The Regulatory Mechanism of miR-574-5p Expression in Cancer
Abstract
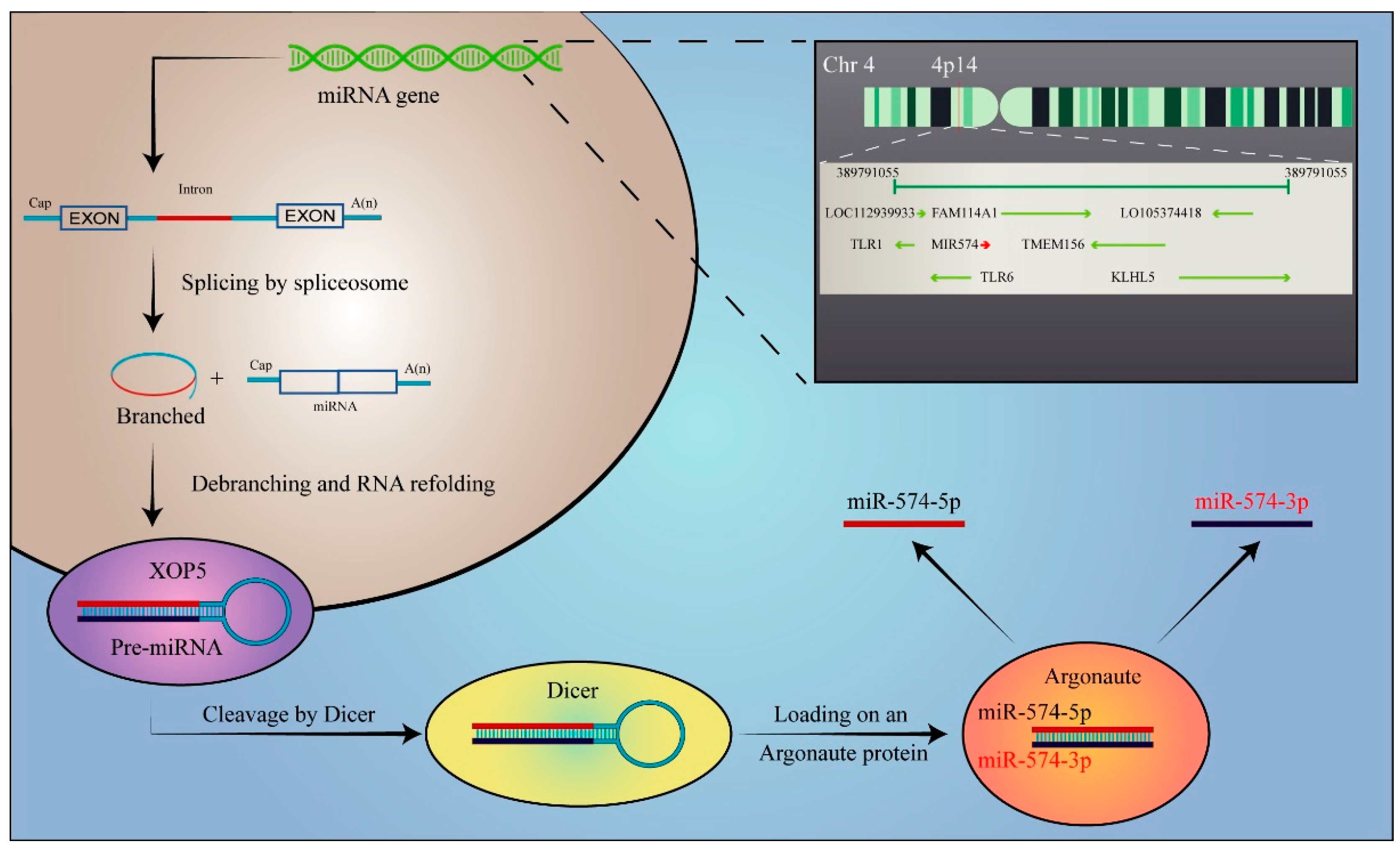
1. Introduction
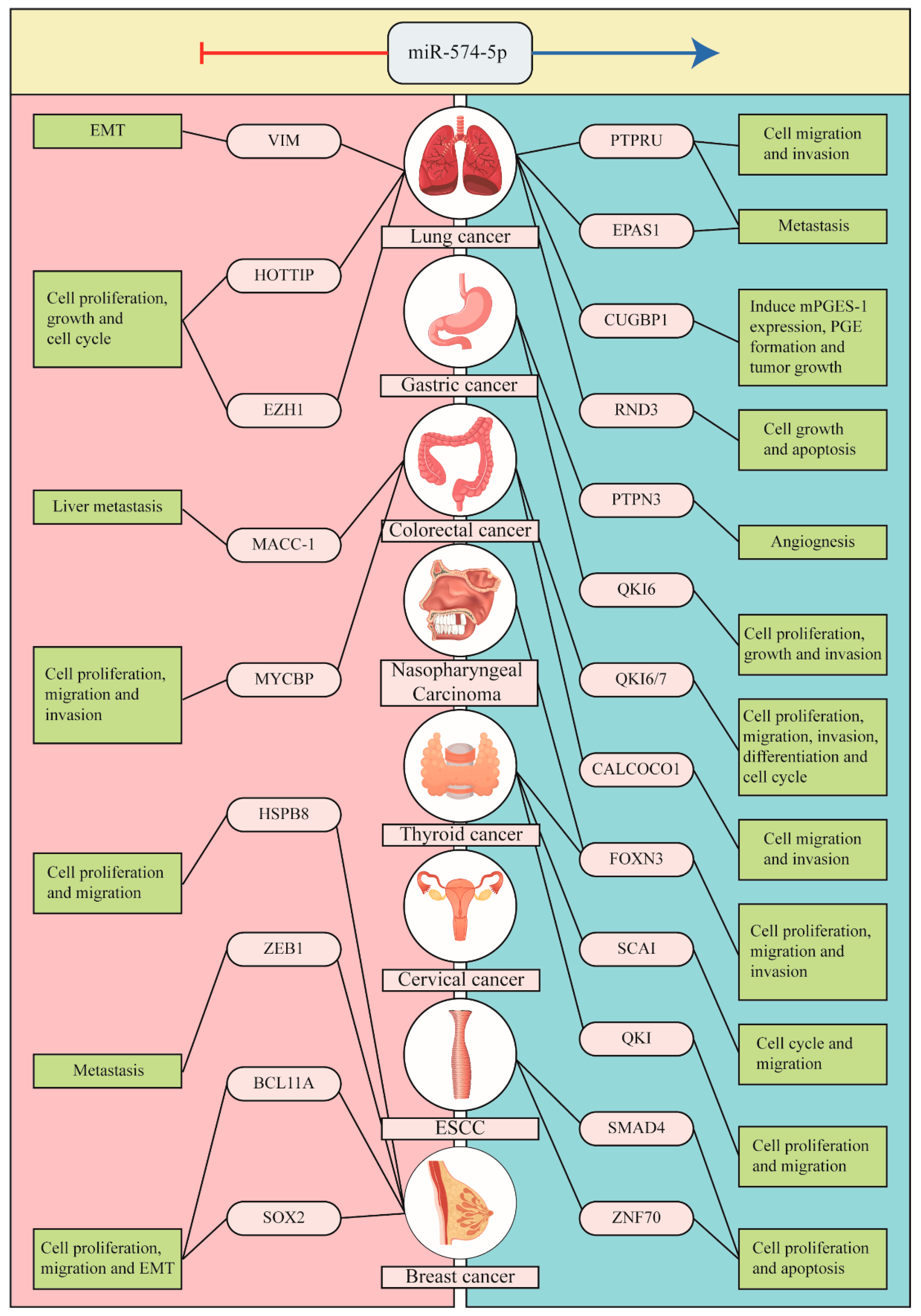
2. The Context-Dependent Role of miR-574-5p in Cancer
2.1. Lung Cancer
2.2. Thyroid Cancer
2.3. Breast Cancer (BC)
2.4. Gastric Cancer (GC)
2.5. Colorectal Cancer (CRC)
2.6. Others
3. The Functional Pathways and Regulatory Mechanisms of miR-574 in Cancer
3.1. Typical Functional Pathways of miR-574-5p in Cancer
3.1.1. Wnt/β-Catenin Pathway
3.1.2. MAPK Pathway
3.2. Other Novel Mechanisms of miR-574-5p Regulation in Tumorigenesis
3.2.1. Arm-Imbalance
3.2.2. Origin-Dependent Role of miR-574-5p
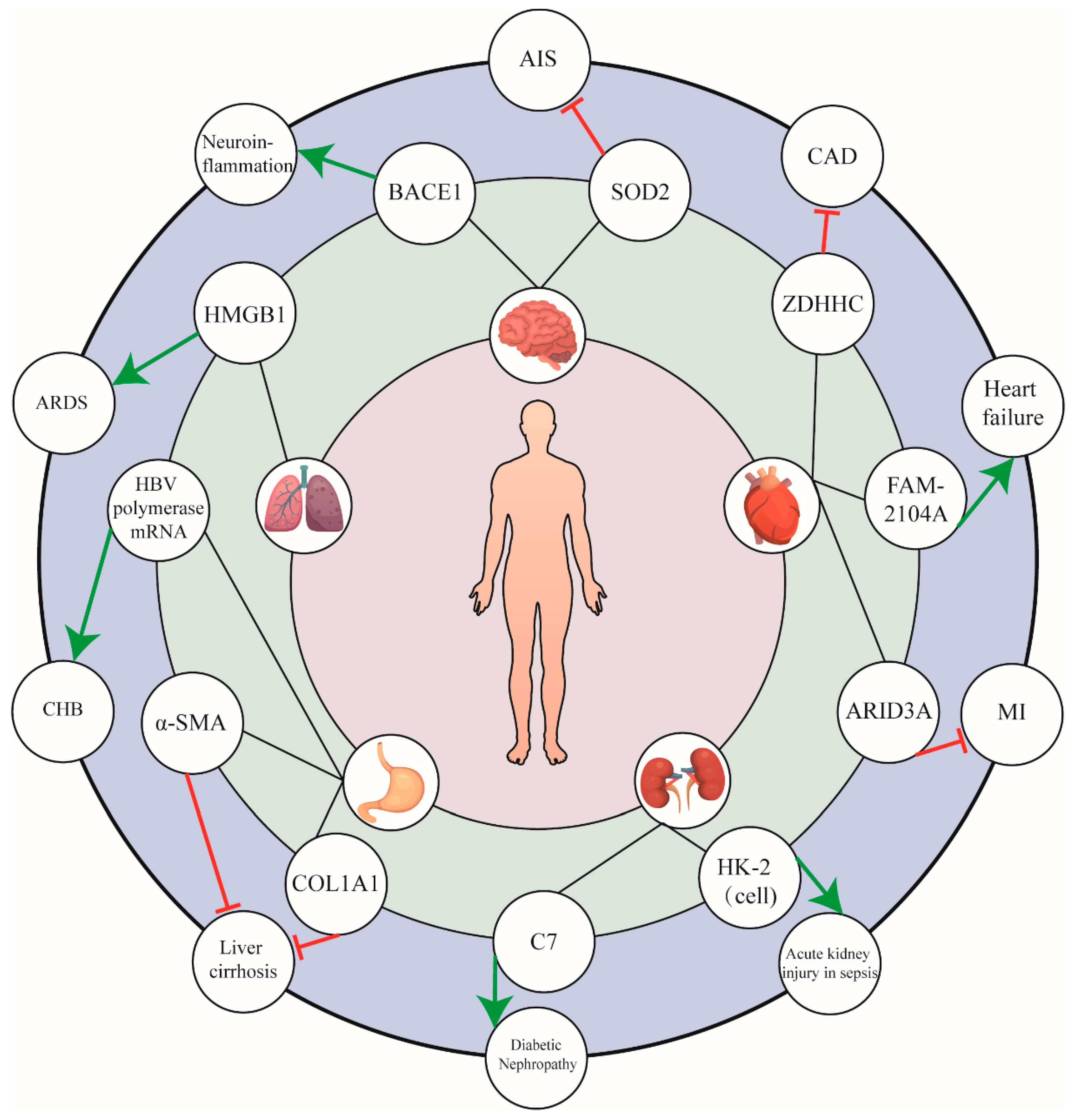
4. miR-574-5p in Other Diseases
5. Clinical Applications of miR-574-5p
5.1. Diagnostic Biomarker
5.2. Prognostic Biomarker
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Saliminejad, K.; Khorram Khorshid, H.; Soleymani Fard, S.; Ghaffari, S. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J. Cell. Physiol. 2019, 234, 5451–5465. [Google Scholar] [CrossRef]
- Lee, R.; Feinbaum, R.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Iaquinta, M.R.; Lanzillotti, C.; Mazziotta, C.; Bononi, I.; Frontini, F.; Mazzoni, E.; Oton-Gonzalez, L.; Rotondo, J.C.; Torreggiani, E.; Tognon, M.; et al. The role of microRNAs in the osteogenic and chondrogenic differentiation of mesenchymal stem cells and bone pathologies. Theranostics 2021, 11, 6573–6591. [Google Scholar] [CrossRef] [PubMed]
- Matsuyama, H.; Suzuki, H. Systems and Synthetic microRNA Biology: From Biogenesis to Disease Pathogenesis. Int. J. Mol. Sci. 2019, 21, 132. [Google Scholar] [CrossRef] [PubMed]
- Bautista-Sánchez, D.; Arriaga-Canon, C.; Pedroza-Torres, A.; De La Rosa-Velázquez, I.; González-Barrios, R.; Contreras-Espinosa, L.; Montiel-Manríquez, R.; Castro-Hernándezet, C.; Fragoso-Ontiveros, V.; Álvarez-Gómez, R.M.; et al. The Promising Role of miR-21 as a Cancer Biomarker and Its Importance in RNA-Based Therapeutics. Mol. Ther. Nucleic Acids 2020, 20, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, E.; Kim, S.Y.; Carmell, M.A.; Murchison, E.P.; Alcorn, H.; Li, M.Z.; Mills, A.A.; Elledge, S.J.; Anderson, K.V.; Hannon, G.J. Dicer is essential for mouse development. Nat. Genet. 2003, 35, 215–217. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Han, J.; Lee, Y.; Yeom, K.; Kim, Y.; Jin, H.; Kim, V. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef]
- Havens, M.A.; Reich, A.A.; Duelli, D.M.; Hastings, M.L. Biogenesis of mammalian microRNAs by a non-canonical processing pathway. Nucleic Acids Res. 2012, 40, 4626–4640. [Google Scholar] [CrossRef]
- Yi, R.; Qin, Y.; Macara, I.; Cullen, B. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003, 17, 3011–3016. [Google Scholar] [CrossRef] [PubMed]
- Medley, J.; Panzade, G.; Zinovyeva, A. microRNA strand selection: Unwinding the rules. Wiley Interdiscip. Rev. RNA 2021, 12, e1627. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Pi, J.; Zou, D.; Wang, X.; Xu, J.; Yu, S.; Zhang, T.; Li, F.; Zhang, X.; Zhao, H.; et al. microRNA arm-imbalance in part from complementary targets mediated decay promotes gastric cancer progression. Nat. Commun. 2019, 10, 4397. [Google Scholar] [CrossRef] [PubMed]
- Yao, P.; Wu, J.; Lindner, D.; Fox, P. Interplay between miR-574-3p and hnRNP L regulates VEGFA mRNA translation and tumorigenesis. Nucleic Acids Res. 2017, 45, 7950–7964. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Herbst, R.; Morgensztern, D.; Boshoff, C. The biology and management of non-small cell lung cancer. Nature 2018, 553, 446–454. [Google Scholar] [CrossRef]
- Stapelfeld, C.; Dammann, C.; Maser, E. Sex-specificity in lung cancer risk. Int. J. Cancer 2020, 146, 2376–2382. [Google Scholar] [CrossRef]
- Li, Q.; Li, X.; Guo, Z.; Xu, F.; Xia, J.; Liu, Z.; Ren, T. MicroRNA-574-5p was pivotal for TLR9 signaling enhanced tumor progression via down-regulating checkpoint suppressor 1 in human lung cancer. PloS ONE 2012, 7, e48278. [Google Scholar] [CrossRef]
- Zhou, R.; Zhou, X.; Yin, Z.; Guo, J.; Hu, T.; Jiang, S.; Liu, L.; Dong, X.; Zhang, S.; Wu, G. MicroRNA-574-5p promotes metastasis of non-small cell lung cancer by targeting PTPRU. Sci. Rep. 2016, 6, 35714. [Google Scholar] [CrossRef]
- Jiang, W.; He, Y.; Ma, Z.; Zhang, Y.; Zhang, C.; Zheng, N.; Tang, X. hsa_circ_0008234 inhibits the progression of lung adenocarcinoma by sponging miR-574-5p. Cell Death Discov. 2021, 7, 123. [Google Scholar] [CrossRef]
- Jakobsson, P.; Thorén, S.; Morgenstern, R.; Samuelsson, B. Identification of human prostaglandin E synthase: A microsomal, glutathione-dependent, inducible enzyme, constituting a potential novel drug target. Proc. Natl. Acad. Sci. USA 1999, 96, 7220–7225. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zhou, Y.; Bai, Y.; Wang, Q.; Bao, J.; Luo, Y.; Guo, Y.; Guo, L. A long non-coding RNA HOTTIP expression is associated with disease progression and predicts outcome in small cell lung cancer patients. Mol. Cancer 2017, 16, 162. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Yi, Y.; Gan, S.; Ye, R.; Huang, C.; Li, M.; Huang, J.; Guo, Y. miR-574-5p mediates epithelial-mesenchymal transition in small cell lung cancer by targeting vimentin via a competitive endogenous RNA network. Oncol. Lett. 2021, 21, 459. [Google Scholar] [CrossRef]
- Zhou, R.; Zhou, X.; Yin, Z.; Guo, J.; Hu, T.; Jiang, S.; Liu, L.; Dong, X.; Zhang, S.; Wu, G. Tumor invasion and metastasis regulated by microRNA-184 and microRNA-574-5p in small-cell lung cancer. Oncotarget 2015, 6, 44609–44622. [Google Scholar] [CrossRef]
- Nikiforov, Y. Thyroid carcinoma: Molecular pathways and therapeutic targets. Mod. Pathol. 2008, 2, S37–S43. [Google Scholar] [CrossRef]
- Molinaro, E.; Romei, C.; Biagini, A.; Sabini, E.; Agate, L.; Mazzeo, S.; Materazzi, G.; Sellari-Franceschini, S.; Ribechini, A.; Torregrossa, L.; et al. Anaplastic thyroid carcinoma: From clinicopathology to genetics and advanced therapies. Nat. Rev. Endocrinol. 2017, 13, 644–660. [Google Scholar] [CrossRef]
- Sugitani, I.; Fujimoto, Y.; Yamamoto, N. Papillary thyroid carcinoma with distant metastases: Survival predictors and the importance of local control. Surgery 2008, 143, 35–42. [Google Scholar] [CrossRef]
- Podnos, Y.; Smith, D.; Wagman, L.; Ellenhorn, J. The implication of lymph node metastasis on survival in patients with well-differentiated thyroid cancer. Am. Surg. 2005, 71, 731–734. [Google Scholar] [CrossRef]
- Pitsava, G.; Stratakis, C.; Faucz, F. PRKAR1A and Thyroid Tumors. Cancers 2021, 13, 3834. [Google Scholar] [CrossRef] [PubMed]
- Fan, M.; Li, X.; Jiang, W.; Huang, Y.; Li, J.; Wang, Z. A long non-coding RNA, PTCSC3, as a tumor suppressor and a target of miRNAs in thyroid cancer cells. Exp. Ther. Med. 2013, 5, 1143–1146. [Google Scholar] [CrossRef] [PubMed]
- Tong, R.; Zhang, J.; Wang, C.; Li, X.; Yu, T.; Wang, L. LncRNA PTCSC3 inhibits the proliferation, invasion and migration of cervical cancer cells via sponging miR-574-5p. Clin. Exp. Pharmacol. Physiol. 2020, 47, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, X.; Xiao, Q.; Wang, Z. MiR-574-5p mediates the cell cycle and apoptosis in thyroid cancer cells via Wnt/β-catenin signaling by repressing the expression of Quaking proteins. Oncol. Lett. 2018, 15, 5841–5848. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Chen, M.; Wan, Y.; Lei, L.; Ruan, H. miR-574-5p Targets FOXN3 to Regulate the Invasion of Nasopharyngeal Carcinoma Cells via Wnt/β-Catenin Pathway. Technol. Cancer Res. Treat. 2020, 19, 1533033820971659. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Wang, M.; Wu, H.; Xiao, M.; Liu, H.; Zhang, D. Loss of FOXN3 in colon cancer activates beta-catenin/TCF signaling and promotes the growth and migration of cancer cells. Oncotarget 2017, 8, 9783–9793. [Google Scholar] [CrossRef]
- Waks, A.G.; Winer, E.P. Breast Cancer Treatment: A Review. JAMA 2019, 321, 288–300. [Google Scholar] [CrossRef]
- Liedtke, C.; Mazouni, C.; Hess, K.R.; André, F.; Tordai, A.; Mejia, J.A.; Symmans, W.F.; Gonzalez-Angulo, A.M.; Hennessy, B.; Green, M.; et al. Response to Neoadjuvant Therapy and Long-Term Survival in Patients With Triple-Negative Breast Cancer. J. Clin. Oncol. 2008, 26, 1275–1281. [Google Scholar] [CrossRef]
- Foulkes, W.D.; Smith, I.E.; Reis-Filho, J.S. Triple-Negative Breast Cancer. N. Engl. J. Med. 2010, 363, 1938–1948. [Google Scholar] [CrossRef]
- Denkert, C.; Liedtke, C.; Tutt, A.; von Minckwitz, G. Molecular alterations in triple-negative breast cancer-the road to new treatment strategies. Lancet 2017, 389, 2430–2442. [Google Scholar] [CrossRef]
- Zhang, K.-J.; Hu, Y.; Luo, N.; Li, X.; Chen, F.-Y.; Yuan, J.-Q.; Guo, L. miR-574-5p attenuates proliferation, migration and EMT in triple-negative breast cancer cells by targeting BCL11A and SOX2 to inhibit the SKIL/TAZ/CTGF axis. Int. J. Oncol. 2020, 56, 1240–1251. [Google Scholar] [CrossRef]
- Wang, P.-S.; Chou, C.-H.; Lin, C.-H.; Yao, Y.-C.; Cheng, H.-C.; Li, H.-Y.; Chuang, Y.-C.; Yang, C.-N.; Ger, L.-P.; Chen, Y.-C.; et al. A novel long non-coding RNA linc-ZNF469-3 promotes lung metastasis through miR-574-5p-ZEB1 axis in triple negative breast cancer. Oncogene 2018, 37, 4662–4678. [Google Scholar] [CrossRef]
- Tseng, L.M.; Hsu, N.C.; Chen, S.C.; Lu, Y.S.; Lin, C.-H.; Chang, D.Y.; Li, H.; Lin, Y.C.; Chang, H.K.; Chao, T.C.; et al. Distant metastasis in triple-negative breast cancer. Neoplasma 2013, 60, 290–294. [Google Scholar] [CrossRef] [PubMed]
- Ajani, J.; D’Amico, T.; Bentrem, D.; Chao, J.; Cooke, D.; Corvera, C.; Das, P.; Enzinger, P.C.; Enzler, T.; Fanta, P.; et al. Gastric Cancer, Version 2.2022, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. JNCCN 2022, 20, 167–192. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhang, R.; Xu, R.; Shang, J.; He, H.; Yang, Q. MicroRNA-574-5p in gastric cancer cells promotes angiogenesis by targeting protein tyrosine phosphatase non-receptor type 3 (PTPN3). Gene 2020, 733, 144383. [Google Scholar] [CrossRef] [PubMed]
- Hisano, Y.; Hla, T. Bioactive lysolipids in cancer and angiogenesis. Pharmacol. Ther. 2019, 193, 91–98. [Google Scholar] [CrossRef] [PubMed]
- De Sanctis, F.; Ugel, S.; Facciponte, J.; Facciabene, A. The dark side of tumor-associated endothelial cells. Semin. Immunol. 2018, 35, 35–47. [Google Scholar] [CrossRef] [PubMed]
- Bruick, R.; McKnight, S. A conserved family of prolyl-4-hydroxylases that modify HIF. Science 2001, 294, 1337–1340. [Google Scholar] [CrossRef]
- Luo, Y.; Liang, M.; Yao, W.; Liu, J.; Niu, Q.; Chen, J.; Liu, Z.; Li, M.; Shi, B.; Pan, J.; et al. Functional role of lncRNA LOC101927497 in N-methyl-N’-nitro-N-nitrosoguanidine-induced malignantly transformed human gastric epithelial cells. Life Sci. 2018, 193, 93–103. [Google Scholar] [CrossRef]
- Liabeuf, D.; Oshima, M.; Stange, D.E.; Sigal, M. Stem Cells, Helicobacter pylori, and Mutational Landscape: Utility of Preclinical Models to Understand Carcinogenesis and to Direct Management of Gastric Cancer. Gastroenterology 2022, 162, 1067–1087. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Sauer, A.G.; Fedewa, S.A.; Butterly, L.F.; Anderson, J.C.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 145–164. [Google Scholar] [CrossRef]
- Kanth, P.; Inadomi, J.M. Screening and prevention of colorectal cancer. BMJ 2021, 374, n1855. [Google Scholar] [CrossRef]
- Ji, S.; Ye, G.; Zhang, J.; Wang, L.; Wang, T.; Wang, Z.; Zhang, T.; Wnag, G.; Guo, Z.; Luo, Y.; et al. miR-574-5p negatively regulates Qki6/7 to impact β-catenin/Wnt signalling and the development of colorectal cancer. Gut 2013, 62, 716–726. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Xiong, D.; Liu, Z.; Li, Y.; Chen, K.; Tang, H. Long noncoding RNA LINC00052 inhibits colorectal cancer metastasis by sponging microRNA-574-5p to modulate CALCOCO1 expression. J. Cell. Biochem. 2019, 120, 17258–17272. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Tang, J.; Chen, J.; Wang, Z. Hsa-miR-574-5p negatively regulates MACC-1 expression to suppress colorectal cancer liver metastasis. Cancer Cell Int. 2014, 14, 47. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Tan, F.; Pei, Q.; Zhou, Z.; Zhou, Y.; Zhang, L.; Wang, D.; Pei, H. Non-coding RNA MFI2-AS1 promotes colorectal cancer cell proliferation, migration and invasion through miR-574-5p/MYCBP axis. Cell Prolif. 2019, 52, e12632. [Google Scholar] [CrossRef]
- Han, G.L.; Wang, J.; Guo, K.; Chen, J.K.; Shang, R.X.; Jiang, T. miRNA-574-5p downregulates ZNF70 and influences the progression of human esophageal squamous cell carcinoma through reactive oxygen species generation and MAPK pathway activation. Anticancer Drugs 2020, 31, 282–291. [Google Scholar] [CrossRef]
- Yang, M.; Liu, R.; Sheng, J.; Liao, J.; Wang, Y.; Pan, E.; Guo, W.; Pu, Y.; Yin, L. Differential expression profiles of microRNAs as potential biomarkers for the early diagnosis of esophageal squamous cell carcinoma. Oncol. Rep. 2013, 29, 169–176. [Google Scholar] [CrossRef]
- Li, J.; Han, X.; Gu, Y.; Wu, J.; Song, J.; Shi, Z.; Chang, H.; Liu, M.; Zhang, Y. LncRNA MTX2-6 Suppresses Cell Proliferation by Acting as ceRNA of miR-574-5p to Accumulate SMAD4 in Esophageal Squamous Cell Carcinoma. Front. Cell Dev. Biol. 2021, 9, 654746. [Google Scholar] [CrossRef]
- Widmann, C.; Gibson, S.; Jarpe, M.B.; Johnson, G.L. Mitogen-activated protein kinase: Conservation of a three-kinase module from yeast to human. Physiol. Rev. 1999, 79, 143–180. [Google Scholar] [CrossRef]
- Cargnello, M.; Roux, P.P. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol. Mol. Biol. Rev. 2011, 75, 50–83. [Google Scholar] [CrossRef]
- Hou, S.; Suresh, P.S.; Qi, X.; Lepp, A.; Mirza, S.P.; Chen, G. p38γ Mitogen-activated protein kinase signals through phosphorylating its phosphatase PTPH1 in regulating ras protein oncogenesis and stress response. J. Biol. Chem. 2012, 287, 27895–27905. [Google Scholar] [CrossRef]
- Takeda, K.; Matsuzawa, A.; Nishitoh, H.; Ichijo, H. Roles of MAPKKK ASK1 in stress-induced cell death. Cell Struct. Funct. 2003, 28, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Matsuzawa, A.; Ichijo, H. Redox control of cell fate by MAP kinase: Physiological roles of ASK1-MAP kinase pathway in stress signaling. Biochim. Biophys. Acta 2008, 1780, 1325–1336. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Sheng, P.; Li, T.; Fields, C.J.; Hiers, N.M.; Wang, Y.; Li, J.; Guardia, C.M.; Licht, J.D.; Xie, M. Widespread microRNA degradation elements in target mRNAs can assist the encoded proteins. Genes Dev. 2021, 35, 1595–1609. [Google Scholar] [CrossRef] [PubMed]
- Ameres, S.L.; Horwich, M.D.; Hung, J.-H.; Xu, J.; Ghildiyal, M.; Weng, Z.; Zamore, P.D. Target RNA-directed trimming and tailing of small silencing RNAs. Science 2010, 328, 1534–1539. [Google Scholar] [CrossRef]
- Saul, M.J.; Baumann, I.; Bruno, A.; Emmerich, A.C.; Wellstein, J.; Ottinger, S.M.; Contursi, A.; Dovizio, M.; Donnini, S.; Tacconelli, S.; et al. miR-574-5p as RNA decoy for CUGBP1 stimulates human lung tumor growth by mPGES-1 induction. FASEB J. 2019, 33, 6933–6947. [Google Scholar] [CrossRef] [PubMed]
- Emmerich, A.C.; Wellstein, J.; Ossipova, E.; Baumann, I.; Lengqvist, J.; Kultima, K.; Jakobsson, P.-J.; Steinhilber, D.; Saul, M.J. Proteomics-Based Characterization of miR-574-5p Decoy to CUGBP1 Suggests Specificity for mPGES-1 Regulation in Human Lung Cancer Cells. Front. Pharmacol. 2020, 11, 196. [Google Scholar] [CrossRef]
- Donzelli, J.; Proestler, E.; Riedel, A.; Nevermann, S.; Hertel, B.; Guenther, A.; Gattenlöhner, S.; Savai, R.; Larsson, K.; Saul, M.J. Small extracellular vesicle-derived miR-574-5p regulates PGE2-biosynthesis via TLR7/8 in lung cancer. J. Extracell. Vesicles 2021, 10, e12143. [Google Scholar] [CrossRef]
- Lai, Z.; Lin, P.; Weng, X.; Su, J.; Chen, Y.; He, Y.; Wu, G.; Wang, J.; Yu, Y.; Zhang, L. MicroRNA-574-5p promotes cell growth of vascular smooth muscle cells in the progression of coronary artery disease. Biomed. Pharmacother. 2018, 97, 162–167. [Google Scholar] [CrossRef]
- Cui, J.; Qi, S.; Liao, R.; Su, D.; Wang, Y.; Xue, S. MiR-574-5p promotes the differentiation of human cardiac fibroblasts via regulating ARID3A. Biochem. Biophys. Res. Commun. 2020, 521, 427–433. [Google Scholar] [CrossRef]
- Wu, J.; Subbaiah, K.C.V.; Jiang, F.; Hedaya, O.; Mohan, A.; Yang, T.; Welle, K.; Ghaemmaghami, S.; Tang, W.H.W.; Small, E.; et al. MicroRNA-574 regulates FAM210A expression and influences pathological cardiac remodeling. EMBO Mol. Med. 2021, 13, e12710. [Google Scholar] [CrossRef]
- Couvillion, M.T.; Soto, I.C.; Shipkovenska, G.; Churchman, L.S. Synchronized mitochondrial and cytosolic translation programs. Nature 2016, 533, 499–503. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Zhou, W.; Rui, Y.; Liu, L.; Chen, B.; Su, X. MicroRNA-574-5p Attenuates Acute Respiratory Distress Syndrome by Targeting HMGB1. Am. J. Respir. Cell Mol. Biol. 2021, 64, 196–207. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Yan, Z.; Hu, Y.; Huang, X.; Pan, C. Complement C7 is Specifically Expressed in Mesangial Cells and is a Potential Diagnostic Biomarker for Diabetic Nephropathy and is Regulated by miR-494-3p and miR-574-5p. Diabetes Metab. Syndr. Obes. 2021, 14, 3077–3088. [Google Scholar] [CrossRef]
- Zhou, X.; Liang, Z.; Qin, S.; Ruan, X.; Jiang, H. Serum-derived miR-574-5p-containing exosomes contribute to liver fibrosis by activating hepatic stellate cells. Mol. Biol. Rep. 2022, 49, 1945–1954. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Wu, D.; Yan, W.; Wang, Y.; You, J.; Wan, X.; Xi, D.; Luo, X.; Han, M.; Ning, Q. Interferon-Induced Macrophage-Derived Exosomes Mediate Antiviral Activity Against Hepatitis B Virus Through miR-574-5p. J. Infect. Dis. 2021, 223, 686–698. [Google Scholar] [CrossRef] [PubMed]
- Ku, T.; Li, B.; Gao, R.; Zhang, Y.; Yan, W.; Ji, X.; Li, G.; Sang, N. NF-κB-regulated microRNA-574-5p underlies synaptic and cognitive impairment in response to atmospheric PM(2.5) aspiration. Part. Fibre Toxicol. 2017, 14, 34. [Google Scholar] [CrossRef] [PubMed]
- Mol, B.W.J.; Roberts, C.T.; Thangaratinam, S.; Magee, L.A.; de Groot, C.J.M.; Hofmeyr, G.J. Pre-eclampsia. Lancet 2016, 387, 999–1011. [Google Scholar] [CrossRef]
- Xu, Y.; Xia, X.; Jiang, Y.; Wu, D.; Wang, S.; Fu, S.; Yang, N.; Zhang, Y.; Sun, L. Down-regulated lncRNA AGAP2-AS1 contributes to pre-eclampsia as a competing endogenous RNA for JDP2 by impairing trophoblastic phenotype. J. Cell. Mol. Med. 2020, 24, 4557–4568. [Google Scholar] [CrossRef]
- Davarinejad, O.; Najafi, S.; Zhaleh, H.; Golmohammadi, F.; Radmehr, F.; Alikhani, M.; Moghadam, R.H.; Rahmati, Y. MiR-574-5P, miR-1827, and miR-4429 as Potential Biomarkers for Schizophrenia. J. Mol. Neurosci. 2022, 72, 226–238. [Google Scholar] [CrossRef]
- Fan, X.; Chen, G.; Ma, F.; Qi, B.; Liang, Y.; Gopng, P.; Meng, C. An lncRNA-miRNA-mRNA-ceRNA network regulates intervertebral disc degeneration: A bioinformatics study based on the dataset analysis. Gen. Physiol. Biophys. 2021, 40, 317–327. [Google Scholar] [CrossRef]
- Wang, F.; Li, Z.; Zhao, M.; Ye, W.; Wu, H.; Liao, Q.; Bu, S.; Zhang, Y. Circulating miRNAs miR-574-5p and miR-3135b are potential metabolic regulators for serum lipids and blood glucose in gestational diabetes mellitus. Gynecol. Endocrinol. 2021, 37, 665–671. [Google Scholar] [CrossRef] [PubMed]
- Hegewald, A.B.; Breitwieser, K.; Ottinger, S.M.; Mobarrez, F.; Korotkova, M.; Rethi, B.; Jakobsson, P.-J.; Catrhina, A.I.; Wähämaa, H.; Saul, M.J. Extracellular miR-574-5p Induces Osteoclast Differentiation via TLR 7/8 in Rheumatoid Arthritis. Front. Immunol. 2020, 11, 585282. [Google Scholar] [CrossRef] [PubMed]
- Berindan-Neagoe, I.; Monroig Pdel, C.; Pasculli, B.; Calin, G.A. MicroRNAome genome: A treasure for cancer diagnosis and therapy. CA Cancer J. Clin. 2014, 64, 311–336. [Google Scholar] [CrossRef] [PubMed]
- Cao, E.A.Z.; Qiu, J.; Yang, G.; Liu, Y.; Luo, W.; You, L.; Zheng, L.; Zhang, T. MiR-135a biogenesis and regulation in malignancy: A new hope for cancer research and therapy. Cancer Biol. Med. 2020, 17, 569–582. [Google Scholar] [CrossRef]
- Swensen, S.J.; Jett, J.R.; Hartman, T.E.; Midthun, D.E.; Mandrekar, S.J.; Hillman, S.L.; Sykes, A.-M.; Aughenbaugh, G.L.; Bungum, A.O.; Allen, K.L. CT screening for lung cancer: Five-year prospective experience. Radiology 2005, 235, 259–265. [Google Scholar] [CrossRef]
- Han, Z.; Li, Y.; Zhang, J.; Guo, C.; Li, Q.; Zhang, X.; Lan, Y.; Gu, W.; Xing, Z.; Liang, L.; et al. Tumor-derived circulating exosomal miR-342-5p and miR-574-5p as promising diagnostic biomarkers for early-stage Lung Adenocarcinoma. Int. J. Med. Sci. 2020, 17, 1428–1438. [Google Scholar] [CrossRef]
- Peng, H.; Wang, J.; Li, J.; Zhao, M.; Huang, S.-K.; Gu, Y.-Y.; Li, Y.; Sun, X.-J.; Yang, L.; Luo, Q.; et al. A circulating non-coding RNA panel as an early detection predictor of non-small cell lung cancer. Life Sci. 2016, 151, 235–242. [Google Scholar] [CrossRef]
- Foss, K.M.; Sima, C.; Ugolini, D.; Neri, M.; Allen, K.E.; Weiss, G.J. miR-1254 and miR-574-5p: Serum-based microRNA biomarkers for early-stage non-small cell lung cancer. J. Thorac. Oncol. 2011, 6, 482–488. [Google Scholar] [CrossRef]
- Zhou, J.; Shao, G.; Chen, X.; Yang, X.; Huang, X.; Peng, P.; Ba, Y.; Zhang, L.; Jehangir, T.; Bu, S.; et al. miRNA 206 and miRNA 574-5p are highly expression in coronary artery disease. Biosci. Rep. 2015, 36, e00295. [Google Scholar] [CrossRef]
- Boileau, A.; Cardenas, C.L.L.; Courtois, A.; Zhang, L.; Rodosthenous, R.S.; Das, S.; Sakalihasan, N.; Michel, J.-B.; Lindsay, M.E.; Devaux, Y. MiR-574-5p: A Circulating Marker of Thoracic Aortic Aneurysm. Int. J. Mol. Sci. 2019, 20, 3924. [Google Scholar] [CrossRef]
- Zheng, D.; Ding, Y.; Ma, Q.; Zhao, L.; Guo, X.; Shen, Y.; He, Y.; Wei, W.; Liu, F. Identification of Serum MicroRNAs as Novel Biomarkers in Esophageal Squamous Cell Carcinoma Using Feature Selection Algorithms. Front. Oncol. 2018, 8, 674. [Google Scholar] [CrossRef]
- Huang, S.-K.; Luo, Q.; Peng, H.; Li, J.; Zhao, M.; Wang, J.; Gu, Y.-Y.; Li, Y.; Yuan, P.; Zhao, G.-H.; et al. A Panel of Serum Noncoding RNAs for the Diagnosis and Monitoring of Response to Therapy in Patients with Breast Cancer. Med. Sci. Monit. 2018, 24, 2476–2488. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Tiwari, A.; Mirzakhani, H.; Wang, A.; Kho, A.; McGeachie, M.; Litonjua, A.; Weiss, S.; Tantisira, K. Circulating MicroRNA: Incident Asthma Prediction and Vitamin D Effect Modification. J. Pers. Med. 2021, 11, 307. [Google Scholar] [CrossRef] [PubMed]
- Boileau, A.; Somoza, A.S.; Dankiewicz, J.; Stammet, P.; Gilje, P.; Erlinge, D.; Hassager, C.; Wise, M.P.; Kuiper, M.; Friberg, H.; et al. Circulating Levels of miR-574-5p Are Associated with Neurological Outcome after Cardiac Arrest in Women: A Target Temperature Management (TTM) Trial Substudy. Dis. Markers 2019, 2019, 1802879. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zhao, L.; Zhang, L.; Qiao, L.; Gao, S. Downregulation of miR-574-5p inhibits HK-2 cell viability and predicts the onset of acute kidney injury in sepsis patients. Ren. Fail. 2021, 43, 942–948. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Meng, K.; Chen, W.; Feng, D.; Jia, Y.; Xie, L. Serum miR-574-5p: A prognostic predictor of sepsis patients. Shock 2012, 37, 263–267. [Google Scholar] [CrossRef]
| Disease | Pathology | Author | Target | Process | Function | Expression of miR | Impact | Pathways |
|---|---|---|---|---|---|---|---|---|
| Lung cancer | small-cell lung cancer (SCLC) | Zhou Rui 2015 | PTPRU, EPAS1 | metastasis | miR-574-5p significantly enhances the metastasis of SCLC and participates in β-catenin signaling by Suppressing protein tyrosine phosphatase receptor type U (PTPRU) or endothelial PAS domain protein 1 (EPAS1). | ↑ | ↓ | Wnt/β-catenin |
| non-small cell lung cancer (NSCLC) | Saul et al., 2019 | CUGBP1 | induce mPGES-1 expression, PGE formation and tumor growth | miR-574-5p induces microsomal prostaglandin E synthase-1 (mPGES-1) expression by preventing CUGBP1 binding to its 3’UTR, leading to enhanced alternative splicing and generation of an mPGES-1 3’UTR isoform, increased mPGES-1 protein expression, and PGE formation | ↑ | ↓ | NA | |
| Emmerich 2020 | CUGBP1 | induce mPGES-1 expression, PGE formation and tumor growth | miR-574-5p as RNA decoy for CUGBP1 stimulates human lung tumor growth by mPGES-1 induction. | ↑ | ↓ | NA | ||
| Zhou Rui 2016 | PTPRU | cell migration and invasion | miR-574-5p enhances the tyrosine phosphorylation of β-catenin by repressing PTPRU expression and promotes the migration and invasion of NSCLC cells. | ↑ | ↓ | Wnt/β-catenin | ||
| Hua Peng 2016 | none | diagnostic biomarker | miR-574-5p can serve as a convenient tool for early NSCLC diagnosis. | ↓ | NA | NA | ||
| Kristen M. 2011 | none | diagnostic biomarker | hsa-miR-574-5p is significantly increased in early-stage NSCLC samples. | ↑ | NA | NA | ||
| lung adenocarcinoma | Zhijun Han 2020 | none | diagnostic biomarker | Circulating exosomal miR-574-5p has potential to serve as novel diagnostic biomarkers for early-stage LA. | ↑ | NA | NA | |
| Gastric cancer | not-mentioned | Zhengyi Zhang 2019 | QKI6 | cell proliferation, growth and invasion | The increase in miR-574-5p/-3p ratio, named miR-574 arm-imbalance, is partially due to the dynamic expression of their highly complementary targets in gastric carcinogenesis; the arm-imbalance of miR-574 is in turn involved and further promotes gastric cancer progression. | ↑ | ↓ | NA |
| not-mentioned | Shu Zhang, 2020 | PTPN3 | angiogenesis | miR-574-5p in gastric cancer cells promotes angiogenesis via enhancing phosphorylation of p44/42 MAPKs by miR-574-5p inhibition of PTPN3 expression. | ↑ | ↓ | MAPKs | |
| not-mentioned | Jianming Li 2014 | none | maintaining the stemness of the gastric CSCs | miR-574-5p shows decreased expression pattern in gastric CSCs | ↓ | ↑ | NA | |
| Colorectal cancer | not-mentioned | Shunlong Ji 2012 | Qki6/7 | cell proliferation, migration, invasion, differentiation and cell cycle | Upregulation of miR-574-5p decreases the expression of Qk6/7 to inhibit b-catenin mRNA and protein expression and inhibit B-catenin /Wnt signal transduction in CRC cells | ↑ | ↓ | Wnt/β-catenin |
| colorectal cancer liver metastasis | Zhe Cui, 2014 | MACC-1 | liver metastasis | hsa-miR-574-5p plays a suppressive role in colorectal cancer liver metastasis by negatively directing MACC-1 expression | ↓ | ↑ | NA | |
| Esophageal squamous cell carcinoma | not-mentioned | Deqiang Zheng 2019 | none | diagnostic and prognostic biomarker | miR574 can distinguish ESCC patients from healthy controls. Moreover, the classifying performance of the miRNA panel can discriminate healthy controls from patients with ESCC stage I-II (AUC > 0.76) and patients with ESCC stage III-IV (AUC > 0.80). | ↑ | ↓ | NA |
| not-mentioned | MIAO YANG 2013 | none | diagnostic biomarker | hsa-miR-574-5p is upregulated in tumor tissues; multiple regression analysis revealed the aberrant expression of hsa-miR-574-5p increases the risk of esophageal cancer. | ↑ | ↓ | NA | |
| not-mentioned | Jie Li 2021 | SMAD4 | cell proliferation and apoptosis | miR-574-5p represses the abundance of ESCC cells, which leads to booming cell growth of ESCC. | ↑ | ↓ | NA | |
| not-mentioned | Guo Liang Han 2020 | ZNF70 | cell proliferation and apoptosis | miR-574-5p serve as a tumor promoter regulating cells proliferation and apoptosis in ESCC through and MAPK pathways. Furthermore, ZNF70 has been proven to be a functional target for miR-574-5p to regulate cells proliferation and apoptosis | ↑ | ↓ | MAPKs | |
| Breast cancer | triple-negative breast cancer | KE-JING ZHANG 2020 | BCL11A, SOX2 | cell proliferation, migration and EMT | miR-574-5p attenuates proliferation, migration, and EMT in triple-negative breast cancer cells by targeting BCL11A and SOX2 to inhibit the SKIL/TAZ/CTGF axis. | ↓ | ↑ | NA |
| not-mentioned | Sheng-kai Huang 2018 | none | diagnostic and prognostic biomarker | miR-574-5p might represent a serum biomarker panel with potential in the diagnosis of breast cancer. | ↑ | NA | ||
| estrogen receptor (ER) positive breast cancer | Margherita Piccolella 2021 | HSPB8 | cell proliferation and migration | Downregulation of miR-574-5p, which binds to HSPB8 ORF, causes increased expression of HSPB8 protein and the proliferation and migration of ER+BC MCF-7 cells. | ↓ | ↑ | NA | |
| Thyroid cancer | not-mentioned | Zhejia Zhang 2018 | QKI | cell cycle and migration | miR-574-5p mediates the cell cycle and apoptosis in thyroid cancer cells via Wnt/β-catenin signaling by repressing the expression of Quaking proteins. | ↑ | ↓ | Wnt/β-catenin |
| not-mentioned | Zhe-JiaZhang 2020 | FOXN3 | cell migration, proliferation, invasion and apoptosis | miR-574-5p directly targets FOXN3 in thyroid cancer cells, which activates Wnt/β-catenin singling pathway and promotes cell migration, proliferation, invasion and apoptosis | ↑ | ↓(pathway activated) | Wnt/β-catenin | |
| papillary thyroid carcinoma (PTC) | Xiaoming Wang 2017 | SCAI | cell proliferation and migration | PTCSC3 absorbs miR-574-5p, and miR-574-5p targets SCAI; SCAI can regulate the activity of Wnt/β-catenin. PTCSC3/miR-574-5p regulates the activity of Wnt/β-catenin via SCAI and mediates cell proliferation and migration of PTC-1. | ↑ | ↓ | Wnt/β-catenin and non-coding RNA | |
| Chordoma | not-mentioned | Emre Can Tuysuza 2019 | MYCBP | cell viability, apoptosis, migration and invasion | miR-574-5p targets MYCBP and inhibits the invasive and migratory phenotype; miR-574-5p decreases viability and increases apoptosis in U-CH1 cells while it has no effect on both viability and apoptosis in MUG-Chor1 cells. | ↓ | ↑ | NA |
| Head and neck squamous cell carcinoma | chemotherapy-induced cell death and nodal metastasis | Marisa Meyers-Needham 2011 | CerS1-2 | cell proliferation and growth | Inhibition of HDAC1 and siRNA-mediated knockdown of miR-574-5p reconstitutes CerS1-2 expression and C18-ceramide generation, which subsequently inhibits cancer cell proliferation. | ↑ | ↓ | NA |
| Disease Type | Author | Target | Disease | Function | Expression of miR in Disease | Impact | Therapeutic Significance |
|---|---|---|---|---|---|---|---|
| Cardiovascular diseases | Jianqing Zhou 2016 | not mentioned | Coronary artery disease (CAD) | miR-574-5p is significantly upregulated in CAD patients, which indicates that it has great potential to provide sensitive and specific diagnostic value for CAD. | ↑ | NA | biomarker |
| Lai Zhongmeng 2018 | ZDHHC | CAD | Upregulation of miR-574-5p enhances cell proliferation and suppresses apoptotic processes in VSMCs through targeting ZDHHC14, suggesting that miR-574-5p is a CAD-related factor that may serve as a potential molecular target for CAD treatment. | ↑ | ↓ | potential target | |
| Wu Jiangbin2021 | FAM2104A | Heart failure | miR-574 targets FAM210A and modulates mitochondrial-encoded protein expression, which may contribute to cardiac remodeling in heart failure. | ↑ | ↑ | protective role | |
| CuJun 2020 | ARID3A | Cardiac fibrosis after myocardial infarction (MI) | miR-574-5p directly targets ARID3A to promote fibroblast-to-myofibroblast differentiation of TGF-β-induced human cardiac fibroblasts (HCFs) | ↑ | ↓ | potential target | |
| Adeline Boileau 2019 | not mentioned | Cardiac arrest | miR-574-5p is associated with neurological outcome after cardiac arrest in women. | ↑ | NA | biomarker | |
| Boileau 2019 | not mentioned | Thoracic aortic aneurysm (TAA) | miR-574-5p may act as a paracrine mediator in TAA pathogenesis and could be secreted in response to Ang II (further research needed) | ↑ | biomarker | ||
| Respiratory diseases | Binchan He 2020 | HMGB1 | Acute respiratory distress syndrome (ARDS) | miR-574 targets HMGB1 to inhibit the inflammatory response via suppression of TLR4/NF-Κb signaling pathway and the NLRP3 inflammasome | ↑ | ↑ | protective role |
| LiJiang 2021 | not mentioned | Asthma | hsa-miR-574-5p is a potential mediator and biomarker of asthma and has excellent prognostic power. | ↑ | NA | biomarker | |
| LI 2017 | not mentioned | Obstructive sleep apnea | miR-574-5p is significantly upregulated in OSA | ↑ | NA | NA | |
| Digestive system disease | Wu Wenyu 2021 | HBV polymerase mRNA | Chronic hepatitis B (CHB) | hsa-miR574-5p downregulates the expression of HBV polymerase mRNA and pgRNA by direct binding to nucleotides 2750-2757 | ↑ | ↑ | protective role; potential target |
| Tan Youwen 2015 | not mentioned | Chronic hepatitis B | miR-574-5p could be a surrogate marker for chronic hepatitis B with persistently normal alanine aminotransferase (ALT) | ↑ | NA | biomarker? | |
| ZhouXia 2021 | α-SMA, COL1A1 | Liver cirrhosis | miR-574-5p in serum exosomes transfers to HSC to activate HSC, which is relevant to liver cirrhosis. | ↑ | ↓ | potential target | |
| Neurological disorders | Tingting Ku 2017 | BACE1 | Neuroinflammation | miR-574-5p directly binds to the 3’ UTR of BACE1 to reduce its role of impairing functional synaptic integrity and spatial learning and memory. Overexpression of miR-574-5p in the hippocampal region decreases BACE1 expression, restores synaptic function, and improves spatial memory and learning following PM2.5 exposure. | ↓ | ↑ | protective role |
| Freischmidt 2013 | not mentioned | Amyotrophic lateral sclerosis (ALS) | TDP-43 binding serum miRNA levels are candidates for an easily accessible biological measure of TDP-43 dysfunction in ALS | ↓ | NA | NA | |
| Xiaobo Yang 2021 | SOD2 | Acute ischemic stroke (AIS) | circPHKA2 protects HBMEC from OGD-induced neurovascular injuries by controlling SOD2 via sponging miR-574-5p. | ↑ | ↓ | NA | |
| Urinary system diseases | GuoHang 2021 | C7 | Diabetic Nephropathy | miR-574-5p can negatively regulate expression of C7. Elevated C7 gene expression level in MES is regulated by miR-494-3p and miR-574-5p in early diabetic nephropathy | ↓ | ↑ | NA |
| Shanshan Liu 2021 | HK-2 (cell) | Acute kidney injury in sepsis | Downregulation of miR-574-5p inhibits HK-2 cell viability and predicts the onset of acute kidney injury in sepsis patients. | ↓ | ↑ | biomarker | |
| Others | Lip 2020 | MKI67 | Preeclampsia | miR-574-5p and miR-1972 decrease the proliferation (probably via decreasing MKI67) and/or migration as well as the tube-formation capacity of endothelial cells | ↑ | ↓ | potential target |
| XuYetao 2020 | JDP2 | Preeclamptic | miR-574 can directly bind to both AGAP2 AS1 and JDP2, and AGAP2 AS1 regulates JDP2 expression via the AGAP2 AS1/miR 574 axis in trophoblasts | ↑ | ↓ | potential target | |
| Belarbi 2018 | EBF1 | Obesity/diabetes | miR-574-5p is associated with human adipose morphology and regulates EBF1 expression in white adipose tissue | ↑ | ↓ | NA | |
| WangFuyan 2021 | Some predicted targets are associated with the metabolism of glucose and lipids and the insulin signaling pathway. | Gestational diabetes mellitus (GDM) | The expression of miR-574-5p is significantly correlated with levels of blood glucose and LDL-C, which indicates that miR-574-5p may serve as a metabolic regulator of glucose and lipids for GDM. | ↓ | NA | biomarker | |
| Davarinejad 2021 | not mentioned | Schizophrenia | hsa-miR-574-5P is suggested as a potential biomarker for diagnosis of schizophrenia. | ↑ | NA | biomarker | |
| Hegewald 2020 | TLR 7/8 | Rheumatoid Arthritis | Extracellular miR-574-5p induces osteoclast differentiation via TLR 7/8 in rheumatoid arthritis. | ↑ | ↓ | potential target | |
| Xutao Fan 2021 | P3H2 | Ontervertebral disc degeneration (IDD) | has-miR-574-5p can suppress the expression on P3H2 in IDD progression | ↓ | ↑ | NA | |
| Wang, Huijuan 2012 | not mentioned | Sepsis | serum miR-574-5p is correlated with the death of sepsis patient | ↓ in nonsurvivors | NA | biomarker | |
| Sabetian 2021 | ACE2 | Male infertility in COVID-19 | Downregulation of miR-574-5p may lead to an increase in ACE2 expression related to male infertility in COVID-19 | ↓ | ↑ | NA |
| Catagory | Author | Disease | Origin | Sample | Tendency | Diagnosis | Prognosis | Combination |
|---|---|---|---|---|---|---|---|---|
| cancer | Zhijun Han 2020 | lung adenocarcinoma | serum | 7 early-stage lung adenocarcinoma patients including pre-operation and post-operation vs. 7 heathy controls | ↑ | Yes | Yes | with miR-342-5p |
| Zhou Rui 2015 | small-cell lung cancer (SCLC) | serum and tissue | a set of 72 SCLC patients (22 limited disease vs. 50 extensive disease) | ↑ | NA | Yes | none | |
| Hua Peng 2016 | non-small cell lung cancer (NSCLC) | serum | training set: 36 NSCLCs vs. 36 controls validation set: 120 NSCLCs vs. 71 controls | ↓ | Yes | Yes | a four non-coding RNAs panel (miR-1254, miR-485-5p, miR-574-5p, and MALAT1) | |
| Kristen M. 2011 | serum | training set: 11 early-stage NSCLCs vs. 11 controls validation set: 22 early-stage NSCLCs vs. 31 controls | ↑ | Yes | NA | with hsa-miR-1254 | ||
| Xiao-Rong Yang 2021 | plasma exosome | 30 EGFR/ALK positive NSCLC patients (16 phase IV patients with bone metastasis and 14 without bone metastasis) vs. 14 healthy donors | ↓ | NA | Yes | with miR-328-3p and miR-423-3p (all belong to cluster B) | ||
| Sheng-kai Huang 2018 | breast cancer | serum | training set: 30 breast cancer patients vs. 30 controls validation set: 128 breast cancer patients vs. 77 controls | ↑ | Yes | Yes (treatment matters) | A panel of ncRNAs, (let-7a, miR-155, miR-574-5p, and MALAT1) | |
| Deqiang Zheng 2019 | esophageal squamous cell carcinoma (ESCC) | serum | 52 ESCC patients vs. 52 age- and sex-matched controls | ↑ | Yes | Yes | with miR-16-5p, miR-451a | |
| MIAO YANG 2013 | tissue | tumor tissue and non-tumor tissue from 138 ESCC patients | ↑ | Yes | NA | a set of RNA profiles (hsa-miR-338-3p, hsa-miR-139-5p, hsa-miR-574-5p, and hsa-miR-601) | ||
| non-cancer | Jianqing Zhou 2016 | coronary artery disease (CAD) | plasma | 67 CAD patients vs. 67 healthy controls | ↑ | Yes | NA | with miR-206 |
| Adeline Boileau 2019 | cardiac arrest | serum | 590 cardiac arrest patients | ↑ | NA | yes (neurological outcome in women) | none | |
| Boileau 2019 | thoracic aortic aneurysm (TAA) | serum and tissue | 19 TAA patients and 19 controls; 28 TAA patients compared to 20 controls | ↑ in serum and ↓ in tissue | Yes | NA | none | |
| LiJiang 2021 | asthma | plasma | training set: 75 participants with recurrent wheezing at 3 years old from the Vitamin D Antenatal Asthma Reduction Trial validation set: 20 participants in Project Viva with recurrent wheezing at 3 years old | ↑ | NA | Yes | with vitamin D level; a set of circulating miRNA including hsa-miR-151a-5p | |
| Shanshan Liu 2021 | acute kidney injury in sepsis | serum | 136 patients with sepsis (58 developed AKI during hospitalization) | ↓ | NA | Yes (acute kidney injury) | none | |
| Wang, Huijuan 2012 | sepsis | serum | training set: 12 surviving and 12 nonsurviving sepsis patients validation set: 66 surviving and 52 nonsurviving sepsis patients | ↑ in survivors | NA | Yes (death) | with miR-297 | |
| WangFuyan 2021 | gestational diabetes mellitus (GDM) | plasma | 53 women with GDM and 46 normal pregnant women. | ↓ | Yes | NA | with miR-3135b | |
| Davarinejad 2021 | Schizophrenia (SCZ) | whole blood | training set: 2 mRNA expression arrays (GSE93987 and GSE38485) and 1 miRNA array (GSE54914) validation set: 40 SCZ patients | ↑ | Yes | NA | with hsa-miR-1827 and hsa-miR-4429 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, W.; Zhao, Y.; Xu, Z.; Wu, X.; Qiao, M.; Zhu, Z.; Zhao, Z. The Regulatory Mechanism of miR-574-5p Expression in Cancer. Biomolecules 2023, 13, 40. https://doi.org/10.3390/biom13010040
Huang W, Zhao Y, Xu Z, Wu X, Qiao M, Zhu Z, Zhao Z. The Regulatory Mechanism of miR-574-5p Expression in Cancer. Biomolecules. 2023; 13(1):40. https://doi.org/10.3390/biom13010040
Chicago/Turabian StyleHuang, Wei, Yifan Zhao, Zhengyi Xu, Xiaoyue Wu, Mingxin Qiao, Zhou Zhu, and Zhihe Zhao. 2023. "The Regulatory Mechanism of miR-574-5p Expression in Cancer" Biomolecules 13, no. 1: 40. https://doi.org/10.3390/biom13010040
APA StyleHuang, W., Zhao, Y., Xu, Z., Wu, X., Qiao, M., Zhu, Z., & Zhao, Z. (2023). The Regulatory Mechanism of miR-574-5p Expression in Cancer. Biomolecules, 13(1), 40. https://doi.org/10.3390/biom13010040






