The Resistome of ESKAPEE Pathogens in Untreated and Treated Wastewater: A Polish Case Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Research Site and Sampling
2.2. DNA Extraction and Sequencing
2.3. Bioinformatic Analysis
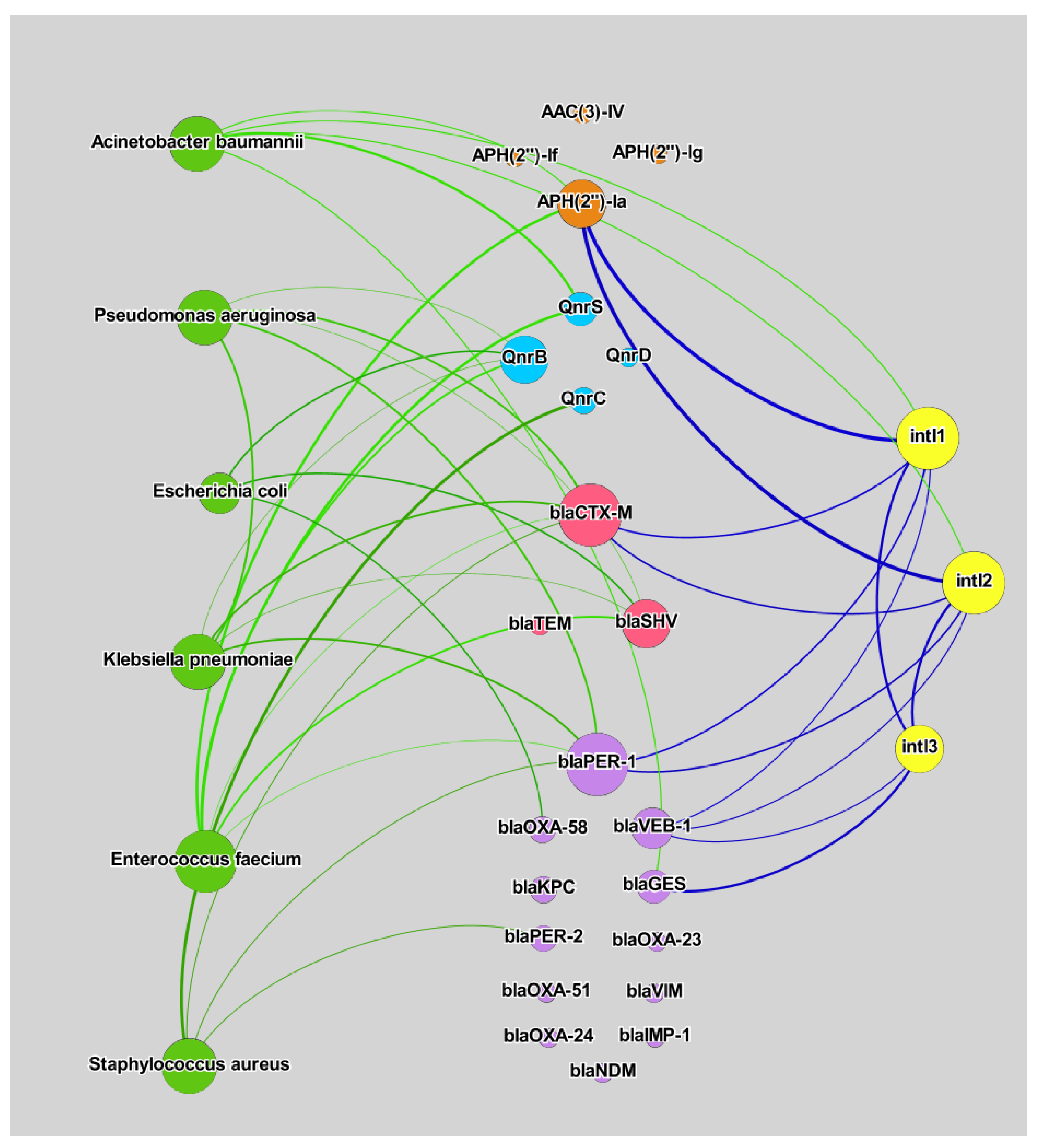
3. Results and Discussion
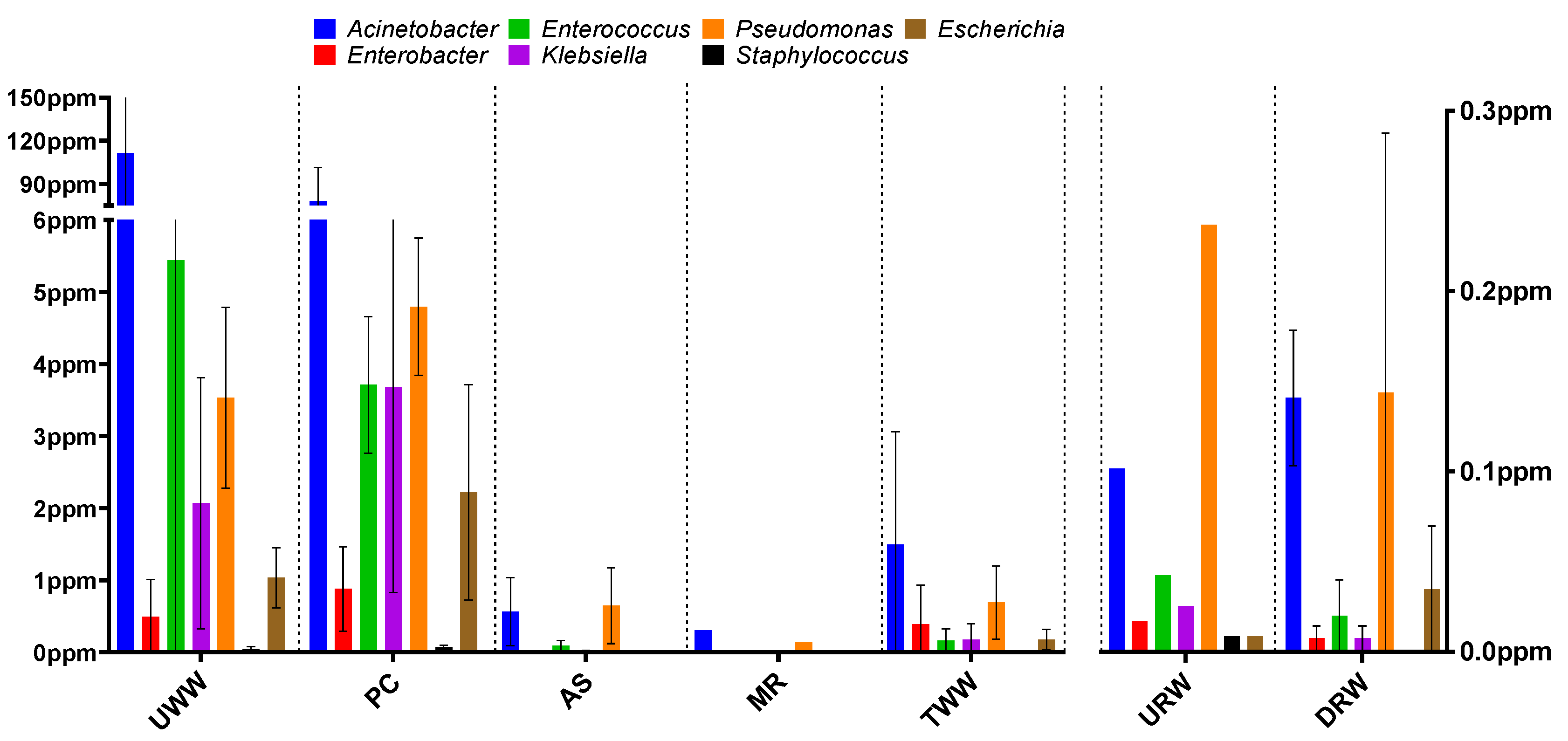
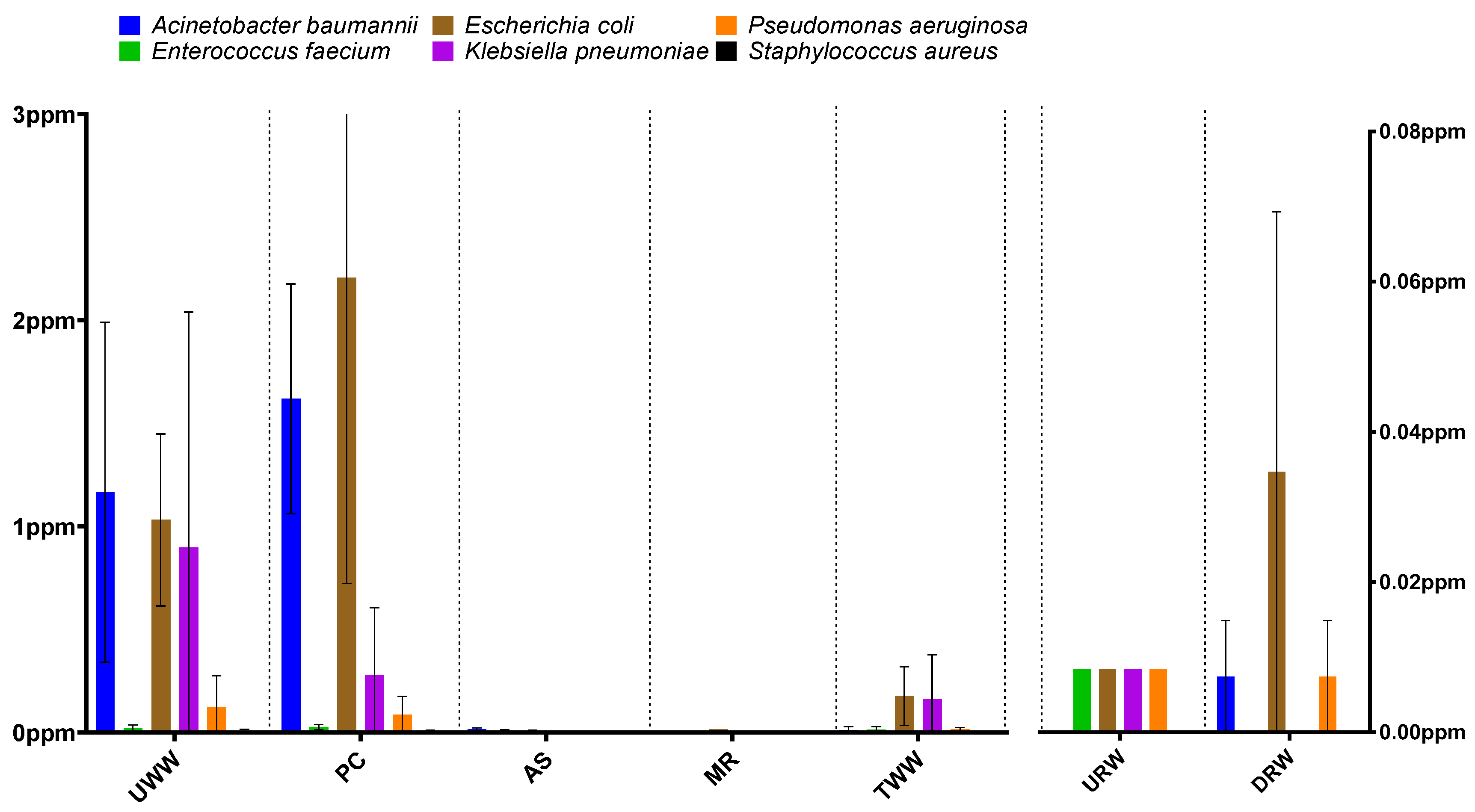
3.1. Taxonomic Structure Analysis
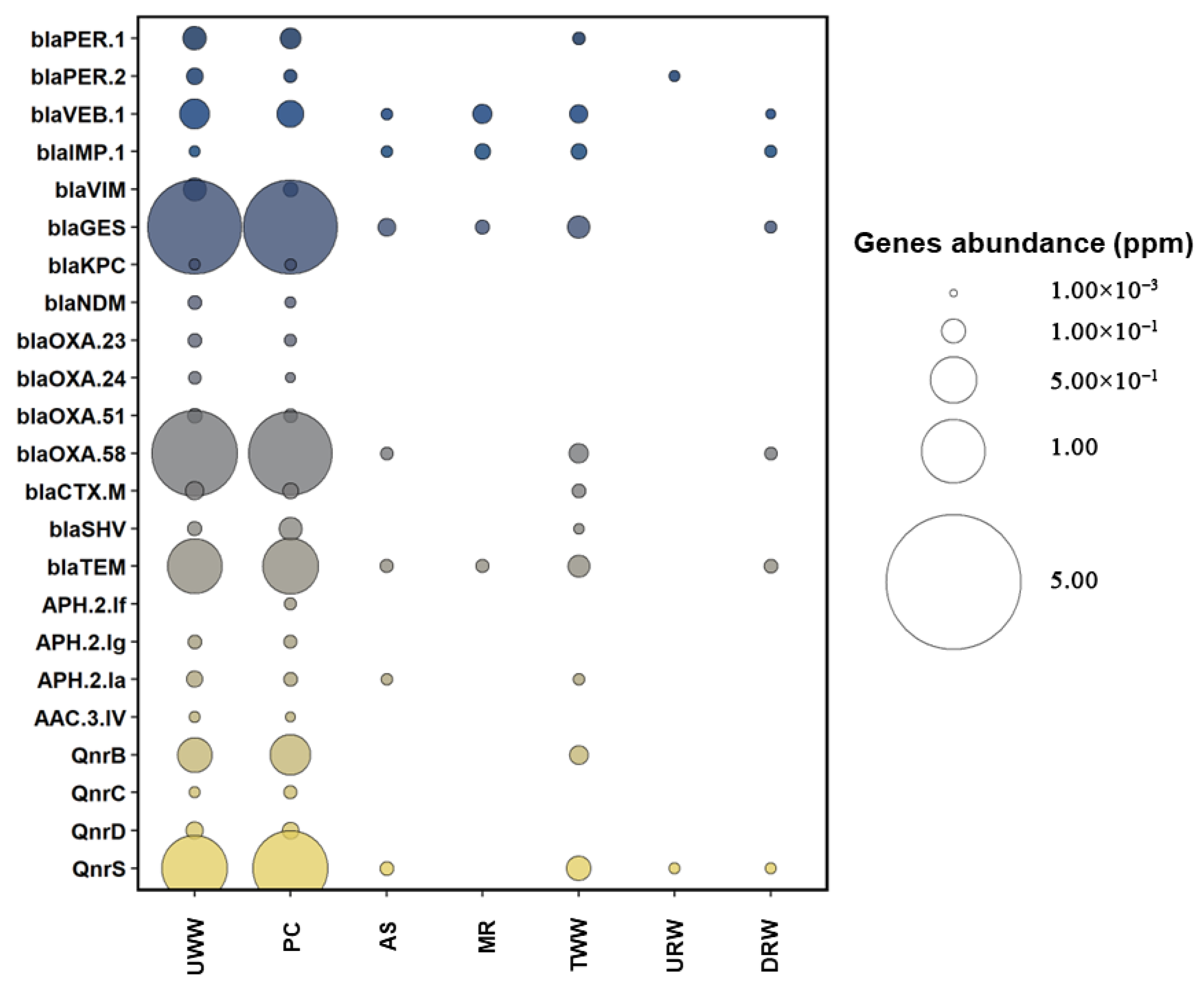
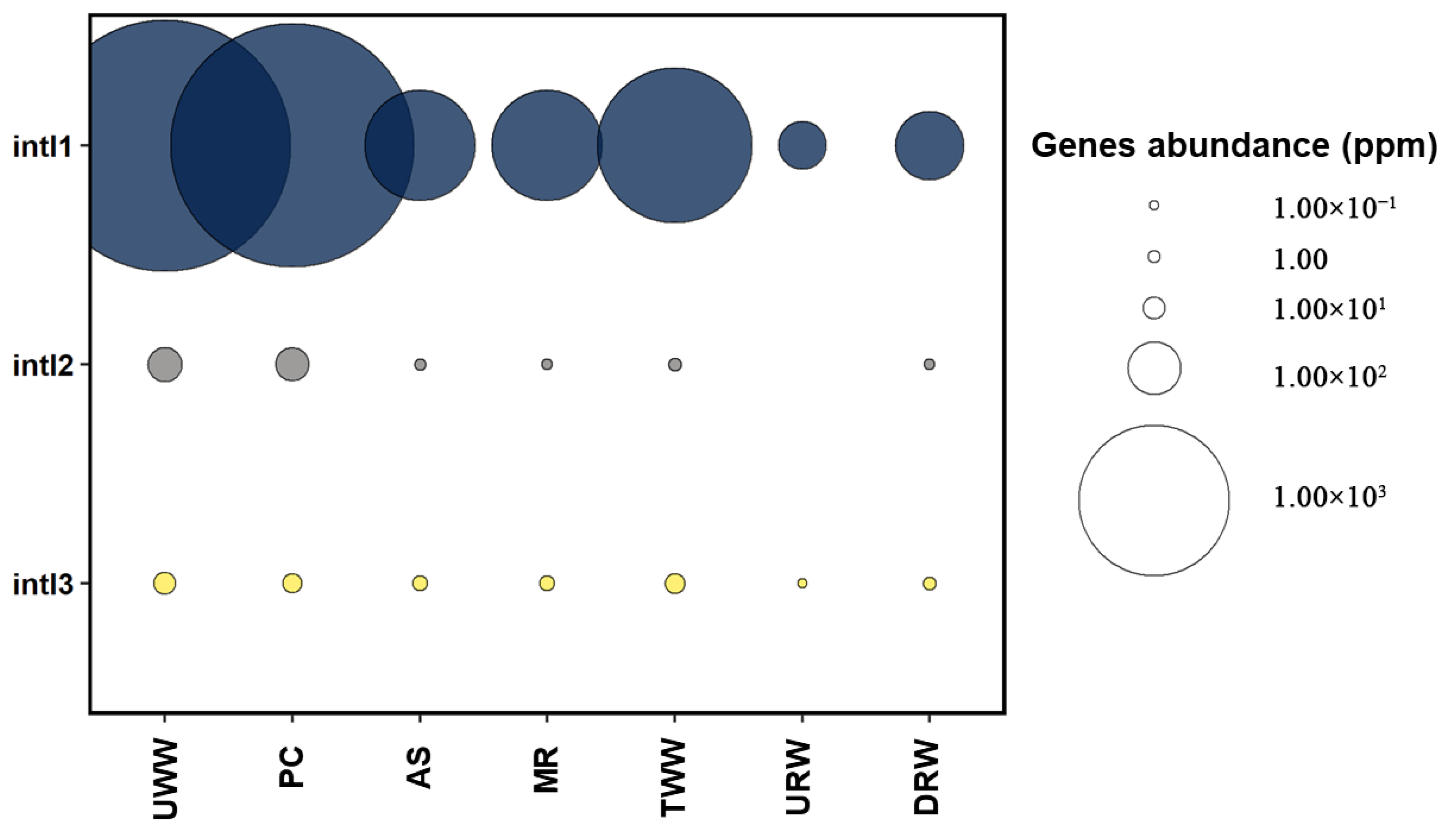
3.2. Prevalence of Antibiotic Resistance Determinants
3.3. Correlation Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nesme, J.; Simonet, P. The soil resistome: A critical review on antibiotic resistance origins, ecology and dissemination potential in telluric bacteria. Environ. Microbiol. 2015, 17, 913–930. [Google Scholar] [CrossRef] [PubMed]
- Timothy, J.F. Antibiotic resistance in Staphylococcus aureus. Curr. Status Future Prospect. FEMS Microbiol. Rev. 2017, 41, 430–449. [Google Scholar]
- Heymann, D.L. The World Health Report 2007: A Safer Future: Global Public Health Security in the 21st Century; World Health Organization: Geneva, Switzerland, 2007. [Google Scholar]
- Marano, R.B.M.; Fernandes, T.; Manaia, C.M.; Nunes, O.; Morrison, D.; Berendonk, T.U.; Kreuzinger, N.; Tenson, T.; Corno, G.; Fatta-Kassinos, D.; et al. A global multinational survey of cefotaxime-resistant coliforms in urban wastewater treatment plants. Environ. Int. 2020, 144, 106035. [Google Scholar] [CrossRef] [PubMed]
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- Berendonk, T.U.; Manaia, C.M.; Merlin, C.; Fatta-Kassinos, D.; Cytryn, E.; Walsh, F.; Bürgmann, H.; Sørum, H.; Norström, M.; Pons, M.N.; et al. Tackling antibiotic resistance: The environmental framework. Nat. Rev. Microbiol. 2015, 13, 310–317. [Google Scholar] [CrossRef]
- Tripathi, V.; Cytryn, E. Impact of anthropogenic activities on the dissemination of antibiotic resistance across ecological boundaries. Essays Biochem. 2017, 61, 11–21. [Google Scholar] [CrossRef][Green Version]
- Santajit, S.; Indrawattana, N. Mechanisms of Antimicrobial Resistance in ESKAPE Pathogens. BioMed Res. Int. 2016, 2016, 1–8. [Google Scholar] [CrossRef]
- Savin, M.; Bierbaum, G.; Hammerl, J.A.; Heinemann, C.; Parcina, M.; Sib, E.; Voigt, A.; Kreyenschmidt, J. Antibiotic-resistant bacteria and antimicrobial residues in wastewater and process water from German pig slaughterhouses and their receiving municipal wastewater treatment plants. Sci. Total Environ. 2020, 727, 138788. [Google Scholar] [CrossRef]
- Grochowalska, A.; Kozioł-Montewka, M.; Oliynyk, O.; Krasij, N. Ventilator-associated pneumonia (VAP) caused by Acinetobacter baumannii in view of the microbial properties of the ESKAPE group in neighbouring countries—Poland and Ukraine. J. Pre-Clin. Clin. Res. 2017, 11, 111–115. [Google Scholar] [CrossRef][Green Version]
- Behroozian, S.; Svensson, S.L.; Davies, J. Kisameet Clay Exhibits Potent Antibacterial Activity against the ESKAPE Pathogens. MBio 2016, 7, e01842-15. [Google Scholar] [CrossRef]
- Bales, P.M.; Renke, E.M.; May, S.L.; Shen, Y.; Nelson, D.C. Purification and Characterization of Biofilm-Associated EPS Exopolysaccharides from ESKAPE Organisms and Other Pathogens. PLoS ONE 2013, 8, e67950. [Google Scholar] [CrossRef] [PubMed]
- ECDC. Antimicrobial resistance in the EU/EEA (EARS-Net). Antimicrob. Resist. EU/EEA 2020, 174, 341. [Google Scholar]
- Smith, A. Baterial resistance to antibiotics. In Hugo and Russell’s Pharmaceutical Microbiology; Wiley-Blackwell: Hoboken, NJ, USA, 2004; pp. 220–232. ISBN 0-632-06467-6. [Google Scholar]
- Indrawattana, N.; Sungkhachat, O.; Sookrung, N.; Chongsa-nguan, M.; Tungtrongchitr, A.; Voravuthikunchai, S.P.; Kong-ngoen, T.; Kurazono, H.; Chaicumpa, W. Staphylococcus aureus Clinical Isolates: Antibiotic Susceptibility, Molecular Characteristics, and Ability to Form Biofilm. BioMed. Res. Int. 2013, 2013, 1–11. [Google Scholar] [CrossRef]
- Appelbaum, P.C. Reduced glycopeptide susceptibility in methicillin-resistant Staphylococcus aureus (MRSA). Int. J. Antimicrob. Agents 2007, 30, 398–408. [Google Scholar] [CrossRef]
- Yong, D.; Toleman, M.A.; Giske, C.G.; Cho, H.S.; Sundman, K.; Lee, K.; Walsh, T.R. Characterization of a New Metallo-β-Lactamase Gene, bla NDM-1, and a Novel Erythromycin Esterase Gene Carried on a Unique Genetic Structure in Klebsiella pneumoniae Sequence Type 14 from India. Antimicrob. Agents Chemother. 2009, 53, 5046–5054. [Google Scholar] [CrossRef] [PubMed]
- Kumarasamy, K.K.; Toleman, M.A.; Walsh, T.R.; Bagaria, J.; Butt, F.; Balakrishnan, R.; Chaudhary, U.; Doumith, M.; Giske, C.G.; Irfan, S.; et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: A molecular, biological, and epidemiological study. Lancet Infect. Dis. 2010, 10, 597–602. [Google Scholar] [CrossRef]
- Bonnin, R.A.; Poirel, L.; Nordmann, P. New Delhi metallo-β-lactamase-producing Acinetobacter baumannii: A novel paradigm for spreading antibiotic resistance genes. Future Microbiol. 2014, 9, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Boucher, H.W.; Talbot, G.H.; Bradley, J.S.; Edwards, J.E.; Gilbert, D.; Rice, L.B.; Scheld, M.; Spellberg, B.; Bartlett, J. Bad bugs, no drugs: No ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009, 48, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Vila, J.; Martí, S.; Sánchez-Céspedes, J. Porins, efflux pumps and multidrug resistance in Acinetobacter baumannii. J. Antimicrob. Chemother. 2007, 59, 1210–1215. [Google Scholar] [CrossRef]
- Vrancianu, C.O.; Pelcaru, C.F.; Alistar, A.; Gheorghe, I.; Marutescu, L.; Popa, M.; Czobor, I.; Gradisteanu, G.; Dobre, E.G.; Chifiriuc, M.C. Escaping from ESKAPE. Clinical significance and antibiotic resistance mechanisms in acinetobacter baumannii: A review. Biointerface Res. Appl. Chem. 2021, 11, 8190–8203. [Google Scholar] [CrossRef]
- Castanheira, M.; Deshpande, L.M.; Mathai, D.; Bell, J.M.; Jones, R.N.; Mendes, R.E. Early Dissemination of NDM-1- and OXA-181-Producing Enterobacteriaceae in Indian Hospitals: Report from the SENTRY Antimicrobial Surveillance Program, 2006–2007. Antimicrob. Agents Chemother. 2011, 55, 1274–1278. [Google Scholar] [CrossRef] [PubMed]
- Pendleton, J.N.; Gorman, S.P.; Gilmore, B.F. Clinical relevance of the ESKAPE pathogens. Expert Rev. Anti. Infect. Ther. 2013, 11, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Laudy, A.E.; Róg, P.; Smolinska-Król, K.; Ćmiel, M.; Søoczynska, A.; Patzer, J.; Dzierzanowska, D.; Wolinowska, R.; Starosciak, B.; Tyski, S. Prevalence of ESBL-producing Pseudomonas aeruginosa isolates in Warsaw, Poland, detected by various phenotypic and genotypic methods. PLoS ONE 2017, 12, e0180121. [Google Scholar] [CrossRef]
- Kot, B.; Wierzchowska, K.; Piechota, M.; Gruzewska, A. Antimicrobial Resistance Patterns in Methicillin-Resistant Staphylococcus aureus from Patients Hospitalized during 2015-2017 in Hospitals in Poland. Med. Princ. Pract. 2020, 29, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Litwin, A.; Fedorowicz, O.; Duszynska, W. Characteristics of microbial factors of healthcare-associated infections including multidrug-resistant pathogens and antibiotic consumption at the university intensive care unit in Poland in the years 2011–2018. Int. J. Environ. Res. Public Health 2020, 17, 6943. [Google Scholar] [CrossRef]
- Zając-Spychała, O.; Wachowiak, J.; Frączkiewicz, J.; Salamonowicz, M.; Kałwak, K.; Gorczyńska, E.; Chybicka, A.; Czyżewski, K.; Dziedzic, M.; Wysocki, M.; et al. Multidrug-resistant bacterial infections in children undergoing haematopoietic stem cell transplantation over a 6-year period: Analysis of the Polish Pediatric Group for Hematopoietic Stem Cell Transplantation. J. Appl. Microbiol. 2020, 128, 292–300. [Google Scholar] [CrossRef]
- Zając-Spychała, O.; Wachowiak, J.; Pieczonka, A.; Siewiera, K.; Frączkiewicz, J.; Kałwak, K.; Gorczyńska, E.; Chybicka, A.; Czyżewski, K.; Jachna-Sawicka, K.; et al. Bacterial infections in pediatric hematopoietic stem cell transplantation recipients: Incidence, epidemiology, and spectrum of pathogens: Report of the Polish Pediatric Group for Hematopoietic Stem Cell Transplantation. Transpl. Infect. Dis. 2016, 18, 690–698. [Google Scholar] [CrossRef]
- Talaga-Ćwiertnia, K.; Krzyściak, P.; Bulanda, M. Do bacteria isolated from ICU patients ESKAPE antibiotic treatment?—In vitro susceptibility of the Enterobacteriaceae family to tigecycline. Anaesthesiol. Intensive Ther. 2017, 49, 210–214. [Google Scholar] [CrossRef]
- Gotkowska-Płachta, A. The Prevalence of Virulent and Multidrug-Resistant Enterococci in River Water and in Treated and Untreated Municipal and Hospital Wastewater. Int. J. Environ. Res. Public Health 2021, 18, 563. [Google Scholar] [CrossRef]
- Rybak, B.; Wawrzyniak, N.; Wolska, L.; Potrykus, M. Escherichia coli and Serratia fonticola ESBLs as a potential source of antibiotics resistance dissemination in the Tricity water reservoirs. Acta Biochim. Pol. 2021, 68, 437–448. [Google Scholar] [CrossRef]
- Hubeny, J.; Ciesielski, S.; Harnisz, M.; Korzeniewska, E.; Dulski, T.; Jałowiecki, Ł.; Płaza, G. Impact of Hospital Wastewater on the Occurrence and Diversity of Beta-Lactamase Genes During Wastewater Treatment with an Emphasis on Carbapenemase Genes: A Metagenomic Approach. Front. Environ. Sci. 2021, 9, 1–12. [Google Scholar] [CrossRef]
- Rolbiecki, D.; Harnisz, M.; Korzeniewska, E.; Buta, M.; Hubeny, J.; Zieliński, W. Detection of carbapenemase-producing, hypervirulent Klebsiella spp. in wastewater and their potential transmission to river water and WWTP employees. Int. J. Hyg. Environ. Health 2021, 237, 113831. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Masella, A.P.; Bartram, A.K.; Truszkowski, J.M.; Brown, D.G.; Neufeld, J.D. PANDAseq: Paired-end assembler for illumina sequences. BMC Bioinform. 2012, 13, 31. [Google Scholar] [CrossRef]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; De Pascale, G.; Ejim, L.; et al. The Comprehensive Antibiotic Resistance Database. Antimicrob. Agents Chemother. 2013, 57, 3348–3357. [Google Scholar] [CrossRef]
- Escudeiro, P.; Pothier, J.; Dionisio, F.; Nogueira, T. Antibiotic Resistance Gene Diversity and Virulence Gene Diversity Are Correlated in Human Gut and Environmental Microbiomes. mSphere 2019, 4, e00135-19. [Google Scholar] [CrossRef]
- Casaburi, G.; Duar, R.M.; Vance, D.P.; Mitchell, R.; Contreras, L.; Frese, S.A.; Smilowitz, J.T.; Underwood, M.A. Early-life gut microbiome modulation reduces the abundance of antibiotic-resistant bacteria. Antimicrob. Resist. Infect. Control 2019, 8, 131. [Google Scholar] [CrossRef]
- Kristiansson, E.; Fick, J.; Janzon, A.; Grabic, R.; Rutgersson, C.; Weijdegård, B.; Söderström, H.; Larsson, D.G.J. Pyrosequencing of Antibiotic-Contaminated River Sediments Reveals High Levels of Resistance and Gene Transfer Elements. PLoS ONE 2011, 6, e17038. [Google Scholar] [CrossRef]
- Zhang, T.; Zhang, X.-X.; Ye, L. Plasmid Metagenome Reveals High Levels of Antibiotic Resistance Genes and Mobile Genetic Elements in Activated Sludge. PLoS ONE 2011, 6, e26041. [Google Scholar] [CrossRef]
- Yang, Y.; Li, B.; Ju, F.; Zhang, T. Exploring Variation of Antibiotic Resistance Genes in Activated Sludge over a Four-Year Period through a Metagenomic Approach. Environ. Sci. Technol. 2013, 47, 10197–10205. [Google Scholar] [CrossRef]
- Su, J.-Q.; An, X.-L.; Li, B.; Chen, Q.-L.; Gillings, M.R.; Chen, H.; Zhang, T.; Zhu, Y.-G. Metagenomics of urban sewage identifies an extensively shared antibiotic resistome in China. Microbiome 2017, 5, 84. [Google Scholar] [CrossRef] [PubMed]
- Tian, Z.; Zhang, Y.; Yu, B.; Yang, M. Changes of resistome, mobilome and potential hosts of antibiotic resistance genes during the transformation of anaerobic digestion from mesophilic to thermophilic. Water Res. 2016, 98, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- Barberán, A.; Bates, S.T.; Casamayor, E.O.; Fierer, N. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 2012, 6, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An Open Source Software for Exploring and Manipulating Networks. In Proceedings of the Third International AAAI Conference on Weblogs and Social Media, San Jose, CA, USA, 17–20 May 2009; pp. 361–362. [Google Scholar] [CrossRef]
- Numberger, D.; Ganzert, L.; Zoccarato, L.; Mühldorfer, K.; Sauer, S.; Grossart, H.-P.; Greenwood, A.D. Characterization of bacterial communities in wastewater with enhanced taxonomic resolution by full-length 16S rRNA sequencing. Sci. Rep. 2019, 9, 9673. [Google Scholar] [CrossRef]
- McLellan, S.L.; Huse, S.M.; Mueller-Spitz, S.R.; Andreishcheva, E.N.; Sogin, M.L. Diversity and population structure of sewage-derived microorganisms in wastewater treatment plant influent. Environ. Microbiol. 2010, 12, 378–392. [Google Scholar] [CrossRef]
- Jäger, T.; Hembach, N.; Elpers, C.; Wieland, A.; Alexander, J.; Hiller, C.; Krauter, G.; Schwartz, T. Reduction of Antibiotic Resistant Bacteria During Conventional and Advanced Wastewater Treatment, and the Disseminated Loads Released to the Environment. Front. Microbiol. 2018, 9, 2599. [Google Scholar] [CrossRef]
- Rafraf, I.D.; Lekunberri, I.; Sànchez-Melsió, A.; Aouni, M.; Borrego, C.M.; Balcázar, J.L. Abundance of antibiotic resistance genes in five municipal wastewater treatment plants in the Monastir Governorate, Tunisia. Environ. Pollut. 2016, 219, 353–358. [Google Scholar] [CrossRef]
- Oliveira, M.; Serrano, I.; Van Harten, S.; Bessa, L.J.; Bernardo, F.; da Costa, P.M. Fecal contamination of wastewater treatment plants in Portugal. Environ. Sci. Pollut. Res. 2016, 23, 14671–14675. [Google Scholar] [CrossRef]
- Niestępski, S.; Harnisz, M.; Ciesielski, S.; Korzeniewska, E.; Osińska, A. Environmental fate of Bacteroidetes, with particular emphasis on Bacteroides fragilis group bacteria and their specific antibiotic resistance genes, in activated sludge wastewater treatment plants. J. Hazard. Mater. 2020, 394, 122544. [Google Scholar] [CrossRef]
- Osińska, A.; Korzeniewska, E.; Harnisz, M.; Niestępski, S. The prevalence and characterization of antibiotic-resistant and virulent Escherichia coli strains in the municipal wastewater system and their environmental fate. Sci. Total Environ. 2017, 577, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Bessa, L.J.; Barbosa-Vasconcelos, A.; Mendes, Â.; Vaz-Pires, P.; Martins da Costa, P. High prevalence of multidrug-resistant Escherichia coli and Enterococcus spp. in river water, upstream and downstream of a wastewater treatment plant. J. Water Health 2014, 12, 426–435. [Google Scholar] [CrossRef] [PubMed]
- Maravić, A.; Skočibušić, M.; Fredotović, Ž.; Šamanić, I.; Cvjetan, S.; Knezović, M.; Puizina, J. Urban riverine environment is a source of multidrug-resistant and ESBL-producing clinically important Acinetobacter spp. Environ. Sci. Pollut. Res. 2016, 23, 3525–3535. [Google Scholar] [CrossRef] [PubMed]
- Hubeny, J.; Korzeniewska, E.; Buta-Hubeny, M.; Zieliński, W.; Rolbiecki, D.; Harnisz, M. Characterization of carbapenem resistance in environmental samples and Acinetobacter spp. isolates from wastewater and river water in Poland. Sci. Total Environ. 2022, 822, 153437. [Google Scholar] [CrossRef] [PubMed]
- Karkman, A.; Johnson, T.A.; Lyra, C.; Stedtfeld, R.D.; Tamminen, M.; Tiedje, J.M.; Virta, M. High-throughput quantification of antibiotic resistance genes from an urban wastewater treatment plant. FEMS Microbiol. Ecol. 2016, 92, fiw014. [Google Scholar] [CrossRef]
- Wang, J.; Chu, L.; Wojnárovits, L.; Takács, E. Occurrence and fate of antibiotics, antibiotic resistant genes (ARGs) and antibiotic resistant bacteria (ARB) in municipal wastewater treatment plant: An overview. Sci. Total Environ. 2020, 744, 140997. [Google Scholar] [CrossRef]
- Pärnänen, K.M.M.; Narciso-Da-Rocha, C.; Kneis, D.; Berendonk, T.U.; Cacace, D.; Do, T.T.; Elpers, C.; Fatta-Kassinos, D.; Henriques, I.; Jaeger, T.; et al. Antibiotic resistance in European wastewater treatment plants mirrors the pattern of clinical antibiotic resistance prevalence. Sci. Adv. 2019, 5, eaau9124. [Google Scholar] [CrossRef]
- ECDC. Antimicrobial consumption in the EU/EEA (ESAC-Net). Eur. Cent. Dis. Prev. Control 2021, 11–14. [Google Scholar]
- Duarte, D.J.; Oldenkamp, R.; Ragas, A.M.J. Modelling environmental antibiotic-resistance gene abundance: A meta-analysis. Sci. Total Environ. 2019, 659, 335–341. [Google Scholar] [CrossRef]
- Tang, J.; Bu, Y.; Zhang, X.-X.; Huang, K.; He, X.; Ye, L.; Shan, Z.; Ren, H. Metagenomic analysis of bacterial community composition and antibiotic resistance genes in a wastewater treatment plant and its receiving surface water. Ecotoxicol. Environ. Saf. 2016, 132, 260–269. [Google Scholar] [CrossRef]
- Zieliński, W.; Hubeny, J.; Buta-Hubeny, M.; Rolbiecki, D.; Harnisz, M.; Paukszto, Ł.; Korzeniewska, E. Metagenomics analysis of probable transmission of determinants of antibiotic resistance from wastewater to the environment—A case study. Sci. Total Environ. 2022, 827, 154354. [Google Scholar] [CrossRef] [PubMed]
- Bondarczuk, K.; Piotrowska-Seget, Z. Microbial diversity and antibiotic resistance in a final effluent-receiving lake. Sci. Total Environ. 2019, 650, 2951–2961. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Ram, B.; Sewwandi, H.; Honda, R.; Chaminda, T. Treatment enhances the prevalence of antibiotic-resistant bacteria and antibiotic resistance genes in the wastewater of Sri Lanka, and India. Environ. Res. 2020, 183. [Google Scholar] [CrossRef]
- Koczura, R.; Mokracka, J.; Jabłońska, L.; Gozdecka, E.; Kubek, M.; Kaznowski, A. Antimicrobial resistance of integron-harboring Escherichia coli isolates from clinical samples, wastewater treatment plant and river water. Sci. Total Environ. 2012, 414, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Korzeniewska, E.; Harnisz, M. Relationship between modification of activated sludge wastewater treatment and changes in antibiotic resistance of bacteria. Sci. Total Environ. 2018, 639, 304–315. [Google Scholar] [CrossRef] [PubMed]
- An, X.-L.; Chen, Q.-L.; Zhu, D.; Zhu, Y.-G.; Gillings, M.R.; Su, J.-Q. Impact of Wastewater Treatment on the Prevalence of Integrons and the Genetic Diversity of Integron Gene Cassettes. Appl. Environ. Microbiol. 2018, 84, e02766-17. [Google Scholar] [CrossRef]
- Wang, M.; Shen, W.; Yan, L.; Wang, X.H.; Xu, H. Stepwise impact of urban wastewater treatment on the bacterial community structure, antibiotic contents, and prevalence of antimicrobial resistance. Environ. Pollut. 2017, 231, 1578–1585. [Google Scholar] [CrossRef]
- Sabri, N.A.; Schmitt, H.; Van Der Zaan, B.; Gerritsen, H.W.; Zuidema, T.; Rijnaarts, H.H.M.; Langenhoff, A.A.M. Prevalence of antibiotics and antibiotic resistance genes in a wastewater effluent-receiving river in the Netherlands. J. Environ. Chem. Eng. 2020, 8, 102245. [Google Scholar] [CrossRef]
- Czekalski, N.; Berthold, T.; Caucci, S.; Egli, A.; Bürgmann, H. Increased Levels of Multiresistant Bacteria and Resistance Genes after Wastewater Treatment and Their Dissemination into Lake Geneva, Switzerland. Front. Microbiol. 2012, 3, 106. [Google Scholar] [CrossRef]
- Osińska, A.; Korzeniewska, E.; Harnisz, M.; Felis, E.; Bajkacz, S.; Jachimowicz, P.; Niestępski, S.; Konopka, I. Small-scale wastewater treatment plants as a source of the dissemination of antibiotic resistance genes in the aquatic environment. J. Hazard. Mater. 2020, 381, 121221. [Google Scholar] [CrossRef]
- Hocquet, D.; Muller, A.; Bertrand, X. What happens in hospitals does not stay in hospitals: Antibiotic-resistant bacteria in hospital wastewater systems. J. Hosp. Infect. 2016, 93, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Paulus, G.K.; Hornstra, L.M.; Alygizakis, N.; Slobodnik, J.; Thomaidis, N.; Medema, G. The impact of on-site hospital wastewater treatment on the downstream communal wastewater system in terms of antibiotics and antibiotic resistance genes. Int. J. Hyg. Environ. Health 2019, 222, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.-Q.; Ying, G.-G.; Pan, C.-G.; Liu, Y.-S.; Zhao, J.-L. Comprehensive Evaluation of Antibiotics Emission and Fate in the River Basins of China: Source Analysis, Multimedia Modeling, and Linkage to Bacterial Resistance. Environ. Sci. Technol. 2015, 49, 6772–6782. [Google Scholar] [CrossRef] [PubMed]
- Roberto, A.A.; Van Gray, J.B.; Engohang-Ndong, J.; Leff, L.G. Distribution and co-occurrence of antibiotic and metal resistance genes in biofilms of an anthropogenically impacted stream. Sci. Total Environ. 2019, 688, 437–449. [Google Scholar] [CrossRef]
- Zieliński, W.; Korzeniewska, E.; Harnisz, M.; Drzymała, J.; Felis, E.; Bajkacz, S. Wastewater treatment plants as a reservoir of integrase and antibiotic resistance genes—An epidemiological threat to workers and environment. Environ. Int. 2021, 156, 106641. [Google Scholar] [CrossRef]
- Bouki, C.; Venieri, D.; Diamadopoulos, E. Detection and fate of antibiotic resistant bacteria in wastewater treatment plants: A review. Ecotoxicol. Environ. Saf. 2013, 91, 1–9. [Google Scholar] [CrossRef]
- Hutinel, M.; Larsson, D.G.J.; Flach, C.-F. Antibiotic resistance genes of emerging concern in municipal and hospital wastewater from a major Swedish city. Sci. Total Environ. 2022, 812, 151433. [Google Scholar] [CrossRef]
- Alexander, J.; Hembach, N.; Schwartz, T. Evaluation of antibiotic resistance dissemination by wastewater treatment plant effluents with different catchment areas in Germany. Sci. Rep. 2020, 10, 8952. [Google Scholar] [CrossRef]
- Sinel, C.; Cacaci, M.; Meignen, P.; Guérin, F.; Davies, B.W.; Sanguinetti, M.; Giard, J.-C.; Cattoir, V. Subinhibitory Concentrations of Ciprofloxacin Enhance Antimicrobial Resistance and Pathogenicity of Enterococcus faecium. Antimicrob. Agents Chemother. 2017, 61, e02763-16. [Google Scholar] [CrossRef]
- Sung, J.Y. Molecular Characterization and Antimicrobial Susceptibility of Biofilm-forming Acinetobacter baumannii Clinical Isolates from Daejeon, Korea. Korean J. Clin. Lab. Sci. 2018, 50, 100–109. [Google Scholar] [CrossRef]
- Asadollahi, P.; Akbari, M.; Soroush, S.; Taherikalani, M.; Asadollahi, K.; Sayehmiri, K.; Maleki, A.; Maleki, M.H.; Karimi, P.; Emaneini, M. Antimicrobial resistance patterns and their encoding genes among Acinetobacter baumannii strains isolated from burned patients. Burns 2012, 38, 1198–1203. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Li, A.-D.; Yin, X.-L.; Zhang, T. The Prevalence of Integrons as the Carrier of Antibiotic Resistance Genes in Natural and Man-Made Environments. Environ. Sci. Technol. 2017, 51, 5721–5728. [Google Scholar] [CrossRef] [PubMed]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17. [Google Scholar] [CrossRef] [PubMed]
- Berglund, B. Environmental dissemination of antibiotic resistance genes and correlation to anthropogenic contamination with antibiotics. Infect. Ecol. Epidemiol. 2015, 5, 28564. [Google Scholar] [CrossRef]
- Deng, Y.; Bao, X.; Ji, L.; Chen, L.; Liu, J.; Miao, J.; Chen, D.; Bian, H.; Li, Y.; Yu, G. Resistance integrons: Class 1, 2 and 3 integrons. Ann. Clin. Microbiol. Antimicrob. 2015, 14, 1–11. [Google Scholar] [CrossRef]
- Huang, L.-Y.; Chen, T.-L.; Lu, P.-L.; Tsai, C.-A.; Cho, W.-L.; Chang, F.-Y.; Fung, C.-P.; Siu, L.K. Dissemination of multidrug-resistant, class 1 integron-carrying Acinetobacter baumannii isolates in Taiwan. Clin. Microbiol. Infect. 2008, 14, 1010–1019. [Google Scholar] [CrossRef]
- Amin, M.; Navidifar, T.; Shooshtari, F.S.; Goodarzi, H. Association of the genes encoding metallo-β-lactamase with the presence of integrons among multidrug-resistant clinical isolates of acinetobacter baumannii. Infect. Drug Resist. 2019, 12, 1171–1180. [Google Scholar] [CrossRef]
- Chen, J.; Li, H.; Yang, J.; Zhan, R.; Chen, A.; Yan, Y. Prevalence and Characterization of Integrons in Multidrug Resistant Acinetobacter baumannii in Eastern China: A Multiple-Hospital Study. Int. J. Environ. Res. Public Health 2015, 12, 10093–10105. [Google Scholar] [CrossRef]
- Ramírez, M.S.; Quiroga, C.; Centrón, D. Novel Rearrangement of a Class 2 Integron in Two Non-Epidemiologically Related Isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 2005, 49, 5179–5181. [Google Scholar] [CrossRef][Green Version]
- Martins, N.; Picão, R.C.; Adams-Sapper, S.; Riley, L.W.; Moreira, B.M. Association of Class 1 and 2 Integrons with Multidrug-Resistant Acinetobacter baumannii International Clones and Acinetobacter nosocomialis Isolates. Antimicrob. Agents Chemother. 2015, 59, 698–701. [Google Scholar] [CrossRef]
- Al-Kadmy, I.M.S.; Ibrahim, S.A.; Al-Saryi, N.; Aziz, S.N.; Besinis, A.; Hetta, H.F. Prevalence of Genes Involved in Colistin Resistance in Acinetobacter baumannii: First Report from Iraq. Microb. Drug Resist. 2020, 26, 616–622. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.Z. Molecular Characterization of Integrons and Their Association with Antibiotic Resistance in Acinetobacter baumannii Isolated from Hospitals in Jeddah. Appl. Biochem. Microbiol. 2021, 57, S64–S70. [Google Scholar] [CrossRef]
- Halaji, M.; Rezaei, A.; Zalipoor, M.; Faghri, J. Investigation of Class I, II, and III Integrons Among Acinetobacter Baumannii Isolates from Hospitalized Patients in Isfahan, Iran. Oman Med. J. 2018, 33, 37–42. [Google Scholar] [CrossRef]
- Shaheli, M.; Baseri Salehi, M.; Bahador, N. The influence of integrons on multidrug resistant acinetobacter spp. Isolated from environment and clinical samples. Trop. Biomed. 2018, 35, 354–364. [Google Scholar] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hubeny, J.; Korzeniewska, E.; Ciesielski, S.; Płaza, G.; Harnisz, M. The Resistome of ESKAPEE Pathogens in Untreated and Treated Wastewater: A Polish Case Study. Biomolecules 2022, 12, 1160. https://doi.org/10.3390/biom12081160
Hubeny J, Korzeniewska E, Ciesielski S, Płaza G, Harnisz M. The Resistome of ESKAPEE Pathogens in Untreated and Treated Wastewater: A Polish Case Study. Biomolecules. 2022; 12(8):1160. https://doi.org/10.3390/biom12081160
Chicago/Turabian StyleHubeny, Jakub, Ewa Korzeniewska, Sławomir Ciesielski, Grażyna Płaza, and Monika Harnisz. 2022. "The Resistome of ESKAPEE Pathogens in Untreated and Treated Wastewater: A Polish Case Study" Biomolecules 12, no. 8: 1160. https://doi.org/10.3390/biom12081160
APA StyleHubeny, J., Korzeniewska, E., Ciesielski, S., Płaza, G., & Harnisz, M. (2022). The Resistome of ESKAPEE Pathogens in Untreated and Treated Wastewater: A Polish Case Study. Biomolecules, 12(8), 1160. https://doi.org/10.3390/biom12081160







