslinks detected by alkaline elution in cultured mammalian cells. Biochem. Pharmacol. 1985, 34, 2741–2747. [Google Scholar] [CrossRef]
- Chen, A.Y.; Liu, L.F. DNA topoisomerases: Essential enzymes and lethal targets. Annu. Rev. Pharmacol. Toxicol. 1994, 34, 191–218. [Google Scholar] [CrossRef]
- Matsumura, Y.; Ananthaswamy, H.N. Toxic effects of ultraviolet radiation on the skin. Toxicol. Appl. Pharmacol. 2004, 195, 298–308. [Google Scholar] [CrossRef]
- Majoros, H.; Borsos, B.N.; Ujfaludi, Z.; Pahi, Z.G.; Morocz, M.; Haracska, L.; Boros, I.M.; Pankotai, T. SerpinB10, a Serine Protease Inhibitor, Is Implicated in UV-Induced Cellular Response. Int. J. Mol. Sci. 2021, 22, 8500. [Google Scholar] [CrossRef]
- Majoros, H.; Ujfaludi, Z.; Borsos, B.N.; Hudacsek, V.V.; Nagy, Z.; Coin, F.; Buzas, K.; Kovacs, I.; Biro, T.; Boros, I.M.; et al. SerpinB2 is involved in cellular response upon UV irradiation. Sci. Rep. 2019, 9, 2753. [Google Scholar] [CrossRef]
- Dedon, P.C.; Goldberg, I.H. Sequence-specific double-strand breakage of DNA by neocarzinostatin involves different chemical mechanisms within a staggered cleavage site. J. Biol. Chem. 1990, 265, 14713–14716. [Google Scholar] [CrossRef]
- Sobell, H.M. Actinomycin and DNA transcription. Proc. Natl. Acad. Sci. USA 1985, 82, 5328–5331. [Google Scholar] [CrossRef]
- Borsos, B.N.; Pantazi, V.; Pahi, Z.G.; Majoros, H.; Ujfaludi, Z.; Berzsenyi, I.; Pankotai, T. The role of p53 in the DNA damage-related ubiquitylation of S2P RNAPII. PLoS ONE 2022, 17, e0267615. [Google Scholar] [CrossRef] [PubMed]
- Huggett, J.; Dheda, K.; Bustin, S.; Zumla, A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005, 6, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Feuer, R.; Vlaic, S.; Arlt, J.; Sawodny, O.; Dahmen, U.; Zanger, U.M.; Thomas, M. LEMming: A Linear Error Model to Normalize Parallel Quantitative Real-Time PCR (qPCR) Data as an Alternative to Reference Gene Based Methods. PLoS ONE 2015, 10, e0135852. [Google Scholar] [CrossRef]
- Pabinger, S.; Rodiger, S.; Kriegner, A.; Vierlinger, K.; Weinhausel, A. A survey of tools for the analysis of quantitative PCR (qPCR) data. Biomol. Detect. Quantif. 2014, 1, 23–33. [Google Scholar] [CrossRef]
- Qiu, W.H.; Zhou, B.S.; Chu, P.G.; Chen, W.G.; Chung, C.; Shih, J.; Hwu, P.; Yeh, C.; Lopez, R.; Yen, Y. Over-expression of fibroblast growth factor receptor 3 in human hepatocellular carcinoma. World J. Gastroenterol. 2005, 11, 5266–5272. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.T.; Baird, K.; Ahn, J.Y.; Meltzer, P.; Lilly, M.; Levis, M.; Small, D. Pim-1 is up-regulated by constitutively activated FLT3 and plays a role in FLT3-mediated cell survival. Blood 2005, 105, 1759–1767. [Google Scholar] [CrossRef]
- Heller, R.A.; Schena, M.; Chai, A.; Shalon, D.; Bedilion, T.; Gilmore, J.; Woolley, D.E.; Davis, R.W. Discovery and analysis of inflammatory disease-related genes using cDNA microarrays. Proc. Natl. Acad. Sci. USA 1997, 94, 2150–2155. [Google Scholar] [CrossRef]
- Wang, Y.; Shen, H.; Pang, L.; Qiu, B.; Yuan, Y.; Guan, X.; Xiang, X. Qihuang Granule protects the retinal pigment epithelium from oxidative stress via regulation of the alternative complement pathway. BMC Complement. Med. Ther. 2023, 23, 55. [Google Scholar] [CrossRef]
- Minogue, B.M.; Richardson, S.M.; Zeef, L.A.; Freemont, A.J.; Hoyland, J.A. Characterization of the human nucleus pulposus cell phenotype and evaluation of novel marker gene expression to define adult stem cell differentiation. Arthritis Rheum. 2010, 62, 3695–3705. [Google Scholar] [CrossRef]
- Bai, Y.; Li, Y.R.; Wang, G.H.; Zhou, X.M.; Zhao, D.M. PrP 106-126 altered PrP mRNA gene expression in mouse microglia BV-2 cells. Virol. Sin. 2010, 25, 440–444. [Google Scholar] [CrossRef] [PubMed]
- Marteau, J.B.; Rigaud, O.; Brugat, T.; Gault, N.; Vallat, L.; Kruhoffer, M.; Orntoft, T.F.; Nguyen-Khac, F.; Chevillard, S.; Merle-Beral, H.; et al. Concomitant heterochromatinisation and down-regulation of gene expression unveils epigenetic silencing of RELB in an aggressive subset of chronic lymphocytic leukemia in males. BMC Med. Genom. 2010, 3, 53. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, S.; Lattuada, D.; Paffoni, A.; Brevini, T.A.; Scarduelli, C.; Bolis, G.; Ragni, G.; Gandolfi, F. Procedure for rapid oocyte selection based on quantitative analysis of cumulus cell gene expression. J. Assist. Reprod. Genet. 2010, 27, 429–434. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Felty, Q.; Yoo, C.; Kennedy, A. Gene expression profile of endothelial cells exposed to estrogenic environmental compounds: Implications to pulmonary vascular lesions. Life Sci. 2010, 86, 919–927. [Google Scholar] [CrossRef] [PubMed]
- Valenti, M.T.; Bertoldo, F.; Dalle Carbonare, L.; Azzarello, G.; Zenari, S.; Zanatta, M.; Balducci, E.; Vinante, O.; Lo Cascio, V. The effect of bisphosphonates on gene expression: GAPDH as a housekeeping or a new target gene? BMC Cancer 2006, 6, 49. [Google Scholar] [CrossRef] [PubMed]
- Arukwe, A. Toxicological housekeeping genes: Do they really keep the house? Environ. Sci. Technol. 2006, 40, 7944–7949. [Google Scholar] [CrossRef]
- Kosir, R.; Acimovic, J.; Golicnik, M.; Perse, M.; Majdic, G.; Fink, M.; Rozman, D. Determination of reference genes for circadian studies in different tissues and mouse strains. BMC Mol. Biol. 2010, 11, 60. [Google Scholar] [CrossRef]
- Anderson, K.C.; Elizur, A. Hepatic reference gene selection in adult and juvenile female Atlantic salmon at normal and elevated temperatures. BMC Res. Notes 2012, 5, 21. [Google Scholar] [CrossRef]
- Xiang-Hong, J.; Yan-Hong, Y.; Han-Jin, X.; Li-Long, A.; Ying-Mei, X.; Pei-Rong, J.; Ming, L. Selection of reference genes for gene expression studies in PBMC from Bama miniature pig under heat stress. Vet. Immunol. Immunopathol. 2011, 144, 160–166. [Google Scholar] [CrossRef]
- Hortopan, G.A.; Dinday, M.T.; Baraban, S.C. Spontaneous seizures and altered gene expression in GABA signaling pathways in a mind bomb mutant zebrafish. J. Neurosci. 2010, 30, 13718–13728. [Google Scholar] [CrossRef]
- Teyssier, J.R.; Ragot, S.; Chauvet-Gelinier, J.C.; Trojak, B.; Bonin, B. Activation of a DeltaFOSB dependent gene expression pattern in the dorsolateral prefrontal cortex of patients with major depressive disorder. J. Affect Disord. 2011, 133, 174–178. [Google Scholar] [CrossRef] [PubMed]
- Perez-Santiago, J.; Diez-Alarcia, R.; Callado, L.F.; Zhang, J.X.; Chana, G.; White, C.H.; Glatt, S.J.; Tsuang, M.T.; Everall, I.P.; Meana, J.J.; et al. A combined analysis of microarray gene expression studies of the human prefrontal cortex identifies genes implicated in schizophrenia. J. Psychiatr. Res. 2012, 46, 1464–1474. [Google Scholar] [CrossRef] [PubMed]
- Morganstern, I.; Liang, S.; Ye, Z.; Karatayev, O.; Leibowitz, S.F. Disturbances in behavior and cortical enkephalin gene expression during the anticipation of ethanol in rats characterized as high drinkers. Alcohol 2012, 46, 559–568. [Google Scholar] [CrossRef] [PubMed]
- Sanz, G.; Jimenez-Marin, A.; Barbancho, M.; Garrido, J.J. Molecular cloning, characterization and gene expression of the full length cDNA encoding the porcine CD11b(alphaM) and chromosomal localization of the porcine CD11a(alphaL)-CD11b(alphaM)-CD11b(alphaD) gene cluster. Vet. Immunol. Immunopathol. 2012, 145, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Goselink, R.M.; van Baal, J.; Widjaja, H.C.; Dekker, R.A.; Zom, R.L.; de Veth, M.J.; van Vuuren, A.M. Effect of rumen-protected choline supplementation on liver and adipose gene expression during the transition period in dairy cattle. J. Dairy Sci. 2013, 96, 1102–1116. [Google Scholar] [CrossRef]
- Ghandhi, S.A.; Ming, L.; Ivanov, V.N.; Hei, T.K.; Amundson, S.A. Regulation of early signaling and gene expression in the alpha-particle and bystander response of IMR-90 human fibroblasts. BMC Med. Genom. 2010, 3, 31. [Google Scholar] [CrossRef]
- Veldhoen, N.; Ikonomou, M.G.; Dubetz, C.; Macpherson, N.; Sampson, T.; Kelly, B.C.; Helbing, C.C. Gene expression profiling and environmental contaminant assessment of migrating Pacific salmon in the Fraser River watershed of British Columbia. Aquat. Toxicol. 2010, 97, 212–225. [Google Scholar] [CrossRef]
- Ranera, B.; Lyahyai, J.; Romero, A.; Vazquez, F.J.; Remacha, A.R.; Bernal, M.L.; Zaragoza, P.; Rodellar, C.; Martin-Burriel, I. Immunophenotype and gene expression profiles of cell surface markers of mesenchymal stem cells derived from equine bone marrow and adipose tissue. Vet. Immunol. Immunopathol. 2011, 144, 147–154. [Google Scholar] [CrossRef]
- Sigl, T.; Meyer, H.H.; Wiedemann, S. Gene expression analysis of protein synthesis pathways in bovine mammary epithelial cells purified from milk during lactation and short-term restricted feeding. J. Anim. Physiol. Anim. Nutr. 2014, 98, 84–95. [Google Scholar] [CrossRef]

| Gene Symbol | Gene Name | Function |
|---|---|---|
| 18S | 18S ribosomal RNA | a part of the ribosomal RNA |
| B2M | Beta-2-microglobulin | a component of the major histocompatibility complex (MHC) class I molecules |
| eEF1α1 | Eukaryotic Translation Elongation Factor 1 Alpha 1 | a key factor in protein synthesis |
| GAPDH | Glyceraldehyde 3-phosphate dehydrogenase | a key factor in glycolysis |
| GUSB | Beta-glucuronidase | encoding a hydrolase that degrades glycosaminoglycans |
| HPRT1 | Hypoxanthine-guanine phosphoribosyltransferase | generation of purine nucleotides via the purine salvage pathway |
| PPIA | Peptidyl-prolyl isomerase A | accelerating the folding of proteins and catalyzing the cis-trans isomerization of proline imidic peptide bonds in oligopeptides |
| TBP | TATA-box binding protein | a transcription factor that functions at the core of the DNA-binding multiprotein factor TFIID |
| Gene | Accession Number | Primer | Sequence (5′-3′) | Primer Length (bp) | Amplicon Length (bp) |
|---|---|---|---|---|---|
| 18S rRNA | NR_145820.1 | forward | AAACGGCTACCACATCCAAG | 20 | 250 |
| reverse | CGCTCCCAAGATCCAACTAC | 20 | |||
| B2M | NM_004048.4 | forward | AGGCTATCCAGCGTACTCCA | 20 | 112 |
| reverse | TTCAATGTCGGATGGATGAA | 20 | |||
| eEF1α1 | NM_001402.6 | forward | TCTGGTTGGAATGGTGACAA | 20 | 141 |
| reverse | ACGAGTTGGTGGTAGGATGC | 20 | |||
| GAPDH | NM_001289745.3 | forward | TCGGAGTCAACGGATTTG | 18 | 220 |
| reverse | TCCTGGAAGATGGTGATGG | 19 | |||
| GUSB | NM_000181.4 | forward | TGCGTAGGGACAAGAACCAC | 20 | 129 |
| reverse | GGGAGGGGTCCAAGGATTTG | 20 | |||
| HPRT1 | NM_000194.3 | forward | GCCCTGGCGTCGTGATTAG | 19 | 140 |
| reverse | TCTCGAGCAAGACGTTCAGT | 20 | |||
| PPIA | NM_001300981.2 | forward | TTCATCTGCACTGCCAAGAC | 20 | 158 |
| reverse | TCGAGTTGTCCACAGTCAGC | 20 | |||
| TBP | NM_001172085.2 | forward | ACTCCACTGTATCCCTCCCC | 20 | 172 |
| reverse | TATATTCGGCGTTTCGGGCA | 20 |
| Treatment | Recommended Genes | Not Recommended Genes |
|---|---|---|
| UVB | 18S rRNA, eEF1α1, GAPDH, GUSB, HPRT1 | B2M, PPIA, TBP |
| NCS | B2M, HPRT1, TBP | 18S rRNA, eEF1α1, GAPDH, GUSB, PPIA |
| ActD | 18S rRNA, B2M, PPIA | eEF1α1, GAPDH, GUSB, HPRT1, TBP |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barta, N.; Ördög, N.; Pantazi, V.; Berzsenyi, I.; Borsos, B.N.; Majoros, H.; Páhi, Z.G.; Ujfaludi, Z.; Pankotai, T. Identifying Suitable Reference Gene Candidates for Quantification of DNA Damage-Induced Cellular Responses in Human U2OS Cell Culture System. Biomolecules 2023, 13, 1523. https://doi.org/10.3390/biom13101523
Barta N, Ördög N, Pantazi V, Berzsenyi I, Borsos BN, Majoros H, Páhi ZG, Ujfaludi Z, Pankotai T. Identifying Suitable Reference Gene Candidates for Quantification of DNA Damage-Induced Cellular Responses in Human U2OS Cell Culture System. Biomolecules. 2023; 13(10):1523. https://doi.org/10.3390/biom13101523
Chicago/Turabian StyleBarta, Nikolett, Nóra Ördög, Vasiliki Pantazi, Ivett Berzsenyi, Barbara N. Borsos, Hajnalka Majoros, Zoltán G. Páhi, Zsuzsanna Ujfaludi, and Tibor Pankotai. 2023. "Identifying Suitable Reference Gene Candidates for Quantification of DNA Damage-Induced Cellular Responses in Human U2OS Cell Culture System" Biomolecules 13, no. 10: 1523. https://doi.org/10.3390/biom13101523
APA StyleBarta, N., Ördög, N., Pantazi, V., Berzsenyi, I., Borsos, B. N., Majoros, H., Páhi, Z. G., Ujfaludi, Z., & Pankotai, T. (2023). Identifying Suitable Reference Gene Candidates for Quantification of DNA Damage-Induced Cellular Responses in Human U2OS Cell Culture System. Biomolecules, 13(10), 1523. https://doi.org/10.3390/biom13101523








