USP39-Mediated Non-Proteolytic Control of ETS2 Suppresses Nuclear Localization and Activity
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Plasmids
2.3. Immunoblotting
2.4. Site-Directed Mutagenesis
2.5. Immunoprecipitation and Pulldown Assays
2.6. RNA Interference
2.7. Deubiquitination Assays
2.8. Cycloheximide Chase Assay
2.9. Luciferase Assays
2.10. Cytoplasmic and Nuclear Fractionation Assays
2.11. Gene Expression Omnibus (GEO) Repository Dataset Analysis
2.12. Statistical Analysis
3. Results
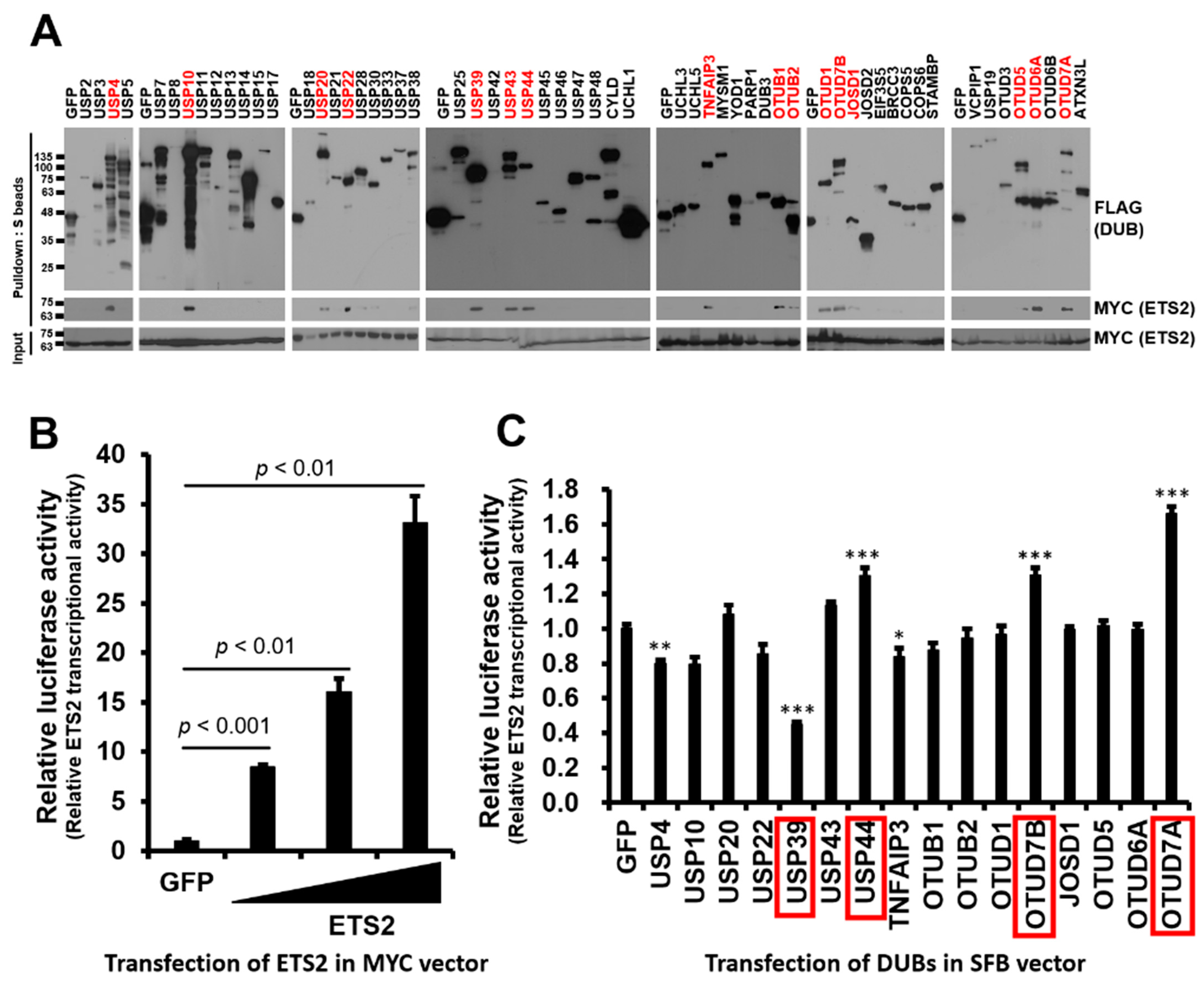
3.1. Identification of DUBs Interacting with ETS2 and Modulating Its Transcriptional Activity
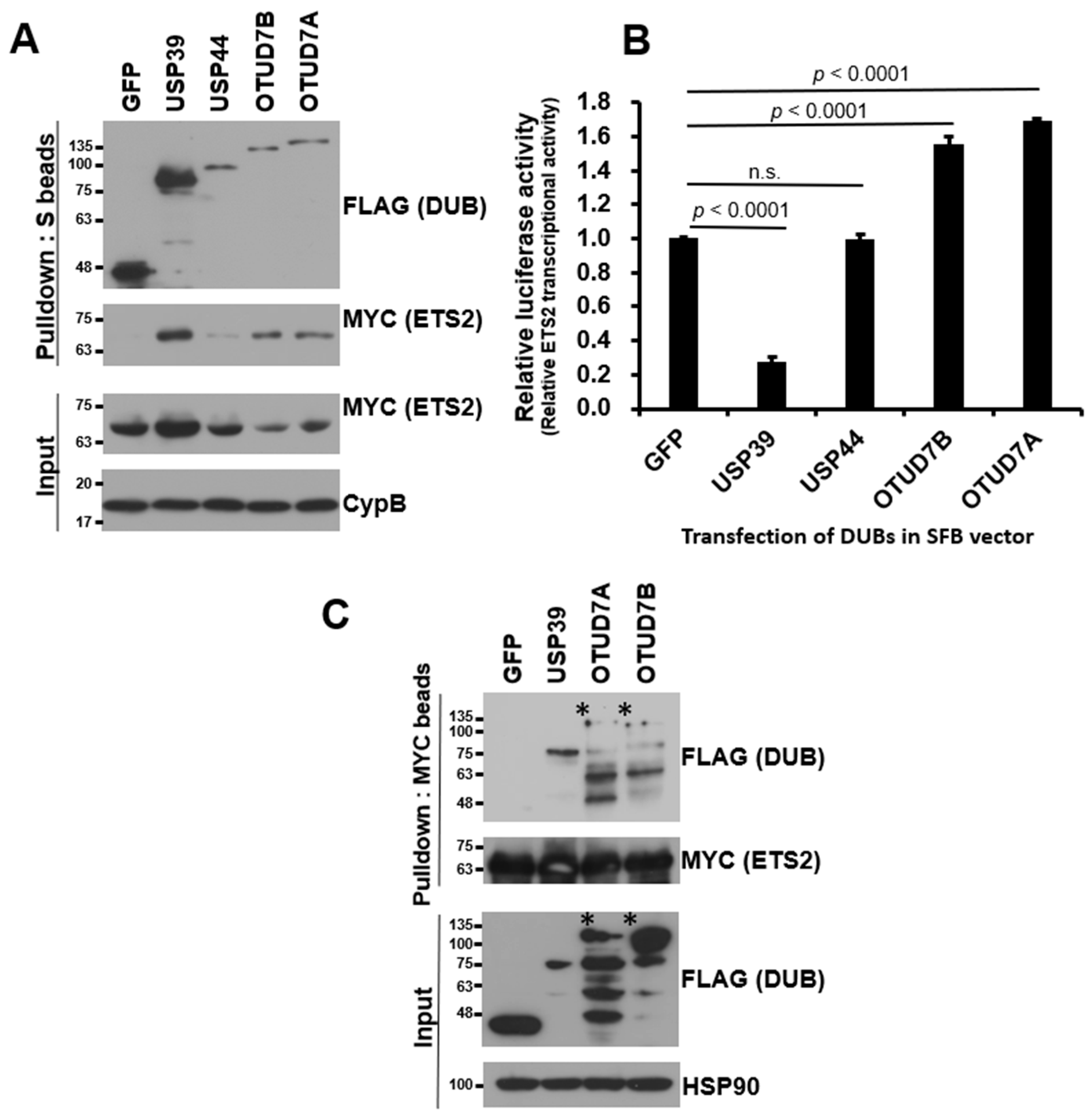
3.2. USP39 Interacts with and Suppresses Transcriptional Activity of ETS2
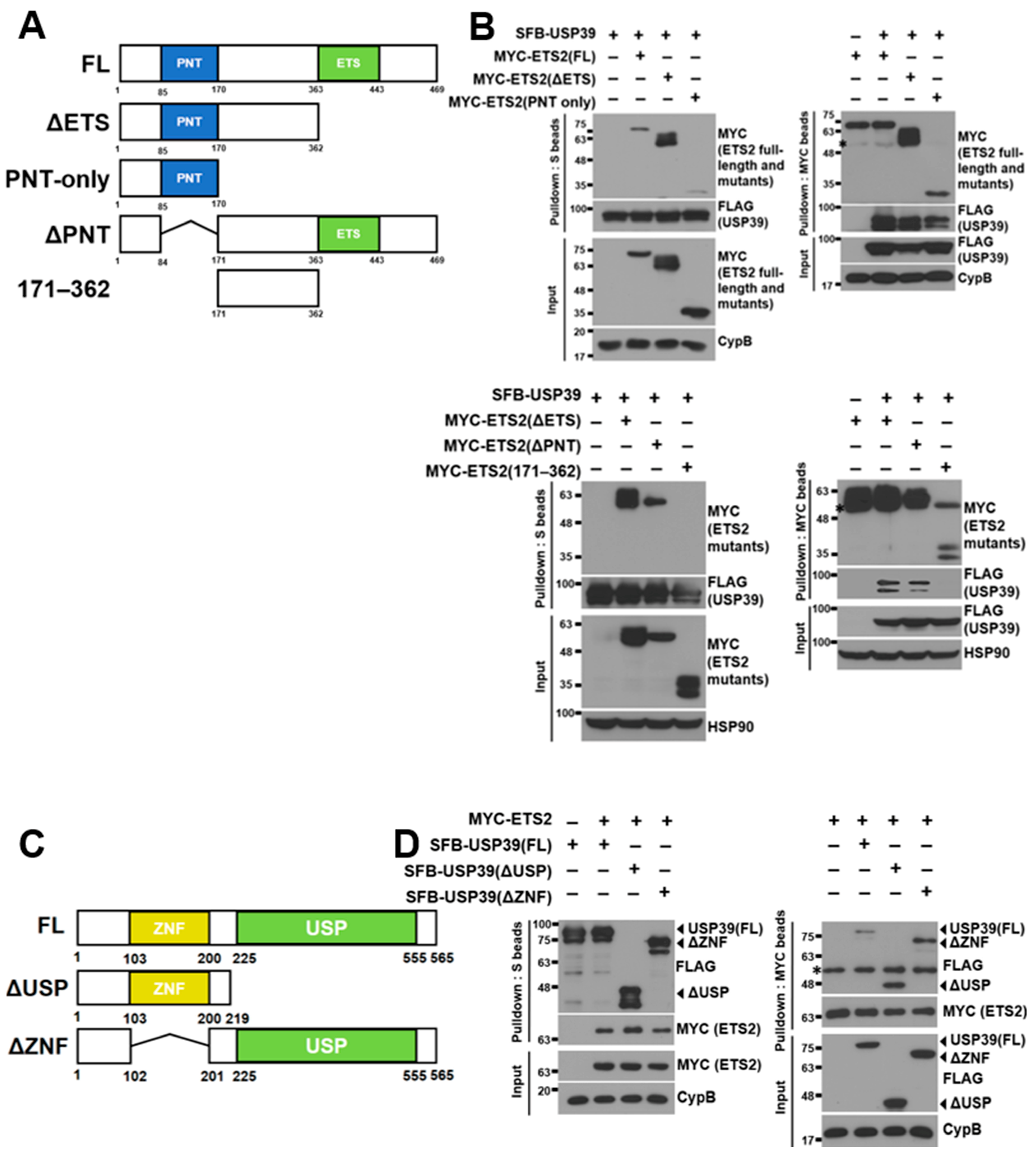
3.3. ETS2 Interacts with USP39 through the Respective Amino-Terminal Regions
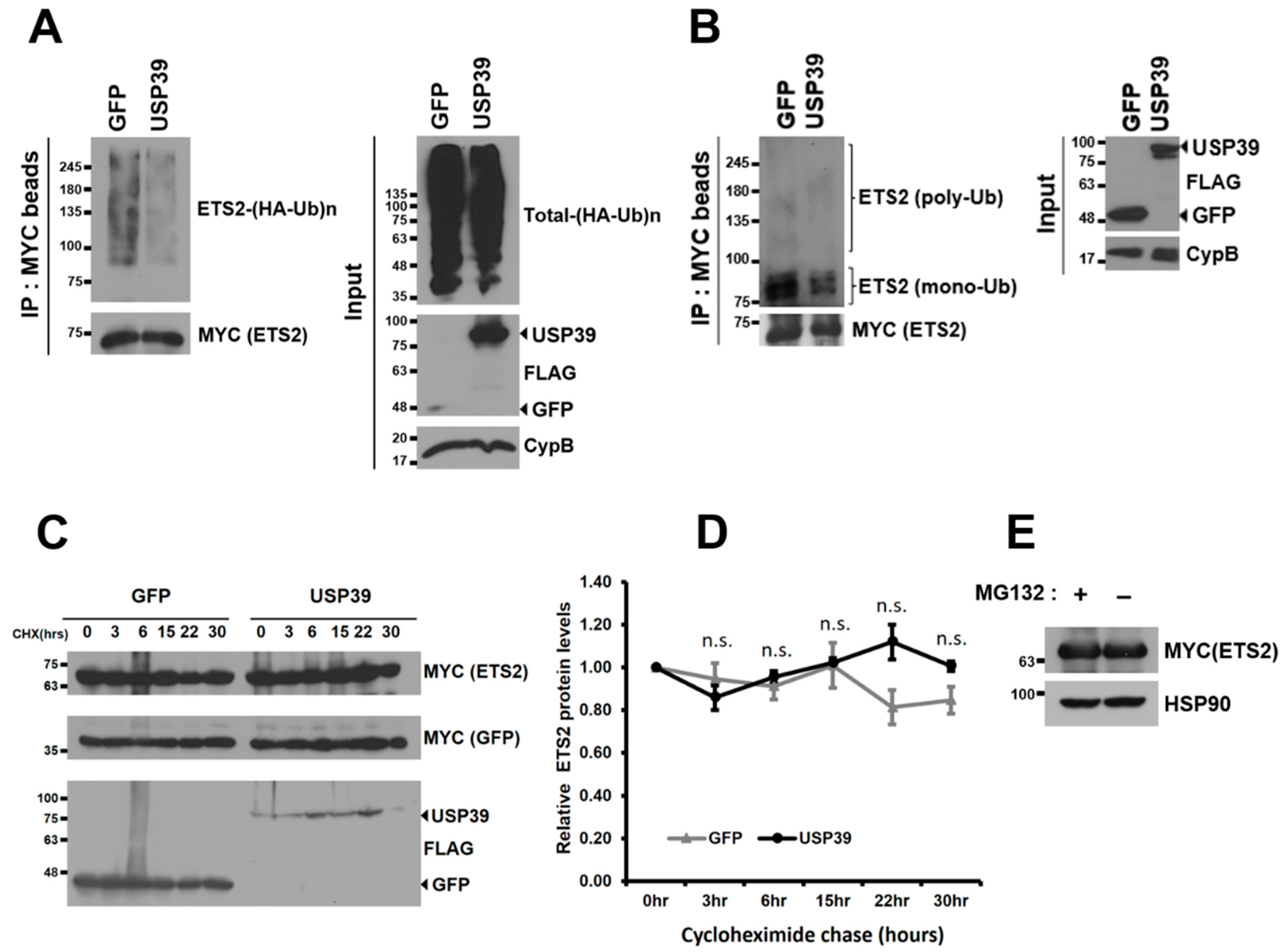
3.4. USP39 Deubiquitinates ETS2 but Does Not Affect Its Protein Stability
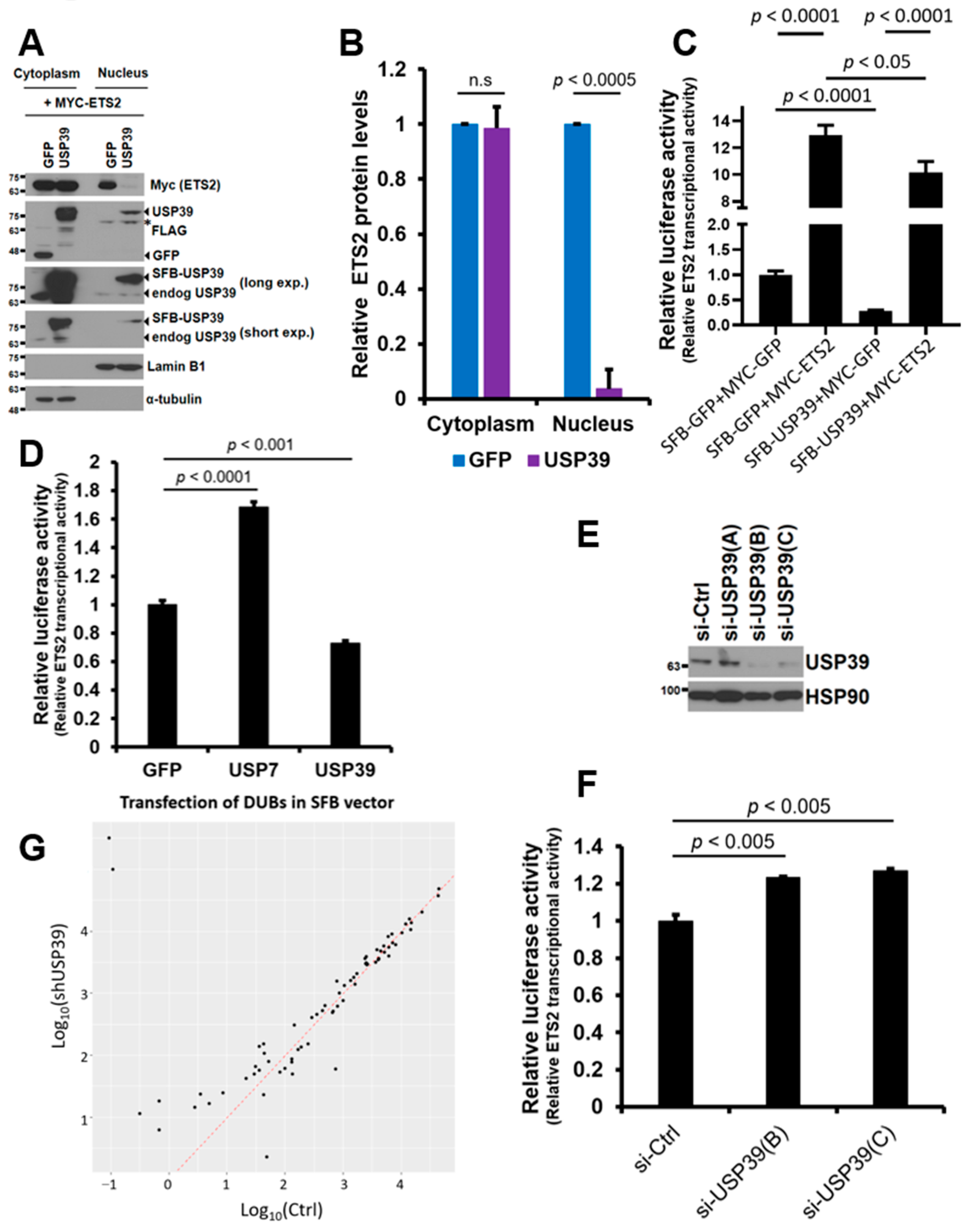
3.5. USP39 Represses the Transcriptional Activity of ETS2 Partly by Suppressing Its Nuclear Localization
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Findlay, V.J.; LaRue, A.C.; Turner, D.P.; Watson, P.M.; Watson, D.K. Understanding the role of ETS-mediated gene regulation in complex biological processes. Adv. Cancer Res. 2013, 119, 1–61. [Google Scholar] [CrossRef]
- Sizemore, G.M.; Pitarresi, J.R.; Balakrishnan, S.; Ostrowski, M.C. The ETS family of oncogenic transcription factors in solid tumours. Nat. Rev. Cancer 2017, 17, 337–351. [Google Scholar] [CrossRef] [PubMed]
- Fry, E.A.; Inoue, K. Aberrant expression of ETS1 and ETS2 proteins in cancer. Cancer Rep. Rev. 2018, 2, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Qi, Y.; Xu, Y.; Wang, M.; Wang, J.; Wang, J.; Yuan, M.; Jia, Y.; Ma, X.; Wang, Y.; et al. lncRNA DLEU2 promotes gastric cancer progression through ETS2 via targeting miR-30a-5p. Cancer Cell Int. 2021, 21, 376. [Google Scholar] [CrossRef] [PubMed]
- Sevilla, L.; Aperlo, C.; Dulic, V.; Chambard, J.C.; Boutonnet, C.; Pasquier, O.; Pognonec, P.; Boulukos, K.E. The Ets2 transcription factor inhibits apoptosis induced by colony-stimulating factor 1 deprivation of macrophages through a Bcl-xL-dependent mechanism. Mol. Cell Biol. 1999, 19, 2624–2634. [Google Scholar] [CrossRef]
- Wu, W.; Zhang, S.; Li, X.; Xue, M.; Cao, S.; Chen, W. Ets-2 regulates cell apoptosis via the Akt pathway, through the regulation of urothelial cancer associated 1, a long non-coding RNA, in bladder cancer cells. PLoS ONE 2013, 8, e73920. [Google Scholar] [CrossRef]
- Xu, D.; Dwyer, J.; Li, H.; Duan, W.; Liu, J.P. Ets2 maintains hTERT gene expression and breast cancer cell proliferation by interacting with c-Myc. J. Biol. Chem. 2008, 283, 23567–23580. [Google Scholar] [CrossRef]
- Munera, J.; Cecena, G.; Jedlicka, P.; Wankell, M.; Oshima, R.G. Ets2 regulates colonic stem cells and sensitivity to tumorigenesis. Stem Cells 2011, 29, 430–439. [Google Scholar] [CrossRef]
- Li, Q.; Yang, L.; Han, K.; Zhu, L.; Zhang, Y.; Ma, S.; Zhang, K.; Yang, B.; Guan, F. Ets2 knockdown inhibits tumorigenesis in esophageal squamous cell carcinoma in vivo and in vitro. Oncotarget 2016, 7, 61458–61468. [Google Scholar] [CrossRef]
- Islas, J.F.; Liu, Y.; Weng, K.C.; Robertson, M.J.; Zhang, S.; Prejusa, A.; Harger, J.; Tikhomirova, D.; Chopra, M.; Iyer, D.; et al. Transcription factors ETS2 and MESP1 transdifferentiate human dermal fibroblasts into cardiac progenitors. Proc. Natl. Acad. Sci. USA 2012, 109, 13016–13021. [Google Scholar] [CrossRef]
- Charlot, C.; Dubois-Pot, H.; Serchov, T.; Tourrette, Y.; Wasylyk, B. A review of post-translational modifications and subcellular localization of Ets transcription factors: Possible connection with cancer and involvement in the hypoxic response. Methods Mol. Biol. 2010, 647, 3–30. [Google Scholar] [CrossRef]
- Fujiwara, S.; Fisher, R.J.; Bhat, N.K.; Diaz de la Espina, S.M.; Papas, T.S. A short-lived nuclear phosphoprotein encoded by the human ets-2 proto-oncogene is stabilized by activation of protein kinase C. Mol. Cell Biol. 1988, 8, 4700–4706. [Google Scholar] [CrossRef] [PubMed]
- Wasylyk, C.; Bradford, A.P.; Gutierrez-Hartmann, A.; Wasylyk, B. Conserved mechanisms of Ras regulation of evolutionary related transcription factors, Ets1 and Pointed P2. Oncogene 1997, 14, 899–913. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Carrero, Z.I.; Kollareddy, M.; Chauhan, K.M.; Ramakrishnan, G.; Martinez, L.A. Mutant p53 protects ETS2 from non-canonical COP1/DET1 dependent degradation. Oncotarget 2016, 7, 12554–12567. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Zhang, Q.; Huang, Y.; Song, J.; Tomaino, R.; Ehrenberger, T.; Lim, E.; Liu, W.; Bronson, R.T.; Bowden, M.; et al. Phosphorylation of ETS1 by Src family kinases prevents its recognition by the COP1 tumor suppressor. Cancer Cell 2014, 26, 222–234. [Google Scholar] [CrossRef]
- Eletr, Z.M.; Wilkinson, K.D. Regulation of proteolysis by human deubiquitinating enzymes. Biochim. Biophys. Acta 2014, 1843, 114–128. [Google Scholar] [CrossRef]
- Potu, H.; Peterson, L.F.; Kandarpa, M.; Pal, A.; Sun, H.; Durham, A.; Harms, P.W.; Hollenhorst, P.C.; Eskiocak, U.; Talpaz, M.; et al. Usp9x regulates Ets-1 ubiquitination and stability to control NRAS expression and tumorigenicity in melanoma. Nat. Commun. 2017, 8, 14449. [Google Scholar] [CrossRef]
- Park, H.B.; Min, Y.; Hwang, S.; Baek, K.H. Suppression of USP7 negatively regulates the stability of ETS proto-oncogene 2 protein. Biomed. Pharmacother. 2023, 162, 114700. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Kim, J. OTUD6A Is an Aurora Kinase A-Specific Deubiquitinase. Int. J. Mol. Sci. 2021, 22, 1936. [Google Scholar] [CrossRef]
- Cao, Q.; Yang, S.; Lv, Q.; Liu, Y.; Li, L.; Wu, X.; Qu, G.; He, X.; Zhang, X.; Sun, S.; et al. Five ETS family members, ELF-1, ETV-4, ETV-3L, ETS-1, and ETS-2 upregulate human leukocyte-associated immunoglobulin-like receptor-1 gene basic promoter activity. Aging 2018, 10, 1390–1401. [Google Scholar] [CrossRef]
- Sharrocks, A.D. The ETS-domain transcription factor family. Nat. Rev. Mol. Cell Biol. 2001, 2, 827–837. [Google Scholar] [CrossRef] [PubMed]
- Mackereth, C.D.; Scharpf, M.; Gentile, L.N.; MacIntosh, S.E.; Slupsky, C.M.; McIntosh, L.P. Diversity in structure and function of the Ets family PNT domains. J. Mol. Biol. 2004, 342, 1249–1264. [Google Scholar] [CrossRef]
- Ye, Y.; Scheel, H.; Hofmann, K.; Komander, D. Dissection of USP catalytic domains reveals five common insertion points. Mol. Biosyst. 2009, 5, 1797–1808. [Google Scholar] [CrossRef] [PubMed]
- Laity, J.H.; Lee, B.M.; Wright, P.E. Zinc finger proteins: New insights into structural and functional diversity. Curr. Opin. Struct. Biol. 2001, 11, 39–46. [Google Scholar] [CrossRef]
- Li, M.; Brooks, C.L.; Wu-Baer, F.; Chen, D.; Baer, R.; Gu, W. Mono- versus polyubiquitination: Differential control of p53 fate by Mdm2. Science 2003, 302, 1972–1975. [Google Scholar] [CrossRef] [PubMed]
- Todi, S.V.; Winborn, B.J.; Scaglione, K.M.; Blount, J.R.; Travis, S.M.; Paulson, H.L. Ubiquitination directly enhances activity of the deubiquitinating enzyme ataxin-3. EMBO J. 2009, 28, 372–382. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wang, Z.; Li, J.; Qin, J.; Song, J.; Li, Y.; Zhao, L.; Zhang, X.; Guo, H.; Shao, C.; et al. Splicing factor USP39 promotes ovarian cancer malignancy through maintaining efficient splicing of oncogenic HMGA2. Cell Death Dis. 2021, 12, 294. [Google Scholar] [CrossRef]
- Shcherbik, N.; Haines, D.S. Ub on the move. J. Cell Biochem. 2004, 93, 11–19. [Google Scholar] [CrossRef]
- Lee, Y.; Piao, H.L.; Kim, J. OTUD7B Activates Wnt Signaling Pathway through the Interaction with LEF1. Biomolecules 2023, 13, 1001. [Google Scholar] [CrossRef]
- van Leuken, R.J.; Luna-Vargas, M.P.; Sixma, T.K.; Wolthuis, R.M.; Medema, R.H. Usp39 is essential for mitotic spindle checkpoint integrity and controls mRNA-levels of aurora B. Cell Cycle 2008, 7, 2710–2719. [Google Scholar] [CrossRef]
- Pinto-Fernandez, A.; Davis, S.; Schofield, A.B.; Scott, H.C.; Zhang, P.; Salah, E.; Mathea, S.; Charles, P.D.; Damianou, A.; Bond, G.; et al. Comprehensive Landscape of Active Deubiquitinating Enzymes Profiled by Advanced Chemoproteomics. Front. Chem. 2019, 7, 592. [Google Scholar] [CrossRef]
- Kim, J.J.; Lee, S.Y.; Hwang, Y.; Kim, S.; Chung, J.M.; Park, S.; Yoon, J.; Yun, H.; Ji, J.H.; Chae, S.; et al. USP39 promotes non-homologous end-joining repair by poly(ADP-ribose)-induced liquid demixing. Nucleic Acids Res. 2021, 49, 11083–11102. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Geng, H.; Liu, G.; Ji, Q.; Cheng, X.; Li, X.; Liu, W.; Thorne, R.F.; Zhang, R.; Liu, X. The Deubiquitinase USP39 Promotes ESCC Tumorigenesis Through Pre-mRNA Splicing of the mTORC2 Component Rictor. Front. Oncol. 2021, 11, 667495. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Ma, J.; Lu, M.; Liu, Z.; Sun, Y.; Chen, H. The Deubiquitinase USP39 Promotes Esophageal Squamous Cell Carcinoma Malignancy as a Splicing Factor. Genes 2022, 13, 819. [Google Scholar] [CrossRef]
- Lygerou, Z.; Christophides, G.; Seraphin, B. A novel genetic screen for snRNP assembly factors in yeast identifies a conserved protein, Sad1p, also required for pre-mRNA splicing. Mol. Cell Biol. 1999, 19, 2008–2020. [Google Scholar] [CrossRef]
- Ruan, G.X.; Li, Y.; Chen, W.; Huang, H.; Zhang, R.; Chen, C.; Lam, K.P.; Xu, S.; Ou, X. The spliceosome component Usp39 controls B cell development by regulating immunoglobulin gene rearrangement. Cell Rep. 2022, 38, 110338. [Google Scholar] [CrossRef]
- Pan, X.W.; Xu, D.; Chen, W.J.; Chen, J.X.; Chen, W.J.; Ye, J.Q.; Gan, S.S.; Zhou, W.; Song, X.; Shi, L.; et al. USP39 promotes malignant proliferation and angiogenesis of renal cell carcinoma by inhibiting VEGF-A(165b) alternative splicing via regulating SRSF1 and SRPK1. Cancer Cell Int. 2021, 21, 486. [Google Scholar] [CrossRef]
- Feng, Z.; Li, K.; Qin, K.; Liang, J.; Shi, M.; Ma, Y.; Zhao, S.; Liang, H.; Han, D.; Shen, B.; et al. The LINC00623/NAT10 signaling axis promotes pancreatic cancer progression by remodeling ac4C modification of mRNA. J. Hematol. Oncol. 2022, 15, 112. [Google Scholar] [CrossRef]
- Xiao, Y.; Chen, X.; Hu, W.; Ma, W.; Di, Q.; Tang, H.; Zhao, X.; Huang, G.; Chen, W. USP39-mediated deubiquitination of Cyclin B1 promotes tumor cell proliferation and glioma progression. Transl. Oncol. 2023, 34, 101713. [Google Scholar] [CrossRef]
- Peng, Y.; Guo, J.; Sun, T.; Fu, Y.; Zheng, H.; Dong, C.; Xiong, S. USP39 Serves as a Deubiquitinase to Stabilize STAT1 and Sustains Type I IFN-Induced Antiviral Immunity. J. Immunol. 2020, 205, 3167–3178. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Lei, Y.; Zhang, G.; Li, X.; Yuan, J.; Li, T.; Zhong, W.; Zhang, Y.; Tan, X.; Song, G. USP39 stabilizes beta-catenin by deubiquitination and suppressing E3 ligase TRIM26 pre-mRNA maturation to promote HCC progression. Cell Death Dis. 2023, 14, 63. [Google Scholar] [CrossRef]
- Wu, J.; Chen, Y.; Geng, G.; Li, L.; Yin, P.; Nowsheen, S.; Li, Y.; Wu, C.; Liu, J.; Zhao, F.; et al. USP39 regulates DNA damage response and chemo-radiation resistance by deubiquitinating and stabilizing CHK2. Cancer Lett. 2019, 449, 114–124. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Liu, Z.; Zhang, E.; Zhang, P.; Wang, Y.; Hang, J.; Li, Q. USP39 promotes tumorigenesis by stabilizing and deubiquitinating SP1 protein in hepatocellular carcinoma. Cell Signal 2021, 85, 110068. [Google Scholar] [CrossRef] [PubMed]
- Quan, J.; Zhao, X.; Xiao, Y.; Wu, H.; Di, Q.; Wu, Z.; Chen, X.; Tang, H.; Zhao, J.; Guan, Y.; et al. USP39 Regulates NF-kappaB-Mediated Inflammatory Responses through Deubiquitinating K48-Linked IkappaBalpha. J. Immunol. 2023, 210, 640–652. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, Y.; Lee, Y.; Kim, J.S.; Zhang, P.; Kim, J. USP39-Mediated Non-Proteolytic Control of ETS2 Suppresses Nuclear Localization and Activity. Biomolecules 2023, 13, 1475. https://doi.org/10.3390/biom13101475
Choi Y, Lee Y, Kim JS, Zhang P, Kim J. USP39-Mediated Non-Proteolytic Control of ETS2 Suppresses Nuclear Localization and Activity. Biomolecules. 2023; 13(10):1475. https://doi.org/10.3390/biom13101475
Chicago/Turabian StyleChoi, Yunsik, Yuri Lee, Jin Seo Kim, Peijing Zhang, and Jongchan Kim. 2023. "USP39-Mediated Non-Proteolytic Control of ETS2 Suppresses Nuclear Localization and Activity" Biomolecules 13, no. 10: 1475. https://doi.org/10.3390/biom13101475
APA StyleChoi, Y., Lee, Y., Kim, J. S., Zhang, P., & Kim, J. (2023). USP39-Mediated Non-Proteolytic Control of ETS2 Suppresses Nuclear Localization and Activity. Biomolecules, 13(10), 1475. https://doi.org/10.3390/biom13101475




