Assessment of Fibrinogen-like 2 (FGL2) in Human Chronic Kidney Disease through Transcriptomics Data Analysis
Abstract
:1. Introduction
2. Results
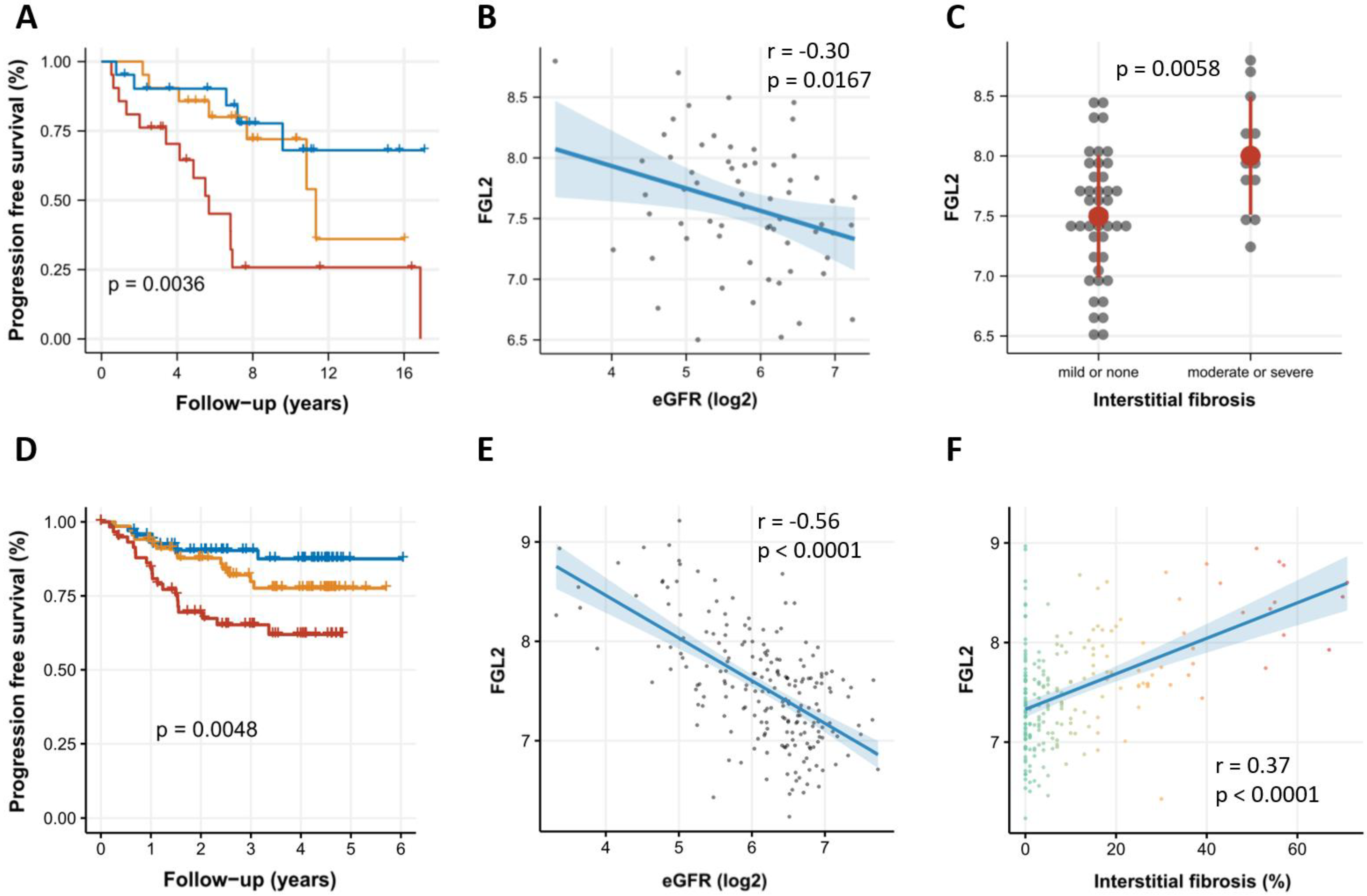
2.1. Association of FGL2 Expression with CKD Progression in the Innsbruck CKD Cohort
2.2. Association of FGL2 with CKD Progression in the NEPTUNE Cohort
2.3. FGL2 Is Significantly Higher Expressed in CKD
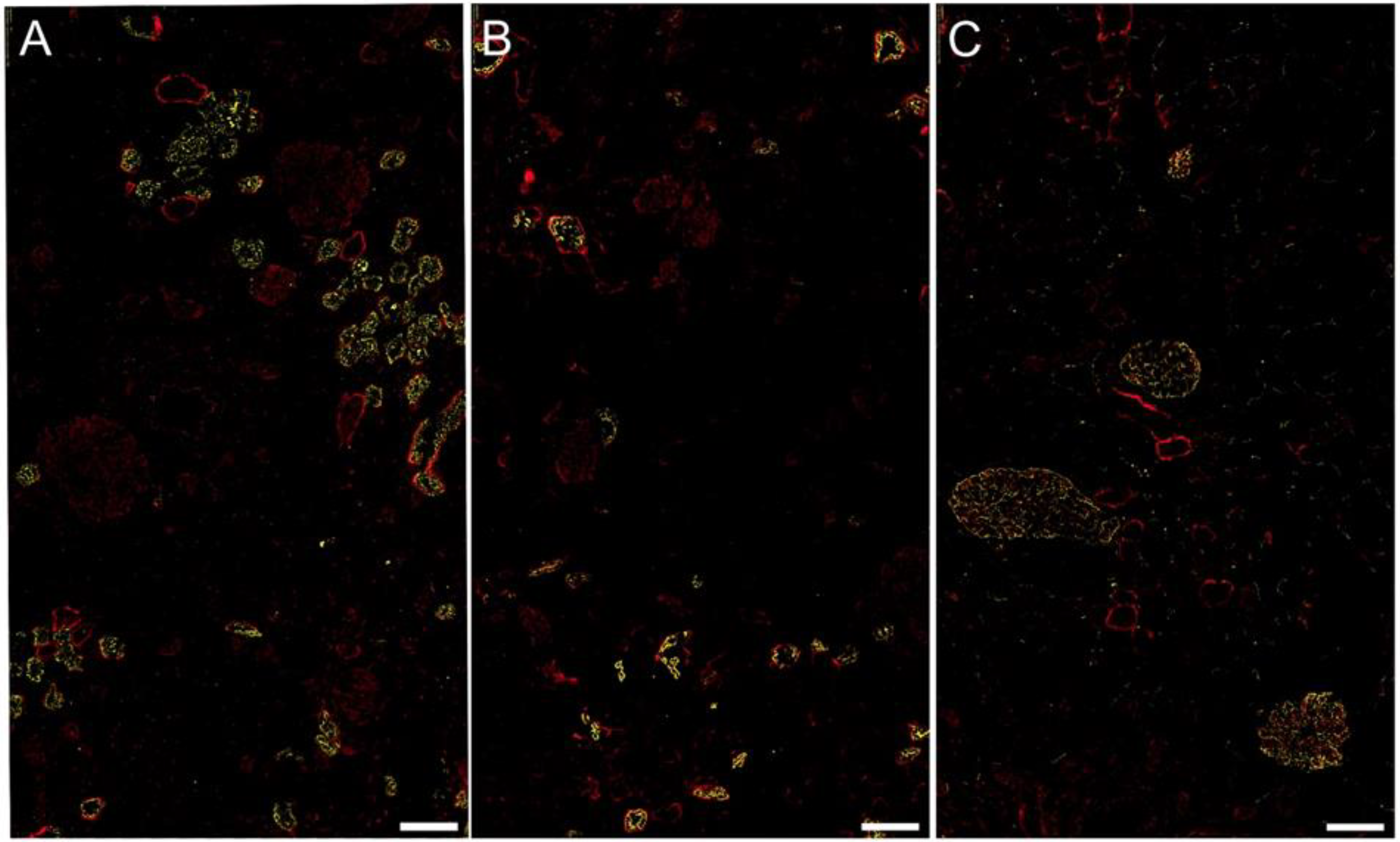
2.4. FGL2 Protein Localization in CKD
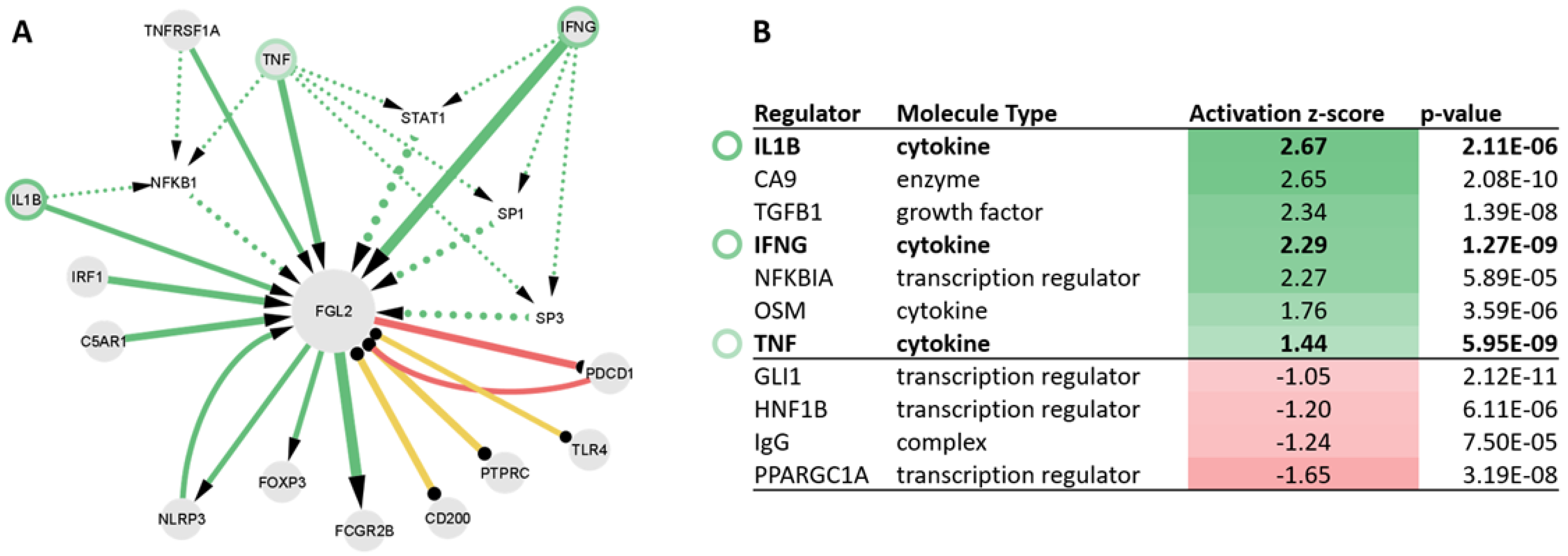
2.5. Upstream Regulators of FGL2 Include TNF, IL1B, and IFNG
3. Discussion
4. Materials and Methods
4.1. FGL2 Outcome Analysis in the Innsbruck CKD Cohort
4.2. FGL2 Outcome Analysis in the NEPTUNE Cohort
4.3. FGL2 mRNA Expression Analysis in CKD Patients as Compared with Healthy Controls
4.4. FGL2 Protein Staining in Human CKD Tissue Samples
4.5. Literature Co-Annotation Analysis of FGL2
4.6. Single Cell RNA Sequencing (scRNA-seq) Analysis
4.7. Ingenuity Upstream Regulator Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bikbov, B.; Purcell, C.A.; Levey, A.S.; Smith, M.; Abdoli, A.; Abebe, M.; Adebayo, O.M.; Afarideh, M.; Agarwal, S.K.; Agudelo-Botero, M.; et al. Global, regional, and national burden of chronic kidney disease, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 2020, 395, 709–733. [Google Scholar] [CrossRef] [Green Version]
- Meng, X.-M.; Nikolic-Paterson, D.J.; Lan, H.Y. Inflammatory processes in renal fibrosis. Nat. Rev. Nephrol. 2014, 10, 493–503. [Google Scholar] [CrossRef]
- Tang, P.M.K.; Nikolic-Paterson, D.J.; Lan, H.Y. Macrophages: Versatile players in renal inflammation and fibrosis. Nat. Rev. Nephrol. 2019, 15, 144–158. [Google Scholar] [CrossRef]
- Tampe, D.; Zeisberg, M. Potential approaches to reverse or repair renal fibrosis. Nat. Rev. Nephrol. 2014, 10, 226–237. [Google Scholar] [CrossRef]
- Klinkhammer, B.M.; Goldschmeding, R.; Floege, J.; Boor, P. Treatment of Renal Fibrosis—Turning Challenges into Opportunities. Adv. Chronic Kidney Dis. 2017, 24, 117–129. [Google Scholar] [CrossRef]
- Yang, G. Physiological functions and clinical implications of fibrinogen-like 2: A review. World J. Clin. Infect. Dis. 2013, 3, 37. [Google Scholar] [CrossRef]
- Hu, J.; Yan, J.; Rao, G.; Latha, K.; Overwijk, W.W.; Heimberger, A.B.; Li, S. The Duality of Fgl2—Secreted Immune Checkpoint Regulator Versus Membrane-Associated Procoagulant: Therapeutic Potential and Implications. Int. Rev. Immunol. 2016, 35, 325–339. [Google Scholar] [CrossRef] [Green Version]
- de Ridder, G.G.; Lundblad, R.L.; Pizzo, S.V. Actions of thrombin in the interstitium. J. Thromb. Haemost. 2016, 14, 40–47. [Google Scholar] [CrossRef] [Green Version]
- Ning, Q.; Sun, Y.; Han, M.; Zhang, L.; Zhu, C.; Zhang, W.; Guo, H.; Li, J.; Yan, W.; Gong, F.; et al. Role of Fibrinogen-Like Protein 2 Prothrombinase/Fibroleukin in Experimental and Human Allograft Rejection. J. Immunol. 2005, 174, 7403–7411. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.; Li, M.; Xu, F.; Li, G.-Q.; Han, B.; He, X.-D.; Li, S.-J.; He, Q.-H.; Lai, X.-Y.; Zhou, S.; et al. Fibrinogen-like protein 2 deficiency aggravates renal fibrosis by facilitating macrophage polarization. Biomed. Pharmacother. 2020, 130, 110468. [Google Scholar] [CrossRef]
- Yuwaraj, S.; Ding, J.W.; Liu, M.; Marsden, P.A.; Levy, G.A. Genomic characterization, localization, and functional expression of FGL2, the human gene encoding fibroleukin: A novel human procoagulant. Genomics 2001, 71, 330–338. [Google Scholar] [CrossRef]
- Stenberg, L.M.; Brown, M.A.; Nilsson, E.; Ljungberg, O.; Stenflo, J. A functional prothrombin gene product is synthesized by human kidney cells. Biochem. Biophys. Res. Commun. 2001, 280, 1036–1041. [Google Scholar] [CrossRef]
- Hewitson, T.D.; Martic, M.; Kelynack, K.J.; Pagel, C.N.; Mackie, E.J.; Becker, G.J. Thrombin is a pro-fibrotic factor for rat renal fibroblasts in vitro. Nephron. Exp. Nephrol. 2005, 101, e42–e49. [Google Scholar] [CrossRef]
- Sörensen, I.; Susnik, N.; Inhester, T.; Degen, J.L.; Melk, A.; Haller, H.; Schmitt, R. Fibrinogen, acting as a mitogen for tubulointerstitial fibroblasts, promotes renal fibrosis. Kidney Int. 2011, 80, 1035–1044. [Google Scholar] [CrossRef] [Green Version]
- Wygrecka, M.; Didiasova, M.; Berscheid, S.; Piskulak, K.; Taborski, B.; Zakrzewicz, D.; Kwapiszewska, G.; Preissner, K.T.; Markart, P. Protease-activated receptors (PAR)-1 and -3 drive epithelial-mesenchymal transition of alveolar epithelial cells—Potential role in lung fibrosis. Thromb. Haemost. 2013, 110, 295–307. [Google Scholar] [CrossRef] [Green Version]
- Bogatkevich, G.S.; Tourkina, E.; Silver, R.M.; Ludwicka-Bradley, A. Thrombin differentiates normal lung fibroblasts to a myofibroblast phenotype via the proteolytically activated receptor-1 and a protein kinase C-dependent pathway. J. Biol. Chem. 2001, 276, 45184–45192. [Google Scholar] [CrossRef] [Green Version]
- Su, G.; Liu, K.; Wang, Y.; Wang, J.; Li, X.; Li, W.; Liao, Y.; Wang, Z. Fibrinogen-like protein 2 expression correlates with microthrombosis in rats with type 2 diabetic nephropathy. J. Biomed. Res. 2011, 25, 120–127. [Google Scholar] [CrossRef] [Green Version]
- Shalev, I.; Liu, H.; Koscik, C.; Bartczak, A.; Javadi, M.; Wong, K.M.; Maknojia, A.; He, W.; Liu, M.F.; Diao, J.; et al. Targeted deletion of fgl2 leads to impaired regulatory T cell activity and development of autoimmune glomerulonephritis. J. Immunol. 2008, 180, 249–260. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Lei, J.; Xie, Q.; Wu, L.; Jin, S.; Guo, B.; Wang, X.; Yan, G.; Zhang, Q.; Zhao, H.; et al. Fibrinogen-like protein 2 controls sepsis catabasis by interacting with resolvin Dp5. Sci. Adv. 2019, 5, eaax0629. [Google Scholar] [CrossRef] [Green Version]
- Bézie, S.; Picarda, E.; Tesson, L.; Renaudin, K.; Durand, J.; Ménoret, S.; Mérieau, E.; Chiffoleau, E.; Guillonneau, C.; Caron, L.; et al. Fibrinogen-Like Protein 2/Fibroleukin Induces Long-Term Allograft Survival in a Rat Model through Regulatory B Cells. PLoS ONE 2015, 10, e0119686. [Google Scholar] [CrossRef]
- Bézie, S.; Ménoret, S.; Tesson, L.; Li, X.-L.; Usal, C.; Anegon, I.; Caron, L. Immunosuppressive role of fibrinogen-like protein 2 (FGL2) in CD8+regulatory T cells-mediated long-term graft survival. J. Transl. Med. 2011, 9, O5. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Z.; Yang, C.; Li, L.; Zhao, T.; Wang, L.; Rong, R.; Yang, B.; Xu, M.; Zhu, T. Increased peripheral and local soluble FGL2 in the recovery of renal ischemia reperfusion injury in a porcine kidney auto-transplantation model. J. Transl. Med. 2014, 12, 53. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Z.; Yang, C.; Wang, L.; Li, L.; Zhao, T.; Hu, L.; Rong, R.; Xu, M.; Zhu, T. The regulatory T cell effector soluble fibrinogen-like protein 2 induces tubular epithelial cell apoptosis in renal transplantation. Exp. Biol. Med. 2014, 239, 193–201. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- The Human Protein Atlas. Available online: https://www.proteinatlas.org/ (accessed on 8 April 2022).
- Rudnicki, M.; Perco, P.; Haene, D.B.; Leierer, J.; Heinzel, A.; Mühlberger, I.; Schweibert, N.; Sunzenauer, J.; Regele, H.; Kronbichler, A.; et al. Renal microRNA- and RNA-profiles in progressive chronic kidney disease. Eur. J. Clin. Investig. 2016, 46, 213–226. [Google Scholar] [CrossRef]
- Perco, P.; Ju, W.; Kerschbaum, J.; Leierer, J.; Menon, R.; Zhu, C.; Kretzler, M.; Mayer, G.; Rudnicki, M.; Nephrotic Syndrome Study Network (NEPTUNE). Identification of dicarbonyl and L-xylulose reductase as a therapeutic target in human chronic kidney disease. JCI Insight 2019, 4, e128120. [Google Scholar] [CrossRef] [Green Version]
- Mariani, L.H.; Martini, S.; Barisoni, L.; Canetta, P.A.; Troost, J.P.; Hodgin, J.B.; Palmer, M.; Rosenberg, A.Z.; Lemley, K.V.; Chien, H.-P.; et al. Interstitial fibrosis scored on whole-slide digital imaging of kidney biopsies is a predictor of outcome in proteinuric glomerulopathies. Nephrol. Dial. Transpl. 2018, 33, 310–318. [Google Scholar] [CrossRef] [Green Version]
- Gadegbeku, C.A.; Gipson, D.S.; Holzman, L.B.; Ojo, A.O.; Song, P.X.K.; Barisoni, L.; Sampson, M.G.; Kopp, J.B.; Lemley, K.V.; Nelson, P.J.; et al. Design of the Nephrotic Syndrome Study Network (NEPTUNE) to evaluate primary glomerular nephropathy by a multidisciplinary approach. Kidney Int. 2013, 83, 749–756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barisoni, L.; Gimpel, C.; Kain, R.; Laurinavicius, A.; Bueno, G.; Zeng, C.; Liu, Z.; Schaefer, F.; Kretzler, M.; Holzman, L.B.; et al. Digital pathology imaging as a novel platform for standardization and globalization of quantitative nephropathology. Clin. Kidney J. 2017, 10, 176–187. [Google Scholar] [CrossRef] [Green Version]
- Berthier, C.C.; Bethunaickan, R.; Gonzalez-Rivera, T.; Nair, V.; Ramanujam, M.; Zhang, W.; Bottinger, E.P.; Segerer, S.; Lindenmeyer, M.; Cohen, C.D.; et al. Cross-Species Transcriptional Network Analysis Defines Shared Inflammatory Responses in Murine and Human Lupus Nephritis. J. Immunol. 2012, 189, 988–1001. [Google Scholar] [CrossRef]
- Ju, W.; Greene, C.S.; Eichinger, F.; Nair, V.; Hodgin, J.B.; Bitzer, M.; Lee, Y.-S.; Zhu, Q.; Kehata, M.; Li, M.; et al. Defining cell-type specificity at the transcriptional level in human disease. Genome Res. 2013, 23, 1862–1873. [Google Scholar] [CrossRef] [Green Version]
- Nakagawa, S.; Nishihara, K.; Miyata, H.; Shinke, H.; Tomita, E.; Kajiwara, M.; Matsubara, T.; Iehara, N.; Igarashi, Y.; Yamada, H.; et al. Molecular Markers of Tubulointerstitial Fibrosis and Tubular Cell Damage in Patients with Chronic Kidney Disease. PLoS ONE 2015, 10, e0136994. [Google Scholar] [CrossRef] [Green Version]
- Neusser, M.A.; Lindenmeyer, M.T.; Moll, A.G.; Segerer, S.; Edenhofer, I.; Sen, K.; Stiehl, D.P.; Kretzler, M.; Gröne, H.J.; Schlöndorff, D.; et al. Human nephrosclerosis triggers a hypoxia-related glomerulopathy. Am. J. Pathol. 2010, 176, 594–607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reich, H.N.; Tritchler, D.; Cattran, D.C.; Herzenberg, A.M.; Eichinger, F.; Boucherot, A.; Henger, A.; Berthier, C.C.; Nair, V.; Cohen, C.D.; et al. A molecular signature of proteinuria in glomerulonephritis. PLoS ONE 2010, 5, e13451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmid, H.; Boucherot, A.; Yasuda, Y.; Henger, A.; Brunner, B.; Eichinger, F.; Nitsche, A.; Kiss, E.; Bleich, M.; Gröne, H.J.; et al. Modular activation of nuclear factor-κB transcriptional programs in human diabetic nephropathy. Diabetes 2006, 55, 2993–3003. [Google Scholar] [CrossRef] [Green Version]
- Woroniecka, K.I.; Park, A.S.D.; Mohtat, D.; Thomas, D.B.; Pullman, J.M.; Susztak, K. Transcriptome analysis of human diabetic kidney disease. Diabetes 2011, 60, 2354–2369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hodgin, J.B.; Borczuk, A.C.; Nasr, S.H.; Markowitz, G.S.; Nair, V.; Martini, S.; Eichinger, F.; Vining, C.; Berthier, C.C.; Kretzler, M.; et al. A Molecular Profile of Focal Segmental Glomerulosclerosis from Formalin-Fixed, Paraffin-Embedded Tissue. Am. J. Pathol. 2010, 177, 1674–1686. [Google Scholar] [CrossRef] [PubMed]
- Ju, W.; Nair, V.; Smith, S.; Zhu, L.; Shedden, K.; Song, P.X.K.; Mariani, L.H.; Eichinger, F.H.; Berthier, C.C.; Randolph, A.; et al. Tissue transcriptome-driven identification of epidermal growth factor as a chronic kidney disease biomarker. Sci. Transl. Med. 2015, 7, 316ra193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menon, R.; Otto, E.A.; Sealfon, R.; Nair, V.; Wong, A.K.; Theesfeld, C.L.; Chen, X.; Wang, Y.; Boppana, A.S.; Luo, J.; et al. SARS-CoV-2 receptor networks in diabetic and COVID-19-associated kidney disease. Kidney Int. 2020, 98, 1502–1518. [Google Scholar] [CrossRef]
- Menez, S.; Ju, W.; Menon, R.; Moledina, D.G.; Thiessen Philbrook, H.; McArthur, E.; Jia, Y.; Obeid, W.; Mansour, S.G.; Koyner, J.L.; et al. Urinary EGF and MCP-1 and risk of CKD after cardiac surgery. JCI Insight 2021, 6, e147464. [Google Scholar] [CrossRef]

| Baseline Characteristics | Cohort (N = 63) | Progressive (N = 26) | Stable (N = 37) |
|---|---|---|---|
| Age (years) | 47.2 ± 17.5 | 55.9 ± 14.4 | 41.1 ± 17.0 |
| eGFR (mL/min/1.73 m2) | 63.3 ± 35.2 | 50.1 ± 32.5 | 72.5 ± 34.5 |
| UPCR (g/g) | 4.4 ± 3.4 | 4.2 ± 2.7 | 4.5 ± 3.8 |
| Follow-up time (years) | 7.0 ± 4.3 | 5.2 ± 3.9 | 8.2 ± 4.2 |
| Female gender (n (%)) | 27 (43) | 7 (27) | 20 (54) |
| Baseline Characteristics | Cohort (N = 239) | FSGS (N = 67) | MCD (N = 55) | MN (N = 47) | Other Diseases (N = 70) |
|---|---|---|---|---|---|
| Age (years) | 32.8 ± 22.8 | 33.9 ± 22.3 | 20.6 ± 20.8 | 47.4 ± 18.3 | 31.6 ± 22 |
| eGFR (mL/min/1.73 m2) | 81.0 ± 37.8 | 75.6 ± 34.7 | 104.4 ± 40.7 | 79.1 ± 27.6 | 69.4 ± 37.7 |
| UPCR (g/g) | 4.5 ± 4.5 | 4.2 ± 5.2 | 3.8 ± 4.1 | 6.1 ± 4.0 | 4.0 ± 4.3 |
| Follow-up time (years) | 2.8 ± 1.5 | 2.8 ± 1.8 | 2.5 ± 1.9 | 2.2 ± 1.9 | 1.8 ± 1.4 |
| Female gender (n (%)) | 86 (36) | 21 (31) | 20 (36) | 19 (40) | 26 (37) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Denicolò, S.; Nair, V.; Leierer, J.; Rudnicki, M.; Kretzler, M.; Mayer, G.; Ju, W.; Perco, P. Assessment of Fibrinogen-like 2 (FGL2) in Human Chronic Kidney Disease through Transcriptomics Data Analysis. Biomolecules 2023, 13, 89. https://doi.org/10.3390/biom13010089
Denicolò S, Nair V, Leierer J, Rudnicki M, Kretzler M, Mayer G, Ju W, Perco P. Assessment of Fibrinogen-like 2 (FGL2) in Human Chronic Kidney Disease through Transcriptomics Data Analysis. Biomolecules. 2023; 13(1):89. https://doi.org/10.3390/biom13010089
Chicago/Turabian StyleDenicolò, Sara, Viji Nair, Johannes Leierer, Michael Rudnicki, Matthias Kretzler, Gert Mayer, Wenjun Ju, and Paul Perco. 2023. "Assessment of Fibrinogen-like 2 (FGL2) in Human Chronic Kidney Disease through Transcriptomics Data Analysis" Biomolecules 13, no. 1: 89. https://doi.org/10.3390/biom13010089





