Autophagy in Atherosclerotic Plaque Cells: Targeting NLRP3 Inflammasome for Self-Rescue
Abstract
1. Introduction
2. The Overview of Autophagy
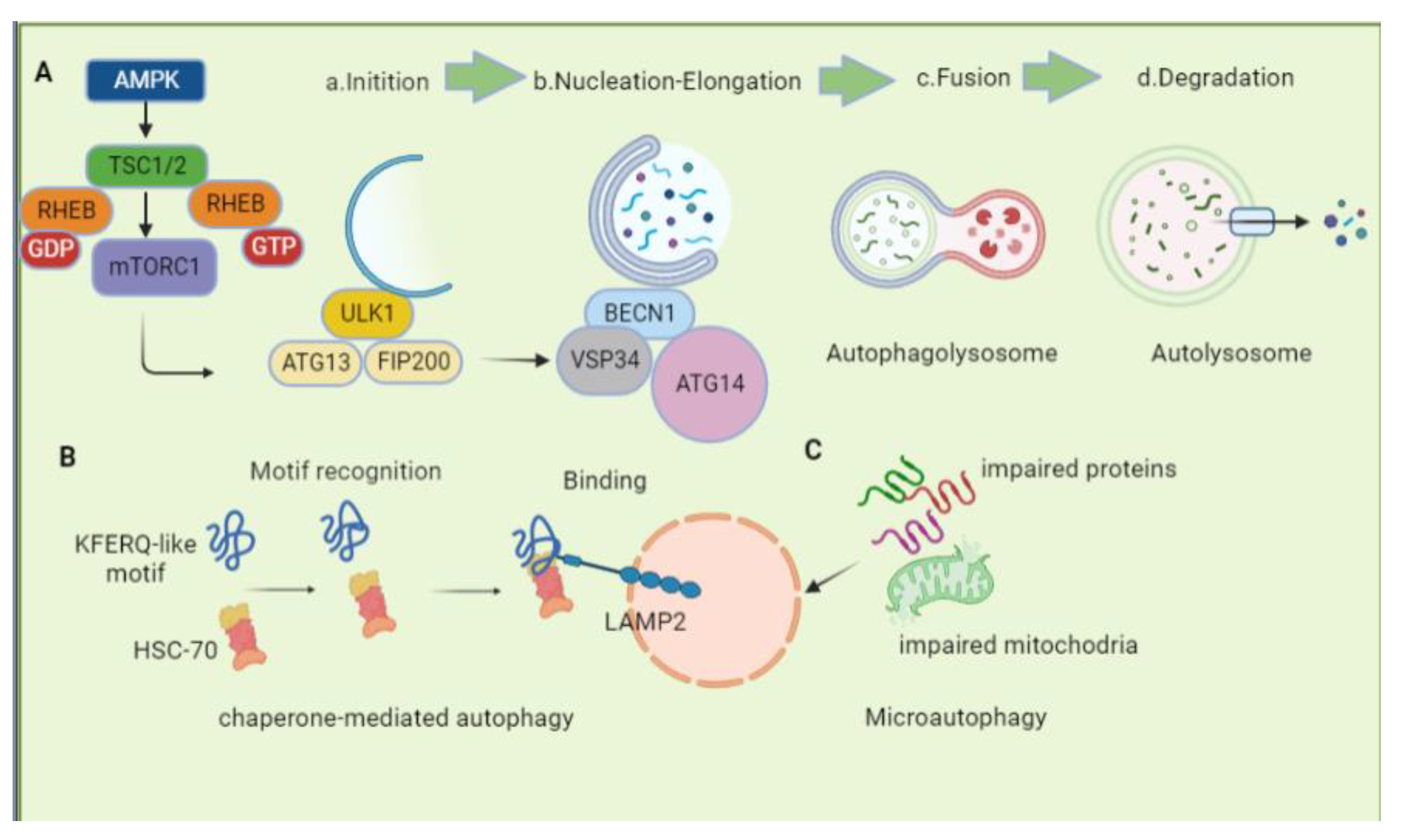
2.1. Macroautophagy
2.1.1. Induction
2.1.2. Nucleation, Elongation, and Maturation
2.1.3. Fusion and Degradation
2.2. Chaperone-Mediated Autophagy
2.3. Microautophagy
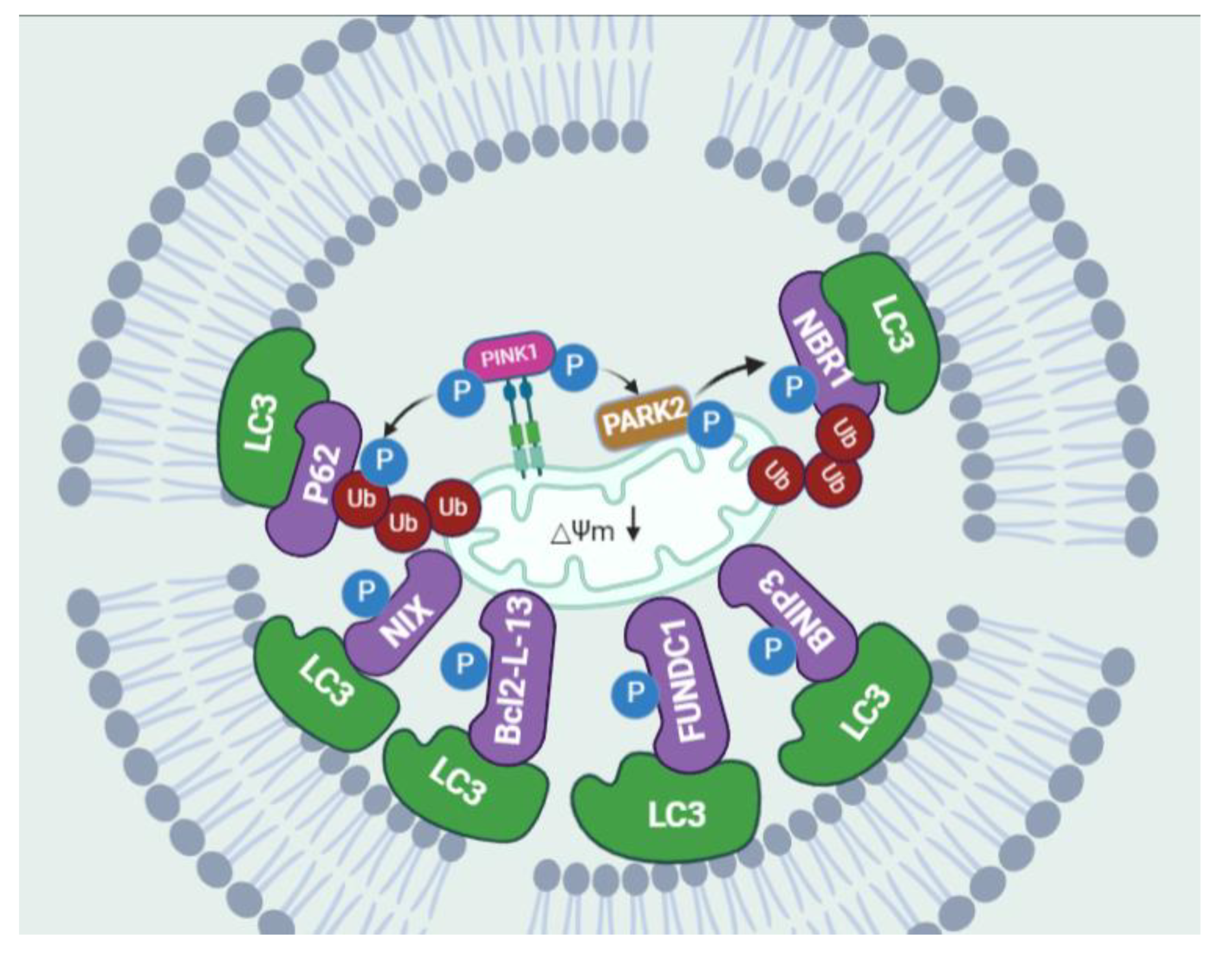
2.4. Selective Autophagy: Mitophagy
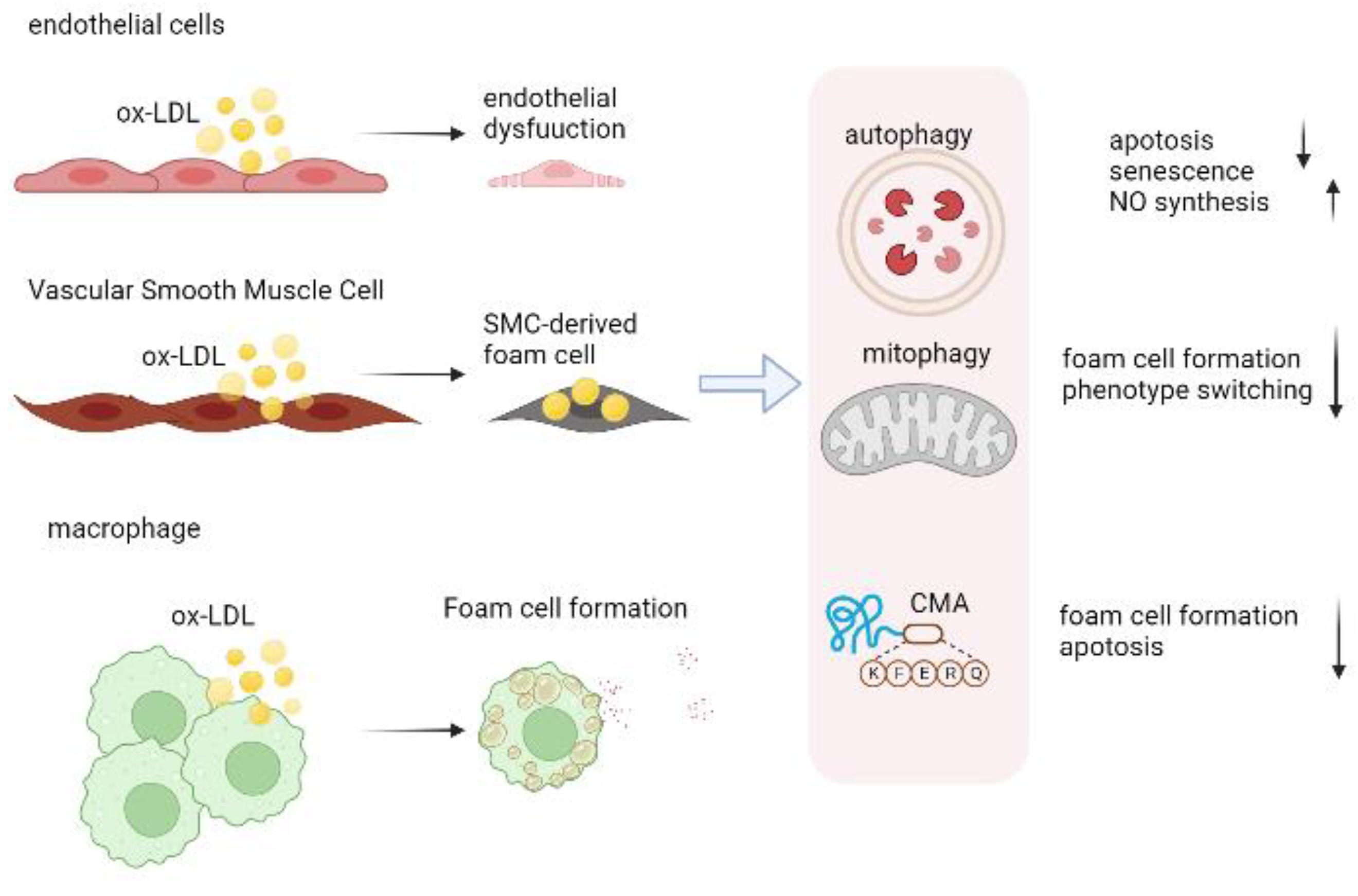
3. Role of Autophagy in Atherosclerosis
3.1. Autophagy in Endothelial Cells
3.2. Autophagy in Vascular Smooth Muscle Cell
3.3. Autophagy in Macrophage
4. CMA in Atherosclerosis
5. Mitophagy in Atherosclerosis
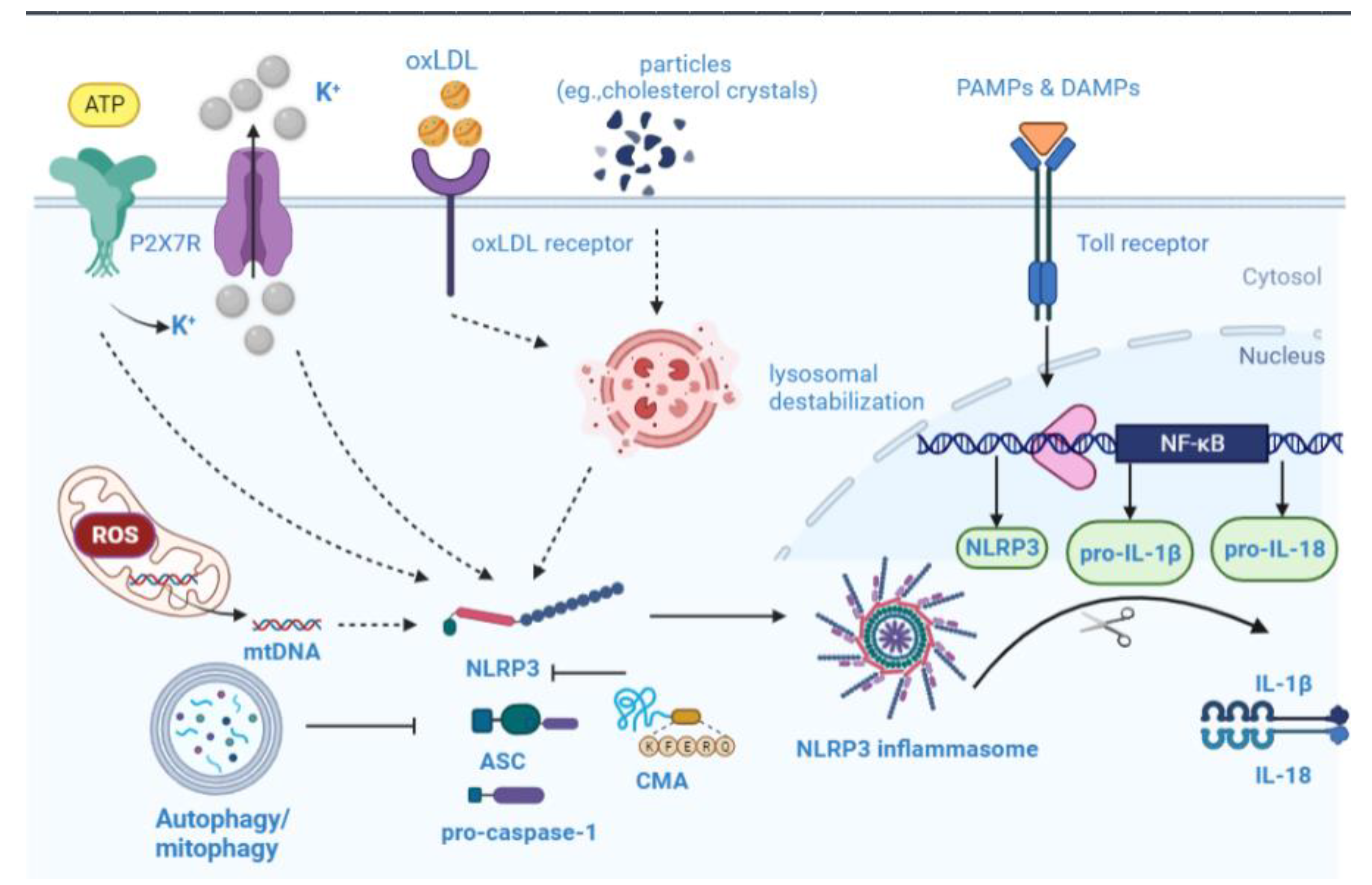
6. Overview of NLRP3 Inflammasome Biology
6.1. NLRP3 Inflammasome Activation
6.2. NLRP3 Inflammasome in Atherosclerosis
7. Autophagy–Inflammasome Interrelation in Atherosclerosis
8. Pharmacological Opportunities in Autophagy–Inflammasome Interplay
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Marchio, P.; Guerra-Ojeda, S.; Vila, J.M.; Aldasoro, M.; Victor, V.M.; Mauricio, M.D. Targeting Early Atherosclerosis: A Focus on Oxidative Stress and Inflammation. Oxid. Med. Cell Longev. 2019, 2019, 8563845. [Google Scholar] [CrossRef] [PubMed]
- Libby, P.; Ridker, P.M.; Hansson, G.K. Progress and challenges in translating the biology of atherosclerosis. Nature 2011, 473, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Allahverdian, S.; Chehroudi, A.C.; McManus, B.M.; Abraham, T.; Francis, G.A. Contribution of intimal smooth muscle cells to cholesterol accumulation and macrophage-like cells in human atherosclerosis. Circulation 2014, 129, 1551–1559. [Google Scholar] [CrossRef] [PubMed]
- Clarke, M.C.; Talib, S.; Figg, N.L.; Bennett, M.R. Vascular smooth muscle cell apoptosis induces interleukin-1-directed inflammation: Effects of hyperlipidemia-mediated inhibition of phagocytosis. Circ. Res. 2010, 106, 363–372. [Google Scholar] [CrossRef]
- Fang, C.; Yin, Y.; Jiang, S.; Zhang, S.; Wang, J.; Wang, Y.; Li, L.; Wang, Y.; Guo, J.; Yu, H.; et al. Increased Vulnerability and Distinct Layered Phenotype at Culprit and Nonculprit Lesions in STEMI Versus NSTEMI. JACC Cardiovasc. Imaging 2022, 15, 672–681. [Google Scholar] [CrossRef]
- Kolte, D.; Libby, P.; Jang, I.K. New Insights into Plaque Erosion as a Mechanism of Acute Coronary Syndromes. JAMA 2021, 325, 1043–1044. [Google Scholar] [CrossRef]
- Bos, D.; Arshi, B.; van den Bouwhuijsen, Q.; Ikram, M.K.; Selwaness, M.; Vernooij, M.W.; Kavousi, M.; van der Lugt, A. Atherosclerotic Carotid Plaque Composition and Incident Stroke and Coronary Events. J. Am. Coll. Cardiol. 2021, 77, 1426–1435. [Google Scholar] [CrossRef]
- Sergin, I.; Bhattacharya, S.; Emanuel, R.; Esen, E.; Stokes, C.J.; Evans, T.D.; Arif, B.; Curci, J.A.; Razani, B. Inclusion bodies enriched for p62 and polyubiquitinated proteins in macrophages protect against atherosclerosis. Sci. Signal. 2016, 9, a2. [Google Scholar] [CrossRef]
- Liu, H.; Cao, Y.; Tong, T.; Shi, J.; Zhang, Y.; Yang, Y.; Liu, C. Autophagy in atherosclerosis: A phenomenon found in human carotid atherosclerotic plaques. Chin. Med. J. 2015, 128, 69–74. [Google Scholar] [CrossRef]
- Madrigal-Matute, J.; de Bruijn, J.; van Kuijk, K.; Riascos-Bernal, D.F.; Diaz, A.; Tasset, I.; Martin-Segura, A.; Gijbels, M.; Sander, B.; Kaushik, S.; et al. Protective role of chaperone-mediated autophagy against atherosclerosis. Proc. Natl. Acad. Sci. USA 2022, 119, e2121133119. [Google Scholar] [CrossRef]
- Qiao, L.; Ma, J.; Zhang, Z.; Sui, W.; Zhai, C.; Xu, D.; Wang, Z.; Lu, H.; Zhang, M.; Zhang, C.; et al. Deficient Chaperone-Mediated Autophagy Promotes Inflammation and Atherosclerosis. Circ. Res. 2021, 129, 1141–1157. [Google Scholar] [CrossRef] [PubMed]
- Epelman, S.; Liu, P.P.; Mann, D.L. Role of innate and adaptive immune mechanisms in cardiac injury and repair. Nat. Rev. Immunol. 2015, 15, 117–129. [Google Scholar] [CrossRef] [PubMed]
- Grebe, A.; Hoss, F.; Latz, E. NLRP3 Inflammasome and the IL-1 Pathway in Atherosclerosis. Circ. Res. 2018, 122, 1722–1740. [Google Scholar] [CrossRef]
- Libby, P. Interleukin-1 Beta as a Target for Atherosclerosis Therapy: Biological Basis of CANTOS and Beyond. J. Am. Coll Cardiol. 2017, 70, 2278–2289. [Google Scholar] [CrossRef] [PubMed]
- Oku, M.; Sakai, Y. Three Distinct Types of Microautophagy Based on Membrane Dynamics and Molecular Machineries. Bioessays 2018, 40, e1800008. [Google Scholar] [CrossRef]
- Feng, Y.; He, D.; Yao, Z.; Klionsky, D.J. The machinery of macroautophagy. Cell Res. 2014, 24, 24–41. [Google Scholar] [CrossRef]
- Schlumpberger, M.; Schaeffeler, E.; Straub, M.; Bredschneider, M.; Wolf, D.H.; Thumm, M. AUT1, a gene essential for autophagocytosis in the yeast Saccharomyces cerevisiae. J. Bacteriol. 1997, 179, 1068–1076. [Google Scholar] [CrossRef]
- Funakoshi, T.; Matsuura, A.; Noda, T.; Ohsumi, Y. Analyses of APG13 gene involved in autophagy in yeast, Saccharomyces cerevisiae. Gene 1997, 192, 207–213. [Google Scholar] [CrossRef]
- Matsuura, A.; Tsukada, M.; Wada, Y.; Ohsumi, Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene 1997, 192, 245–250. [Google Scholar] [CrossRef]
- Losier, T.T.; Akuma, M.; McKee-Muir, O.C.; LeBlond, N.D.; Suk, Y.; Alsaadi, R.M.; Guo, Z.; Reshke, R.; Sad, S.; Campbell-Valois, F.X.; et al. AMPK Promotes Xenophagy through Priming of Autophagic Kinases upon Detection of Bacterial Outer Membrane Vesicles. Cell Rep. 2019, 26, 2150–2165. [Google Scholar] [CrossRef]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef] [PubMed]
- Dibble, C.C.; Elis, W.; Menon, S.; Qin, W.; Klekota, J.; Asara, J.M.; Finan, P.M.; Kwiatkowski, D.J.; Murphy, L.O.; Manning, B.D. TBC1D7 is a third subunit of the TSC1-TSC2 complex upstream of mTORC1. Mol. Cell 2012, 47, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Bujak, A.L.; Crane, J.D.; Lally, J.S.; Ford, R.J.; Kang, S.J.; Rebalka, I.A.; Green, A.E.; Kemp, B.E.; Hawke, T.J.; Schertzer, J.D.; et al. AMPK activation of muscle autophagy prevents fasting-induced hypoglycemia and myopathy during aging. Cell Metab. 2015, 21, 883–890. [Google Scholar] [CrossRef] [PubMed]
- Inoki, K.; Zhu, T.; Guan, K.L. TSC2 mediates cellular energy response to control cell growth and survival. Cell 2003, 115, 577–590. [Google Scholar] [CrossRef] [PubMed]
- Gwinn, D.M.; Shackelford, D.B.; Egan, D.F.; Mihaylova, M.M.; Mery, A.; Vasquez, D.S.; Turk, B.E.; Shaw, R.J. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell 2008, 30, 214–226. [Google Scholar] [CrossRef]
- Hosokawa, N.; Sasaki, T.; Iemura, S.; Natsume, T.; Hara, T.; Mizushima, N. Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy 2009, 5, 973–979. [Google Scholar] [CrossRef]
- Ganley, I.G.; Lam, D.H.; Wang, J.; Ding, X.; Chen, S.; Jiang, X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 2009, 284, 12297–12305. [Google Scholar] [CrossRef]
- Hosokawa, N.; Hara, T.; Kaizuka, T.; Kishi, C.; Takamura, A.; Miura, Y.; Iemura, S.; Natsume, T.; Takehana, K.; Yamada, N.; et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell 2009, 20, 1981–1991. [Google Scholar] [CrossRef]
- Dikic, I.; Elazar, Z. Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 2018, 19, 349–364. [Google Scholar] [CrossRef]
- Yamamoto, H.; Fujioka, Y.; Suzuki, S.W.; Noshiro, D.; Suzuki, H.; Kondo-Kakuta, C.; Kimura, Y.; Hirano, H.; Ando, T.; Noda, N.N.; et al. The Intrinsically Disordered Protein Atg13 Mediates Supramolecular Assembly of Autophagy Initiation Complexes. Dev. Cell 2016, 38, 86–99. [Google Scholar] [CrossRef]
- Kawamata, T.; Kamada, Y.; Kabeya, Y.; Sekito, T.; Ohsumi, Y. Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol. Biol. Cell 2008, 19, 2039–2050. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Overmeyer, J.H.; Maltese, W.A. Functional specificity of the mammalian Beclin-Vps34 PI 3-kinase complex in macroautophagy versus endocytosis and lysosomal enzyme trafficking. J. Cell Sci. 2006, 119, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Fimia, G.M.; Stoykova, A.; Romagnoli, A.; Giunta, L.; Di Bartolomeo, S.; Nardacci, R.; Corazzari, M.; Fuoco, C.; Ucar, A.; Schwartz, P.; et al. Ambra1 regulates autophagy and development of the nervous system. Nature 2007, 447, 1121–1125. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Lee, J.S.; Inn, K.S.; Gack, M.U.; Li, Q.; Roberts, E.A.; Vergne, I.; Deretic, V.; Feng, P.; Akazawa, C.; et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat. Cell Biol. 2008, 10, 776–787. [Google Scholar] [CrossRef]
- Molejon, M.I.; Ropolo, A.; Re, A.L.; Boggio, V.; Vaccaro, M.I. The VMP1-Beclin 1 interaction regulates autophagy induction. Sci. Rep. 2013, 3, 1055. [Google Scholar] [CrossRef]
- Kishi-Itakura, C.; Koyama-Honda, I.; Itakura, E.; Mizushima, N. Ultrastructural analysis of autophagosome organization using mammalian autophagy-deficient cells. J. Cell Sci. 2014, 127, 4089–4102. [Google Scholar] [CrossRef]
- Levine, B.; Kroemer, G. Biological Functions of Autophagy Genes: A Disease Perspective. Cell 2019, 176, 11–42. [Google Scholar] [CrossRef]
- Tsuboyama, K.; Koyama-Honda, I.; Sakamaki, Y.; Koike, M.; Morishita, H.; Mizushima, N. The ATG conjugation systems are important for degradation of the inner autophagosomal membrane. Science 2016, 354, 1036–1041. [Google Scholar] [CrossRef]
- Weidberg, H.; Shvets, E.; Shpilka, T.; Shimron, F.; Shinder, V.; Elazar, Z. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 2010, 29, 1792–1802. [Google Scholar] [CrossRef]
- Jahn, R.; Scheller, R.H. SNAREs--engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 2006, 7, 631–643. [Google Scholar] [CrossRef]
- Langemeyer, L.; Frohlich, F.; Ungermann, C. Rab GTPase Function in Endosome and Lysosome Biogenesis. Trends Cell Biol. 2018, 28, 957–970. [Google Scholar] [CrossRef] [PubMed]
- Yu, I.M.; Hughson, F.M. Tethering factors as organizers of intracellular vesicular traffic. Annu. Rev. Cell Dev. Biol. 2010, 26, 137–156. [Google Scholar] [CrossRef] [PubMed]
- Moreau, K.; Renna, M.; Rubinsztein, D.C. Connections between SNAREs and autophagy. Trends Biochem. Sci. 2013, 38, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Huang, J.; Geng, J.; Nair, U.; Klionsky, D.J. Atg22 recycles amino acids to link the degradative and recycling functions of autophagy. Mol. Biol. Cell 2006, 17, 5094–5104. [Google Scholar] [CrossRef]
- Cuervo, A.M.; Wong, E. Chaperone-mediated autophagy: Roles in disease and aging. Cell Res. 2014, 24, 92–104. [Google Scholar] [CrossRef]
- Kaushik, S.; Cuervo, A.M. Chaperone-mediated autophagy: A unique way to enter the lysosome world. Trends Cell Biol. 2012, 22, 407–417. [Google Scholar] [CrossRef]
- Chiang, H.L.; Terlecky, S.R.; Plant, C.P.; Dice, J.F. A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins. Science 1989, 246, 382–385. [Google Scholar] [CrossRef]
- Lv, L.; Li, D.; Zhao, D.; Lin, R.; Chu, Y.; Zhang, H.; Zha, Z.; Liu, Y.; Li, Z.; Xu, Y.; et al. Acetylation targets the M2 isoform of pyruvate kinase for degradation through chaperone-mediated autophagy and promotes tumor growth. Mol. Cell 2011, 42, 719–730. [Google Scholar] [CrossRef]
- Thompson, L.M.; Aiken, C.T.; Kaltenbach, L.S.; Agrawal, N.; Illes, K.; Khoshnan, A.; Martinez-Vincente, M.; Arrasate, M.; O’Rourke, J.G.; Khashwji, H.; et al. IKK phosphorylates Huntingtin and targets it for degradation by the proteasome and lysosome. J. Cell Biol. 2009, 187, 1083–1099. [Google Scholar] [CrossRef]
- Cuervo, A.M.; Dice, J.F. Unique properties of lamp2a compared to other lamp2 isoforms. J. Cell Sci. 2000, 113 Pt 24, 4441–4450. [Google Scholar] [CrossRef]
- Cuervo, A.M.; Dice, J.F. Regulation of lamp2a levels in the lysosomal membrane. Traffic 2000, 1, 570–583. [Google Scholar] [CrossRef] [PubMed]
- Roberts, P.; Moshitch-Moshkovitz, S.; Kvam, E.; O’Toole, E.; Winey, M.; Goldfarb, D.S. Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol. Biol. Cell 2003, 14, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Lemasters, J.J. Variants of mitochondrial autophagy: Types 1 and 2 mitophagy and micromitophagy (Type 3). Redox Biol. 2014, 2, 749–754. [Google Scholar] [CrossRef] [PubMed]
- Kirchner, P.; Bourdenx, M.; Madrigal-Matute, J.; Tiano, S.; Diaz, A.; Bartholdy, B.A.; Will, B.; Cuervo, A.M. Proteome-wide analysis of chaperone-mediated autophagy targeting motifs. PLoS Biol. 2019, 17, e3000301. [Google Scholar] [CrossRef] [PubMed]
- Sahu, R.; Kaushik, S.; Clement, C.C.; Cannizzo, E.S.; Scharf, B.; Follenzi, A.; Potolicchio, I.; Nieves, E.; Cuervo, A.M.; Santambrogio, L. Microautophagy of cytosolic proteins by late endosomes. Dev. Cell. 2011, 20, 131–139. [Google Scholar] [CrossRef]
- Mejlvang, J.; Olsvik, H.; Svenning, S.; Bruun, J.A.; Abudu, Y.P.; Larsen, K.B.; Brech, A.; Hansen, T.E.; Brenne, H.; Hansen, T.; et al. Starvation induces rapid degradation of selective autophagy receptors by endosomal microautophagy. J. Cell Biol. 2018, 217, 3640–3655. [Google Scholar] [CrossRef]
- Priault, M.; Salin, B.; Schaeffer, J.; Vallette, F.M.; di Rago, J.P.; Martinou, J.C. Impairing the bioenergetic status and the biogenesis of mitochondria triggers mitophagy in yeast. Cell Death Differ. 2005, 12, 1613–1621. [Google Scholar] [CrossRef]
- Birgisdottir, A.B.; Lamark, T.; Johansen, T. The LIR motif—Crucial for selective autophagy. J. Cell Sci. 2013, 126, 3237–3247. [Google Scholar] [CrossRef]
- Liu, L.; Sakakibara, K.; Chen, Q.; Okamoto, K. Receptor-mediated mitophagy in yeast and mammalian systems. Cell Res. 2014, 24, 787–795. [Google Scholar] [CrossRef]
- Durcan, T.M.; Fon, E.A. The three ‘P’s of mitophagy: PARKIN, PINK1, and post-translational modifications. Genes Dev. 2015, 29, 989–999. [Google Scholar] [CrossRef]
- Aerts, L.; Craessaerts, K.; De Strooper, B.; Morais, V.A. PINK1 kinase catalytic activity is regulated by phosphorylation on serines 228 and 402. J. Biol. Chem. 2015, 290, 2798–2811. [Google Scholar] [CrossRef] [PubMed]
- Narendra, D.P.; Jin, S.M.; Tanaka, A.; Suen, D.F.; Gautier, C.A.; Shen, J.; Cookson, M.R.; Youle, R.J. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010, 8, e1000298. [Google Scholar] [CrossRef]
- Kondapalli, C.; Kazlauskaite, A.; Zhang, N.; Woodroof, H.I.; Campbell, D.G.; Gourlay, R.; Burchell, L.; Walden, H.; Macartney, T.J.; Deak, M.; et al. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol. 2012, 2, 120080. [Google Scholar] [CrossRef]
- Geisler, S.; Holmstrom, K.M.; Skujat, D.; Fiesel, F.C.; Rothfuss, O.C.; Kahle, P.J.; Springer, W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 2010, 12, 119–131. [Google Scholar] [CrossRef]
- Chen, Y.; Dorn, G.N. PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science 2013, 340, 471–475. [Google Scholar] [CrossRef] [PubMed]
- Lazarou, M.; Sliter, D.A.; Kane, L.A.; Sarraf, S.A.; Wang, C.; Burman, J.L.; Sideris, D.P.; Fogel, A.I.; Youle, R.J. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 2015, 524, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Thomas, R.L.; Kubli, D.A.; Gustafsson, A.B. Bnip3-mediated defects in oxidative phosphorylation promote mitophagy. Autophagy 2011, 7, 775–777. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, T.E.; Johnson, L.R.; Kang, H.H.; Sun, J.C. BNIP3- and BNIP3L-Mediated Mitophagy Promotes the Generation of Natural Killer Cell Memory. Immunity 2015, 43, 331–342. [Google Scholar] [CrossRef]
- Murakawa, T.; Okamoto, K.; Omiya, S.; Taneike, M.; Yamaguchi, O.; Otsu, K. A Mammalian Mitophagy Receptor, Bcl2-L-13, Recruits the ULK1 Complex to Induce Mitophagy. Cell Rep. 2019, 26, 338–345. [Google Scholar] [CrossRef]
- Wu, W.; Li, W.; Chen, H.; Jiang, L.; Zhu, R.; Feng, D. FUNDC1 is a novel mitochondrial-associated-membrane (MAM) protein required for hypoxia-induced mitochondrial fission and mitophagy. Autophagy 2016, 12, 1675–1676. [Google Scholar] [CrossRef]
- Zhang, J.; Ney, P.A. NIX induces mitochondrial autophagy in reticulocytes. Autophagy 2008, 4, 354–356. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Lu, Q.; Wang, Q.; Ding, Y.; Ma, Z.; Mao, X.; Huang, K.; Xie, Z.; Zou, M.H. Binding of FUN14 Domain Containing 1 With Inositol 1,4,5-Trisphosphate Receptor in Mitochondria-Associated Endoplasmic Reticulum Membranes Maintains Mitochondrial Dynamics and Function in Hearts in Vivo. Circulation 2017, 136, 2248–2266. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Lin, C.; Wu, K.; Jiang, L.; Wang, X.; Li, W.; Zhuang, H.; Zhang, X.; Chen, H.; Li, S.; et al. FUNDC1 regulates mitochondrial dynamics at the ER-mitochondrial contact site under hypoxic conditions. EMBO J. 2016, 35, 1368–1384. [Google Scholar] [CrossRef]
- Schrijvers, D.M.; De Meyer, G.R.; Martinet, W. Autophagy in atherosclerosis: A potential drug target for plaque stabilization. Arter. Thromb. Vasc. Biol. 2011, 31, 2787–2791. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.Q.; Zhong, L.; Zhang, L.; Ji, X.P.; Zhang, M.; Zhao, Y.X.; Zhang, C.; Zhang, Y. Oral rapamycin attenuates inflammation and enhances stability of atherosclerotic plaques in rabbits independent of serum lipid levels. Br. J. Pharmacol. 2009, 156, 941–951. [Google Scholar] [CrossRef]
- Xie, Y.; You, S.J.; Zhang, Y.L.; Han, Q.; Cao, Y.J.; Xu, X.S.; Yang, Y.P.; Li, J.; Liu, C.F. Protective role of autophagy in AGE-induced early injury of human vascular endothelial cells. Mol. Med. Rep. 2011, 4, 459–464. [Google Scholar] [CrossRef]
- Hua, Y.; Zhang, J.; Liu, Q.; Su, J.; Zhao, Y.; Zheng, G.; Yang, Z.; Zhuo, D.; Ma, C.; Fan, G. The Induction of Endothelial Autophagy and Its Role in the Development of Atherosclerosis. Front. Cardiovasc. Med. 2022, 9, 831847. [Google Scholar] [CrossRef]
- Santovito, D.; Egea, V.; Bidzhekov, K.; Natarelli, L.; Mourao, A.; Blanchet, X.; Wichapong, K.; Aslani, M.; Brunssen, C.; Horckmans, M.; et al. Noncanonical inhibition of caspase-3 by a nuclear microRNA confers endothelial protection by autophagy in atherosclerosis. Sci. Transl. Med. 2020, 12, eaaz2294. [Google Scholar] [CrossRef]
- Torisu, K.; Singh, K.K.; Torisu, T.; Lovren, F.; Liu, J.; Pan, Y.; Quan, A.; Ramadan, A.; Al-Omran, M.; Pankova, N.; et al. Intact endothelial autophagy is required to maintain vascular lipid homeostasis. Aging Cell 2016, 15, 187–191. [Google Scholar] [CrossRef]
- Kheloufi, M.; Vion, A.C.; Hammoutene, A.; Poisson, J.; Lasselin, J.; Devue, C.; Pic, I.; Dupont, N.; Busse, J.; Stark, K.; et al. Endothelial autophagic flux hampers atherosclerotic lesion development. Autophagy 2018, 14, 173–175. [Google Scholar] [CrossRef]
- Bharath, L.P.; Mueller, R.; Li, Y.; Ruan, T.; Kunz, D.; Goodrich, R.; Mills, T.; Deeter, L.; Sargsyan, A.; Anandh, B.P.; et al. Impairment of autophagy in endothelial cells prevents shear-stress-induced increases in nitric oxide bioavailability. Can. J. Physiol. Pharmacol. 2014, 92, 605–612. [Google Scholar] [CrossRef] [PubMed]
- Robichaud, S.; Rasheed, A.; Pietrangelo, A.; Doyoung, K.A.; Boucher, D.M.; Emerton, C.; Vijithakumar, V.; Gharibeh, L.; Fairman, G.; Mak, E.; et al. Autophagy Is Differentially Regulated in Leukocyte and Nonleukocyte Foam Cells During Atherosclerosis. Circ. Res. 2022, 130, 831–847. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Jiang, X.; Cui, X.; Wang, J.; Liu, S.; Li, H.; Yang, J.; Zhang, C.; Zhang, W. Smooth muscle-specific HuR knockout induces defective autophagy and atherosclerosis. Cell Death Dis. 2021, 12, 385. [Google Scholar] [CrossRef] [PubMed]
- Chaulet, H.; Desgranges, C.; Renault, M.A.; Dupuch, F.; Ezan, G.; Peiretti, F.; Loirand, G.; Pacaud, P.; Gadeau, A.P. Extracellular nucleotides induce arterial smooth muscle cell migration via osteopontin. Circ. Res. 2001, 89, 772–778. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, J.; Li, Y.; Singh, P.; Cao, L.; Xu, L.J.; Li, D.; Wang, Y.; Xie, Z.; Gui, Y.; et al. Sonic hedgehog promotes autophagy of vascular smooth muscle cells. Am. J. Physiol. Heart Circ. Physiol. 2012, 303, H1319–H1331. [Google Scholar] [CrossRef] [PubMed]
- Salabei, J.K.; Hill, B.G. Implications of autophagy for vascular smooth muscle cell function and plasticity. Free Radic Biol. Med. 2013, 65, 693–703. [Google Scholar] [CrossRef] [PubMed]
- Pi, S.; Mao, L.; Chen, J.; Shi, H.; Liu, Y.; Guo, X.; Li, Y.; Zhou, L.; He, H.; Yu, C.; et al. The P2RY12 receptor promotes VSMC-derived foam cell formation by inhibiting autophagy in advanced atherosclerosis. Autophagy 2021, 17, 980–1000. [Google Scholar] [CrossRef]
- Razani, B.; Feng, C.; Coleman, T.; Emanuel, R.; Wen, H.; Hwang, S.; Ting, J.P.; Virgin, H.W.; Kastan, M.B.; Semenkovich, C.F. Autophagy links inflammasomes to atherosclerotic progression. Cell Metab. 2012, 15, 534–544. [Google Scholar] [CrossRef]
- Liu, B.; Zhang, B.; Guo, R.; Li, S.; Xu, Y. Enhancement in efferocytosis of oxidized low-density lipoprotein-induced apoptotic RAW264.7 cells through Sirt1-mediated autophagy. Int. J. Mol. Med. 2014, 33, 523–533. [Google Scholar] [CrossRef]
- Martinet, W.; Schrijvers, D.M.; De Meyer, G.R. Necrotic cell death in atherosclerosis. Basic Res. Cardiol. 2011, 106, 749–760. [Google Scholar] [CrossRef]
- Mallat, Z.; Hugel, B.; Ohan, J.; Leseche, G.; Freyssinet, J.M.; Tedgui, A. Shed membrane microparticles with procoagulant potential in human atherosclerotic plaques: A role for apoptosis in plaque thrombogenicity. Circulation 1999, 99, 348–353. [Google Scholar] [CrossRef] [PubMed]
- Kojima, Y.; Downing, K.; Kundu, R.; Miller, C.; Dewey, F.; Lancero, H.; Raaz, U.; Perisic, L.; Hedin, U.; Schadt, E.; et al. Cyclin-dependent kinase inhibitor 2B regulates efferocytosis and atherosclerosis. J. Clin. Investig. 2014, 124, 1083–1097. [Google Scholar] [CrossRef]
- Tabas, I. Macrophage death and defective inflammation resolution in atherosclerosis. Nat. Rev. Immunol. 2010, 10, 36–46. [Google Scholar] [CrossRef]
- Kounakis, K.; Chaniotakis, M.; Markaki, M.; Tavernarakis, N. Emerging Roles of Lipophagy in Health and Disease. Front. Cell Dev. Biol. 2019, 7, 185. [Google Scholar] [CrossRef]
- Ouimet, M.; Franklin, V.; Mak, E.; Liao, X.; Tabas, I.; Marcel, Y.L. Autophagy regulates cholesterol efflux from macrophage foam cells via lysosomal acid lipase. Cell Metab. 2011, 13, 655–667. [Google Scholar] [CrossRef]
- Ouimet, M.; Ediriweera, H.; Afonso, M.S.; Ramkhelawon, B.; Singaravelu, R.; Liao, X.; Bandler, R.C.; Rahman, K.; Fisher, E.A.; Rayner, K.J.; et al. microRNA-33 Regulates Macrophage Autophagy in Atherosclerosis. Arter. Thromb. Vasc. Biol. 2017, 37, 1058–1067. [Google Scholar] [CrossRef] [PubMed]
- Yao, F.; Lv, Y.C.; Zhang, M.; Xie, W.; Tan, Y.L.; Gong, D.; Cheng, H.P.; Liu, D.; Li, L.; Liu, X.Y.; et al. Apelin-13 impedes foam cell formation by activating Class III PI3K/Beclin-1-mediated autophagic pathway. Biochem. Biophys. Res. Commun. 2015, 466, 637–643. [Google Scholar] [CrossRef] [PubMed]
- Brichkina, A.; Bulavin, D.V. WIP-ing out atherosclerosis with autophagy. Autophagy 2012, 8, 1545–1547. [Google Scholar] [CrossRef][Green Version]
- Zhou, H.; Shi, C.; Hu, S.; Zhu, H.; Ren, J.; Chen, Y. BI1 is associated with microvascular protection in cardiac ischemia reperfusion injury via repressing Syk-Nox2-Drp1-mitochondrial fission pathways. Angiogenesis 2018, 21, 599–615. [Google Scholar] [CrossRef]
- Wang, J.; Toan, S.; Zhou, H. New insights into the role of mitochondria in cardiac microvascular ischemia/reperfusion injury. Angiogenesis 2020, 23, 299–314. [Google Scholar] [CrossRef]
- Zheng, J.; Lu, C. Oxidized LDL Causes Endothelial Apoptosis by Inhibiting Mitochondrial Fusion and Mitochondria Autophagy. Front. Cell Dev. Biol. 2020, 8, 600950. [Google Scholar] [CrossRef] [PubMed]
- Dai, D.F.; Karunadharma, P.P.; Chiao, Y.A.; Basisty, N.; Crispin, D.; Hsieh, E.J.; Chen, T.; Gu, H.; Djukovic, D.; Raftery, D.; et al. Altered proteome turnover and remodeling by short-term caloric restriction or rapamycin rejuvenate the aging heart. Aging Cell 2014, 13, 529–539. [Google Scholar] [CrossRef] [PubMed]
- Mai, S.; Muster, B.; Bereiter-Hahn, J.; Jendrach, M. Autophagy proteins LC3B, ATG5 and ATG12 participate in quality control after mitochondrial damage and influence lifespan. Autophagy 2012, 8, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.R.; Zhou, Y.J.; Yang, J.Q.; Liu, F.; Wu, X.P.; Sha, Y. Melatonin Attenuates Calcium Deposition from Vascular Smooth Muscle Cells by Activating Mitochondrial Fusion and Mitophagy via an AMPK/OPA1 Signaling Pathway. Oxid. Med. Cell Longev. 2020, 2020, 5298483. [Google Scholar] [CrossRef] [PubMed]
- Swiader, A.; Nahapetyan, H.; Faccini, J.; D’Angelo, R.; Mucher, E.; Elbaz, M.; Boya, P.; Vindis, C. Mitophagy acts as a safeguard mechanism against human vascular smooth muscle cell apoptosis induced by atherogenic lipids. Oncotarget 2016, 7, 28821–28835. [Google Scholar] [CrossRef]
- He, L.; Zhou, Q.; Huang, Z.; Xu, J.; Zhou, H.; Lv, D.; Lu, L.; Huang, S.; Tang, M.; Zhong, J.; et al. PINK1/Parkin-mediated mitophagy promotes apelin-13-induced vascular smooth muscle cell proliferation by AMPKalpha and exacerbates atherosclerotic lesions. J. Cell Physiol. 2019, 234, 8668–8682. [Google Scholar] [CrossRef]
- Ma, S.; Chen, J.; Feng, J.; Zhang, R.; Fan, M.; Han, D.; Li, X.; Li, C.; Ren, J.; Wang, Y.; et al. Melatonin Ameliorates the Progression of Atherosclerosis via Mitophagy Activation and NLRP3 Inflammasome Inhibition. Oxid. Med. Cell Longev. 2018, 2018, 9286458. [Google Scholar] [CrossRef]
- Duan, M.; Chen, H.; Yin, L.; Zhu, X.; Novak, P.; Lv, Y.; Zhao, G.; Yin, K. Mitochondrial apolipoprotein A-I binding protein alleviates atherosclerosis by regulating mitophagy and macrophage polarization. Cell Commun. Signal. 2022, 20, 60. [Google Scholar] [CrossRef]
- Choi, S.H.; Agatisa-Boyle, C.; Gonen, A.; Kim, A.; Kim, J.; Alekseeva, E.; Tsimikas, S.; Miller, Y.I. Intracellular AIBP (Apolipoprotein A-I Binding Protein) Regulates Oxidized LDL (Low-Density Lipoprotein)-Induced Mitophagy in Macrophages. Arter. Thromb. Vasc. Biol. 2021, 41, e82–e96. [Google Scholar] [CrossRef]
- De Meyer, G.R.; Grootaert, M.O.; Michiels, C.F.; Kurdi, A.; Schrijvers, D.M.; Martinet, W. Autophagy in vascular disease. Circ. Res. 2015, 116, 468–479. [Google Scholar] [CrossRef]
- Kurz, T.; Terman, A.; Brunk, U.T. Autophagy, ageing and apoptosis: The role of oxidative stress and lysosomal iron. Arch. Biochem. Biophys. 2007, 462, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, S.; Pluddemann, A.; Gordon, S. Macrophage pattern recognition receptors in immunity, homeostasis and self tolerance. Adv. Exp. Med. Biol. 2009, 653, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Schroder, K.; Tschopp, J. The inflammasomes. Cell 2010, 140, 821–832. [Google Scholar] [CrossRef]
- Martinon, F.; Burns, K.; Tschopp, J. The inflammasome: A molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol. Cell 2002, 10, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Kelley, N.; Jeltema, D.; Duan, Y.; He, Y. The NLRP3 Inflammasome: An Overview of Mechanisms of Activation and Regulation. Int. J. Mol. Sci. 2019, 20, 3328. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Pang, H.; Li, J.; Luo, S.; Huang, G.; Li, X.; Xie, Z.; Zhou, Z. The NLRP3 Inflammasome and Its Role in T1DM. Front. Immunol. 2020, 11, 1595. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, D.; Vande, W.L.; Lamkanfi, M. Therapeutic modulation of inflammasome pathways. Immunol. Rev. 2020, 297, 123–138. [Google Scholar] [CrossRef] [PubMed]
- Meyers, A.K.; Zhu, X. The NLRP3 Inflammasome: Metabolic Regulation and Contribution to Inflammaging. Cells 2020, 9, 1808. [Google Scholar] [CrossRef]
- Takahashi, M. Cell-Specific Roles of NLRP3 Inflammasome in Myocardial Infarction. J. Cardiovasc. Pharmacol. 2019, 74, 188–193. [Google Scholar] [CrossRef]
- Kanneganti, T.D.; Lamkanfi, M.; Kim, Y.G.; Chen, G.; Park, J.H.; Franchi, L.; Vandenabeele, P.; Nunez, G. Pannexin-1-mediated recognition of bacterial molecules activates the cryopyrin inflammasome independent of Toll-like receptor signaling. Immunity 2007, 26, 433–443. [Google Scholar] [CrossRef]
- Hornung, V.; Bauernfeind, F.; Halle, A.; Samstad, E.O.; Kono, H.; Rock, K.L.; Fitzgerald, K.A.; Latz, E. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat. Immunol. 2008, 9, 847–856. [Google Scholar] [CrossRef]
- Martinon, F.; Petrilli, V.; Mayor, A.; Tardivel, A.; Tschopp, J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature 2006, 440, 237–241. [Google Scholar] [CrossRef] [PubMed]
- Shimada, K.; Crother, T.R.; Karlin, J.; Dagvadorj, J.; Chiba, N.; Chen, S.; Ramanujan, V.K.; Wolf, A.J.; Vergnes, L.; Ojcius, D.M.; et al. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity 2012, 36, 401–414. [Google Scholar] [CrossRef]
- Zhong, Z.; Liang, S.; Sanchez-Lopez, E.; He, F.; Shalapour, S.; Lin, X.J.; Wong, J.; Ding, S.; Seki, E.; Schnabl, B.; et al. New mitochondrial DNA synthesis enables NLRP3 inflammasome activation. Nature 2018, 560, 198–203. [Google Scholar] [CrossRef] [PubMed]
- Heid, M.E.; Keyel, P.A.; Kamga, C.; Shiva, S.; Watkins, S.C.; Salter, R.D. Mitochondrial reactive oxygen species induces NLRP3-dependent lysosomal damage and inflammasome activation. J. Immunol. 2013, 191, 5230–5238. [Google Scholar] [CrossRef]
- Paramel, V.G.; Folkersen, L.; Strawbridge, R.J.; Halvorsen, B.; Yndestad, A.; Ranheim, T.; Krohg-Sorensen, K.; Skjelland, M.; Espevik, T.; Aukrust, P.; et al. NLRP3 Inflammasome Expression and Activation in Human Atherosclerosis. J. Am. Heart Assoc. 2016, 5, e003031. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Xie, W.L.; Kong, W.W.; Chen, D.; Qu, P. Expression of the NLRP3 Inflammasome in Carotid Atherosclerosis. J. Stroke Cereb. Dis. 2015, 24, 2455–2466. [Google Scholar] [CrossRef]
- Usui, F.; Shirasuna, K.; Kimura, H.; Tatsumi, K.; Kawashima, A.; Karasawa, T.; Hida, S.; Sagara, J.; Taniguchi, S.; Takahashi, M. Critical role of caspase-1 in vascular inflammation and development of atherosclerosis in Western diet-fed apolipoprotein E-deficient mice. Biochem. Biophys. Res. Commun. 2012, 425, 162–168. [Google Scholar] [CrossRef]
- Duewell, P.; Kono, H.; Rayner, K.J.; Sirois, C.M.; Vladimer, G.; Bauernfeind, F.G.; Abela, G.S.; Franchi, L.; Nunez, G.; Schnurr, M.; et al. NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature 2010, 464, 1357–1361. [Google Scholar] [CrossRef]
- Janoudi, A.; Shamoun, F.E.; Kalavakunta, J.K.; Abela, G.S. Cholesterol crystal induced arterial inflammation and destabilization of atherosclerotic plaque. Eur. Heart J. 2016, 37, 1959–1967. [Google Scholar] [CrossRef]
- Rajamaki, K.; Lappalainen, J.; Oorni, K.; Valimaki, E.; Matikainen, S.; Kovanen, P.T.; Eklund, K.K. Cholesterol crystals activate the NLRP3 inflammasome in human macrophages: A novel link between cholesterol metabolism and inflammation. PLoS ONE 2010, 5, e11765. [Google Scholar] [CrossRef] [PubMed]
- Samstad, E.O.; Niyonzima, N.; Nymo, S.; Aune, M.H.; Ryan, L.; Bakke, S.S.; Lappegard, K.T.; Brekke, O.L.; Lambris, J.D.; Damas, J.K.; et al. Cholesterol crystals induce complement-dependent inflammasome activation and cytokine release. J. Immunol. 2014, 192, 2837–2845. [Google Scholar] [CrossRef] [PubMed]
- Freigang, S.; Ampenberger, F.; Spohn, G.; Heer, S.; Shamshiev, A.T.; Kisielow, J.; Hersberger, M.; Yamamoto, M.; Bachmann, M.F.; Kopf, M. Nrf2 is essential for cholesterol crystal-induced inflammasome activation and exacerbation of atherosclerosis. Eur. J. Immunol. 2011, 41, 2040–2051. [Google Scholar] [CrossRef] [PubMed]
- Rhoads, J.P.; Lukens, J.R.; Wilhelm, A.J.; Moore, J.L.; Mendez-Fernandez, Y.; Kanneganti, T.D.; Major, A.S. Oxidized Low-Density Lipoprotein Immune Complex Priming of the Nlrp3 Inflammasome Involves TLR and FcgammaR Cooperation and Is Dependent on CARD9. J. Immunol. 2017, 198, 2105–2114. [Google Scholar] [CrossRef]
- Kotla, S.; Singh, N.K.; Rao, G.N. ROS via BTK-p300-STAT1-PPARgamma signaling activation mediates cholesterol crystals-induced CD36 expression and foam cell formation. Redox Biol. 2017, 11, 350–364. [Google Scholar] [CrossRef]
- Guo, F.X.; Wu, Q.; Li, P.; Zheng, L.; Ye, S.; Dai, X.Y.; Kang, C.M.; Lu, J.B.; Xu, B.M.; Xu, Y.J.; et al. The role of the LncRNA-FA2H-2-MLKL pathway in atherosclerosis by regulation of autophagy flux and inflammation through mTOR-dependent signaling. Cell Death Differ. 2019, 26, 1670–1687. [Google Scholar] [CrossRef]
- Shi, C.S.; Shenderov, K.; Huang, N.N.; Kabat, J.; Abu-Asab, M.; Fitzgerald, K.A.; Sher, A.; Kehrl, J.H. Activation of autophagy by inflammatory signals limits IL-1beta production by targeting ubiquitinated inflammasomes for destruction. Nat. Immunol. 2012, 13, 255–263. [Google Scholar] [CrossRef]
- Kimura, T.; Jain, A.; Choi, S.W.; Mandell, M.A.; Schroder, K.; Johansen, T.; Deretic, V. TRIM-mediated precision autophagy targets cytoplasmic regulators of innate immunity. J. Cell Biol. 2015, 210, 973–989. [Google Scholar] [CrossRef]
- Emanuel, R.; Sergin, I.; Bhattacharya, S.; Turner, J.; Epelman, S.; Settembre, C.; Diwan, A.; Ballabio, A.; Razani, B. Induction of lysosomal biogenesis in atherosclerotic macrophages can rescue lipid-induced lysosomal dysfunction and downstream sequelae. Arter. Thromb. Vasc. Biol. 2014, 34, 1942–1952. [Google Scholar] [CrossRef]
- Chen, M.L.; Zhu, X.H.; Ran, L.; Lang, H.D.; Yi, L.; Mi, M.T. Trimethylamine-N-Oxide Induces Vascular Inflammation by Activating the NLRP3 Inflammasome Through the SIRT3-SOD2-mtROS Signaling Pathway. J. Am. Heart Assoc. 2017, 6, e006347. [Google Scholar] [CrossRef]
- Liu, P.; Huang, G.; Wei, T.; Gao, J.; Huang, C.; Sun, M.; Zhu, L.; Shen, W. Sirtuin 3-induced macrophage autophagy in regulating NLRP3 inflammasome activation. Biochim. Biophys Acta Mol. Basis Dis. 2018, 1864, 764–777. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Yoon, J.H.; Ryu, J.H. Mitophagy: A balance regulator of NLRP3 inflammasome activation. BMB Rep. 2016, 49, 529–535. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Yazdi, A.S.; Menu, P.; Tschopp, J. A role for mitochondria in NLRP3 inflammasome activation. Nature 2011, 469, 221–225. [Google Scholar] [CrossRef]
- Nakahira, K.; Haspel, J.A.; Rathinam, V.A.; Lee, S.J.; Dolinay, T.; Lam, H.C.; Englert, J.A.; Rabinovitch, M.; Cernadas, M.; Kim, H.P.; et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat. Immunol. 2011, 12, 222–230. [Google Scholar] [CrossRef]
- Jin, Y.; Liu, Y.; Xu, L.; Xu, J.; Xiong, Y.; Peng, Y.; Ding, K.; Zheng, S.; Yang, N.; Zhang, Z.; et al. Novel role for caspase 1 inhibitor VX765 in suppressing NLRP3 inflammasome assembly and atherosclerosis via promoting mitophagy and efferocytosis. Cell Death Dis. 2022, 13, 512. [Google Scholar] [CrossRef] [PubMed]
- Schneider, J.L.; Suh, Y.; Cuervo, A.M. Deficient chaperone-mediated autophagy in liver leads to metabolic dysregulation. Cell Metab. 2014, 20, 417–432. [Google Scholar] [CrossRef]
- Kaushik, S.; Cuervo, A.M. Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nat. Cell Biol. 2015, 17, 759–770. [Google Scholar] [CrossRef] [PubMed]
- Madrigal-Matute, J.; Cuervo, A.M.; Sluimer, J.C. Chaperone-mediated autophagy protects against atherosclerosis. Autophagy 2022, 18, 2505–2507. [Google Scholar] [CrossRef]
- Cheng, Y.; Pan, X.; Wang, J.; Li, X.; Yang, S.; Yin, R.; Ma, A.; Zhu, X. Fucoidan Inhibits NLRP3 Inflammasome Activation by Enhancing p62/SQSTM1-Dependent Selective Autophagy to Alleviate Atherosclerosis. Oxid. Med. Cell Longev. 2020, 2020, 3186306. [Google Scholar] [CrossRef]
- Iacano, A.J.; Lewis, H.; Hazen, J.E.; Andro, H.; Smith, J.D.; Gulshan, K. Miltefosine increases macrophage cholesterol release and inhibits NLRP3-inflammasome assembly and IL-1beta release. Sci Rep. 2019, 9, 11128. [Google Scholar] [CrossRef]
- Shen, L.; Yang, Y.; Ou, T.; Key, C.C.; Tong, S.H.; Sequeira, R.C.; Nelson, J.M.; Nie, Y.; Wang, Z.; Boudyguina, E.; et al. Dietary PUFAs attenuate NLRP3 inflammasome activation via enhancing macrophage autophagy. J. Lipid Res. 2017, 58, 1808–1821. [Google Scholar] [CrossRef] [PubMed]
- Abderrazak, A.; Couchie, D.; Mahmood, D.F.; Elhage, R.; Vindis, C.; Laffargue, M.; Mateo, V.; Buchele, B.; Ayala, M.R.; El, G.M.; et al. Anti-inflammatory and antiatherogenic effects of the NLRP3 inflammasome inhibitor arglabin in ApoE2.Ki mice fed a high-fat diet. Circulation 2015, 131, 1061–1070. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Zhan, H.; Shao, Y.; Xiong, Y.; Zeng, L.; Zhang, C.; Liu, Z.; Huang, Z.; Su, H.; Yang, Z. 13-Methylberberine improves endothelial dysfunction by inhibiting NLRP3 inflammasome activation via autophagy induction in human umbilical vein endothelial cells. Chin. Med. 2020, 15, 8. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y.; Maejima, Y.; Nakagama, S.; Shiheido-Watanabe, Y.; Tamura, N.; Sasano, T. Rivaroxaban, a Direct Oral Factor Xa Inhibitor, Attenuates Atherosclerosis by Alleviating Factor Xa-PAR2-Mediated Autophagy Suppression. JACC Basic Transl. Sci. 2021, 6, 964–980. [Google Scholar] [CrossRef] [PubMed]
- Diao, Y. Clematichinenoside AR Alleviates Foam Cell Formation and the Inflammatory Response in Ox-LDL-Induced RAW264.7 Cells by Activating Autophagy. Inflammation 2021, 44, 758–768. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Xu, L.W.; Che, X.Y.; Xiao, Q.Q.; Pu, J.; Shao, Q.; He, B. Atorvastatin Inhibits Inflammatory Response, Attenuates Lipid Deposition, and Improves the Stability of Vulnerable Atherosclerotic Plaques by Modulating Autophagy. Front. Pharmacol. 2018, 9, 438. [Google Scholar] [CrossRef]
| Compounds | Mechanisms of Autophagy Induction | Antiatherosclerotic Effects | References |
|---|---|---|---|
| Fucoidan | Autophagy (direct) | ↓NLRP3 inflammasome ↓lipid accumulation | [149] |
| Miltefosine | AMPK/ULK1 | ↓mtROS ↑mitochondrial membrane potential ↓NLRP3 inflammasome ↑cholesterol release | [150] |
| PUFAs | Autophagy (direct) | ↓NLRP3 inflammasome ↓IL-1β secretion caspase-1 cleavage | [151] |
| Arglabin | Autophagy (direct) | ↓IL-1β and IL-18 ↓plasma lipids ↑anti-Inflammatory M2 Phenotype | [152] |
| Melatonin | Sirt3/FOXO3a/Parkin | ↓mtROS ↓NLRP3 inflammasome ↓plaque size and vulnerability | [107] |
| VX765 | Parkin | ↓NLRP3 inflammasome assembly ↓foam cell formation ↑efferocytosis | [145] |
| 13-Methylberberine | Autophagy (direct) | ↓ROS ↓NLRP3 inflammasome | [153] |
| Rivaroxaban | Protease-activated receptor 2 | ↓NLRP3 inflammasome atherosclerotic plaques↓ | [154] |
| Clematichinenoside AR | Autophagy (direct) | ↓NLRP3 inflammasome ↑ABCA1/ABCG1 ↑cholesterol efflux ↓ foam cell formation | [155] |
| Atorvastatin | mTOR | ↓NLRP3 inflammasome ↓lipid deposition ↓foam cell formation | [156] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Zhu, X.; Wei, Y. Autophagy in Atherosclerotic Plaque Cells: Targeting NLRP3 Inflammasome for Self-Rescue. Biomolecules 2023, 13, 15. https://doi.org/10.3390/biom13010015
Li X, Zhu X, Wei Y. Autophagy in Atherosclerotic Plaque Cells: Targeting NLRP3 Inflammasome for Self-Rescue. Biomolecules. 2023; 13(1):15. https://doi.org/10.3390/biom13010015
Chicago/Turabian StyleLi, Xuelian, Xianjie Zhu, and Yumiao Wei. 2023. "Autophagy in Atherosclerotic Plaque Cells: Targeting NLRP3 Inflammasome for Self-Rescue" Biomolecules 13, no. 1: 15. https://doi.org/10.3390/biom13010015
APA StyleLi, X., Zhu, X., & Wei, Y. (2023). Autophagy in Atherosclerotic Plaque Cells: Targeting NLRP3 Inflammasome for Self-Rescue. Biomolecules, 13(1), 15. https://doi.org/10.3390/biom13010015





