Conformational Plasticity of Centrin 1 from Toxoplasma gondii in Binding to the Centrosomal Protein SFI1
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Protein Production and Peptide Synthesis
2.3. Size Exclusion Chromatography
2.4. Isothermal Titration Calorimetry Experiments
2.5. Circular Dichroism Spectroscopy
2.6. Fluorescence Spectroscopy
2.7. Nuclear Magnetic Resonance Spectroscopy
3. Results
3.1. Energetics of TgCEN1 Binding to TgSFI1 Repeats
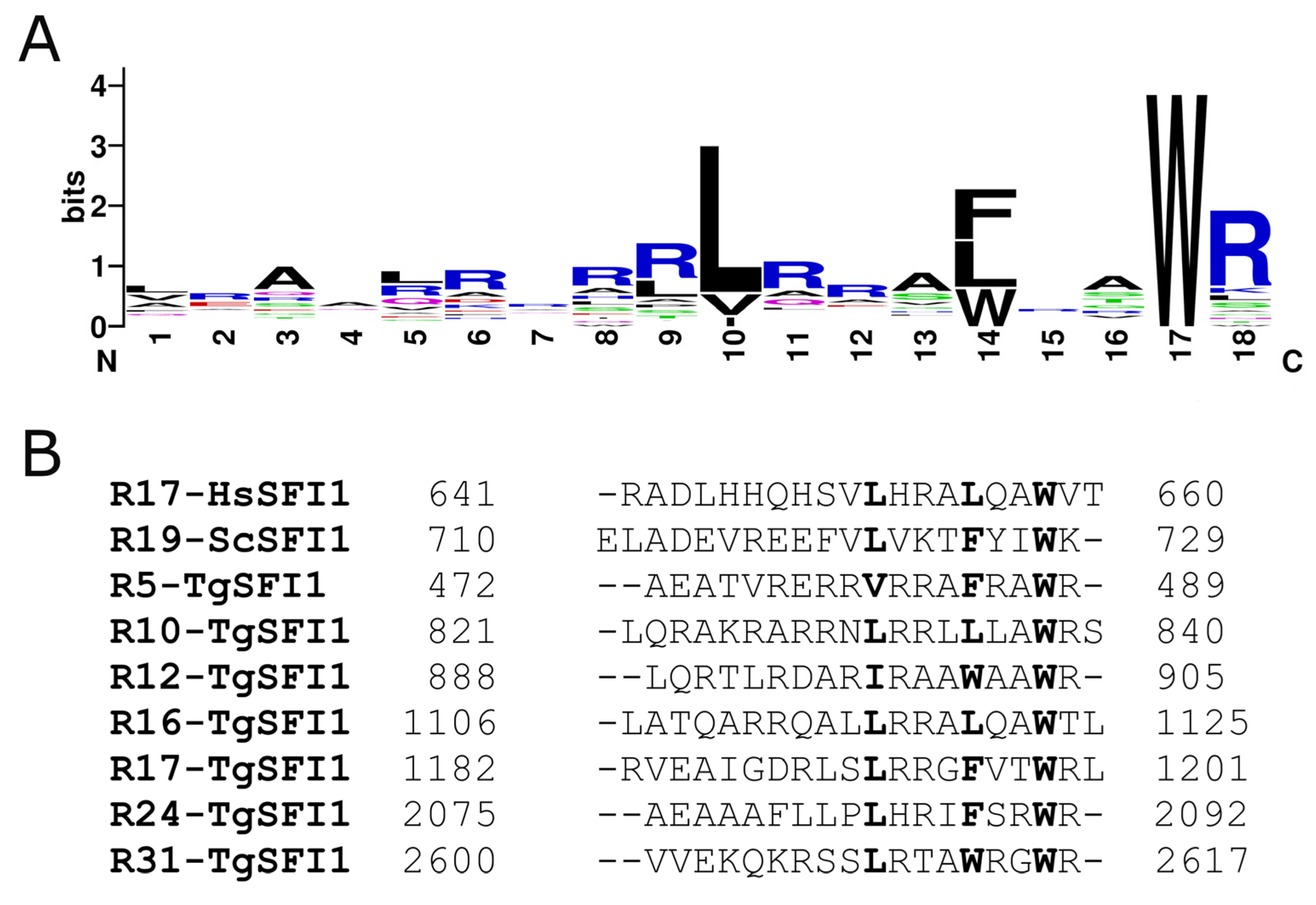
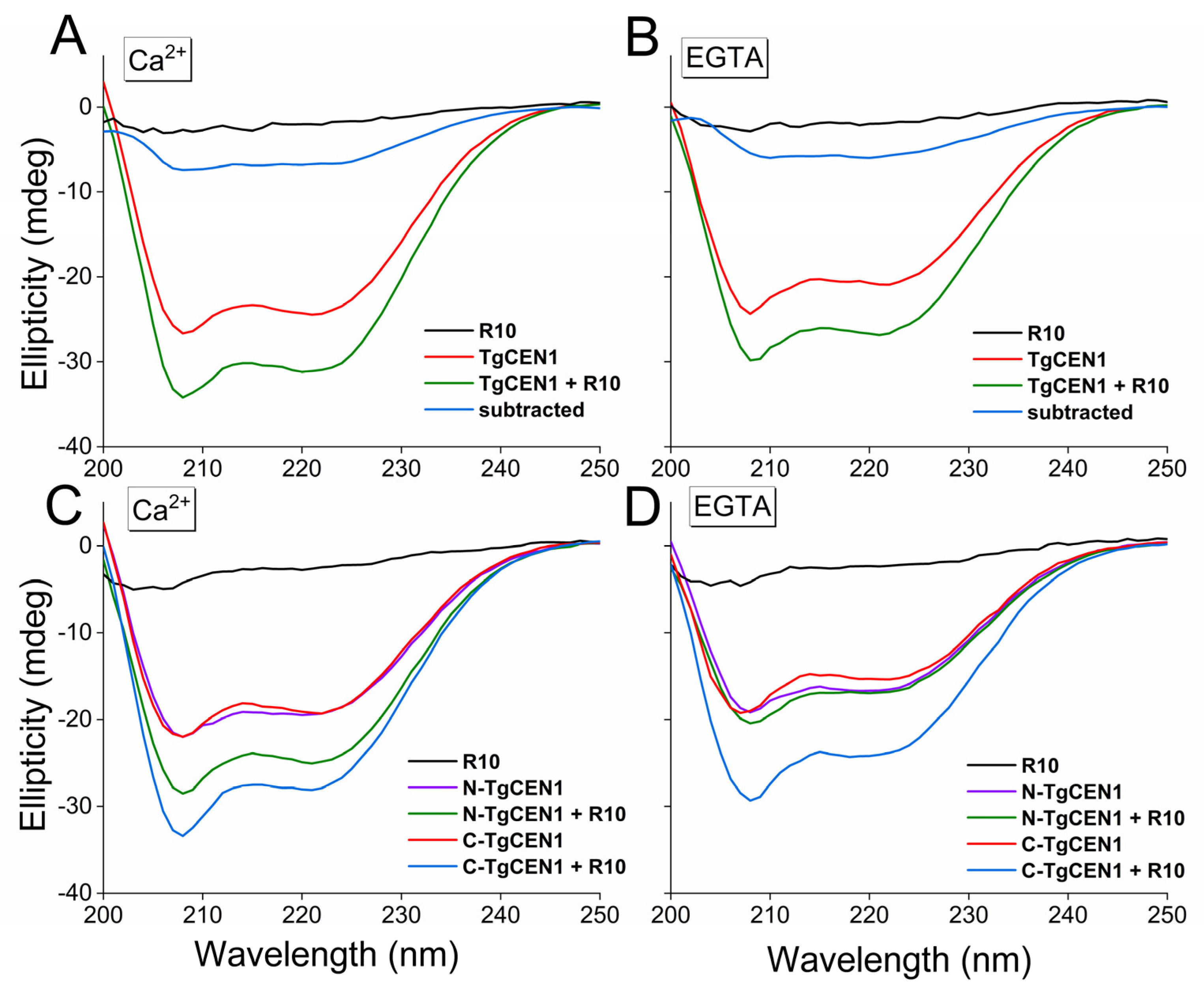
3.2. Conformational Features of the Interaction
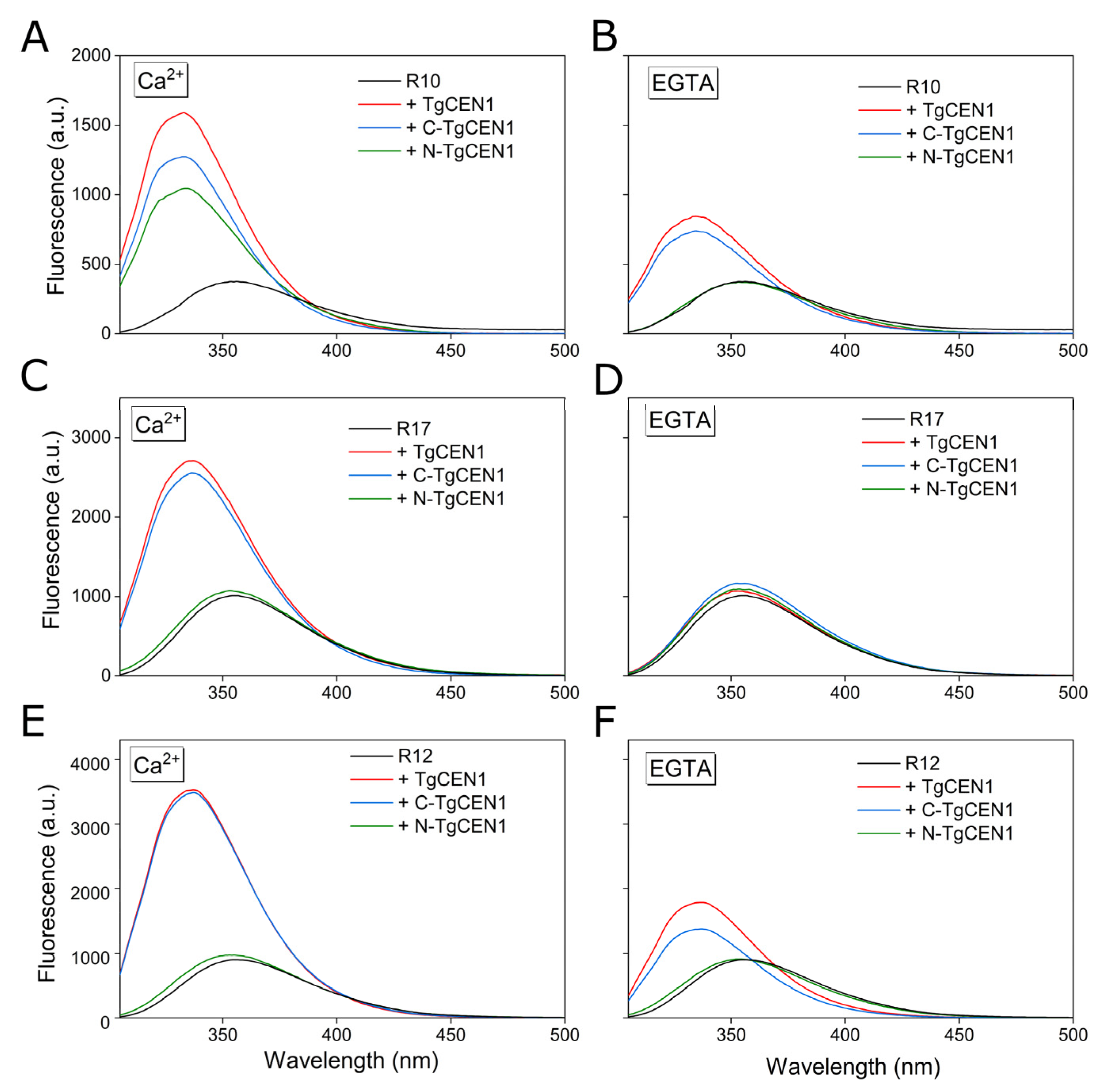
3.3. Trp Residue of TgSFI1 Repeats Is Embedded in TgCEN1 Hydrophobic Pocket
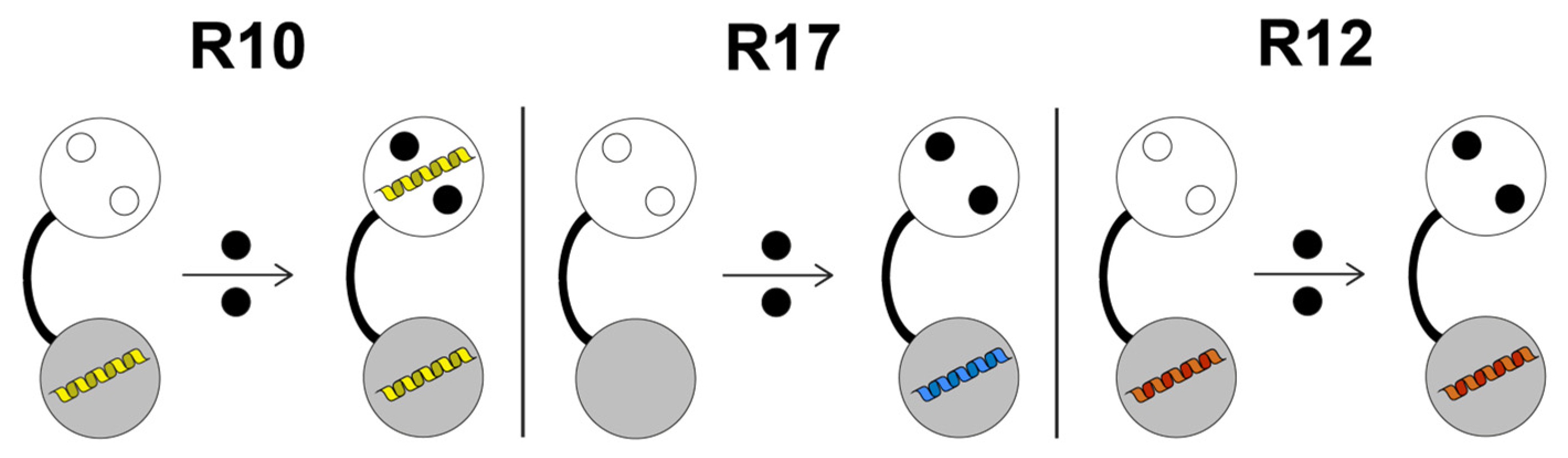
4. Discussion
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Paoletti, A.; Moudjou, M.; Paintrand, M.; Salisbury, J.L.; Bornens, M. Most of centrin in animal cells is not centrosome-associated and centrosomal centrin is confined to the distal lumen of centrioles. J. Cell Sci. 1996, 109 Pt 13, 3089–3102. [Google Scholar] [CrossRef] [PubMed]
- Salisbury, J.L.; Baron, A.T.; Sanders, M.A. The centrin-based cytoskeleton of Chlamydomonas reinhardtii: Distribution in interphase and mitotic cells. J. Cell Biol. 1988, 107, 635–641. [Google Scholar] [CrossRef] [PubMed]
- Salisbury, J.L. A mechanistic view on the evolutionary origin for centrin-based control of centriole duplication. J. Cell. Physiol. 2007, 213, 420–428. [Google Scholar] [CrossRef] [PubMed]
- Nishi, R.; Okuda, Y.; Watanabe, E.; Mori, T.; Iwai, S.; Masutani, C.; Sugasawa, K.; Hanaoka, F. Centrin 2 stimulates nucleotide excision repair by interacting with xeroderma pigmentosum group C protein. Mol. Cell Biol. 2005, 25, 5664–5674. [Google Scholar] [CrossRef] [PubMed]
- Jani, D.; Lutz, S.; Marshall, N.J.; Fischer, T.; Kohler, A.; Ellisdon, A.M.; Hurt, E.; Stewart, M. Sus1, Cdc31, and the Sac3 CID region form a conserved interaction platform that promotes nuclear pore association and mRNA export. Mol. Cell 2009, 33, 727–737. [Google Scholar] [CrossRef]
- Pedretti, M.; Conter, C.; Dominici, P.; Astegno, A. SAC3B is a target of CML19, the centrin 2 of Arabidopsis thaliana. Biochem. J. 2020, 477, 173–189. [Google Scholar] [CrossRef]
- Liang, L.; Flury, S.; Kalck, V.; Hohn, B.; Molinier, J. CENTRIN2 interacts with the Arabidopsis homolog of the human XPC protein (AtRAD4) and contributes to efficient synthesis-dependent repair of bulky DNA lesions. Plant Mol. Biol. 2006, 61, 345–356. [Google Scholar] [CrossRef]
- Miron, S.; Durand, D.; Chilom, C.; Perez, J.; Craescu, C.T. Binding of calcium, magnesium, and target peptides to Cdc31, the centrin of yeast Saccharomyces cerevisiae. Biochemistry 2011, 50, 6409–6422. [Google Scholar] [CrossRef]
- Baum, P.; Furlong, C.; Byers, B. Yeast gene required for spindle pole body duplication: Homology of its product with Ca2+-binding proteins. Proc. Natl. Acad. Sci. USA 1986, 83, 5512–5516. [Google Scholar] [CrossRef]
- Huang, B.; Mengersen, A.; Lee, V.D. Molecular cloning of cDNA for caltractin, a basal body-associated Ca2+-binding protein: Homology in its protein sequence with calmodulin and the yeast CDC31 gene product. J. Cell Biol. 1988, 107, 133–140. [Google Scholar] [CrossRef]
- La Verde, V.; Trande, M.; D’Onofrio, M.; Dominici, P.; Astegno, A. Binding of calcium and target peptide to calmodulin-like protein CML19, the centrin 2 of Arabidopsis thaliana. Int. J. Biol. Macromol. 2018, 108, 1289–1299. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Sanz, J.; Yang, A.; Blouquit, Y.; Duchambon, P.; Assairi, L.; Craescu, C.T. Binding of human centrin 2 to the centrosomal protein hSfi1. FEBS J. 2006, 273, 4504–4515. [Google Scholar] [CrossRef] [PubMed]
- Kilmartin, J.V. Sfi1p has conserved centrin-binding sites and an essential function in budding yeast spindle pole body duplication. J. Cell Biol. 2003, 162, 1211–1221. [Google Scholar] [CrossRef] [PubMed]
- Jani, D.; Lutz, S.; Hurt, E.; Laskey, R.A.; Stewart, M.; Wickramasinghe, V.O. Functional and structural characterization of the mammalian TREX-2 complex that links transcription with nuclear messenger RNA export. Nucleic Acids Res. 2012, 40, 4562–4573. [Google Scholar] [CrossRef] [PubMed]
- Araki, M.; Masutani, C.; Takemura, M.; Uchida, A.; Sugasawa, K.; Kondoh, J.; Ohkuma, Y.; Hanaoka, F. Centrosome protein centrin 2/caltractin 1 is part of the xeroderma pigmentosum group C complex that initiates global genome nucleotide excision repair. J. Biol. Chem. 2001, 276, 18665–18672. [Google Scholar] [CrossRef] [PubMed]
- Gogendeau, D.; Klotz, C.; Arnaiz, O.; Malinowska, A.; Dadlez, M.; de Loubresse, N.G.; Ruiz, F.; Koll, F.; Beisson, J. Functional diversification of centrins and cell morphological complexity. J. Cell Sci. 2008, 121, 65–74. [Google Scholar] [CrossRef][Green Version]
- Pedretti, M.; Bombardi, L.; Conter, C.; Favretto, F.; Dominici, P.; Astegno, A. Structural Basis for the Functional Diversity of Centrins: A Focus on Calcium Sensing Properties and Target Recognition. Int. J. Mol. Sci. 2021, 22, 12173. [Google Scholar] [CrossRef]
- Díaz Casas, A.; Chazin, W.J.; Pastrana-Ríos, B. Prp40 Homolog A Is a Novel Centrin Target. Biophys. J. 2017, 112, 2529–2539. [Google Scholar] [CrossRef][Green Version]
- Conter, C.; Bombardi, L.; Pedretti, M.; Favretto, F.; Di Matteo, A.; Dominici, P.; Astegno, A. The interplay of self-assembly and target binding in centrin 1 from Toxoplasma gondii. Biochem. J. 2021, 478, 2571–2587. [Google Scholar] [CrossRef]
- Popescu, A.; Miron, S.; Blouquit, Y.; Duchambon, P.; Christova, P.; Craescu, C.T. Xeroderma pigmentosum group C protein possesses a high affinity binding site to human centrin 2 and calmodulin. J. Biol. Chem. 2003, 278, 40252–40261. [Google Scholar] [CrossRef]
- Hu, H.; Sheehan, J.H.; Chazin, W.J. The Mode of Action of Centrin: Binding of Ca2+ and a peptide fragment of Kar1p to the c-terminal domain. J. Biol. Chem. 2004, 279, 50895–50903. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Chazin, W.J. Unique features in the C-terminal domain provide caltractin with target specificity. J. Mol. Biol. 2003, 330, 473–484. [Google Scholar] [CrossRef]
- Troilo, F.; Pedretti, M.; Travaglini-Allocatelli, C.; Astegno, A.; Di Matteo, A. Rapid kinetics of calcium dissociation from plant calmodulin and calmodulin-like proteins and effect of target peptides. Biochem. Biophys. Res. Commun. 2022, 590, 103–108. [Google Scholar] [CrossRef]
- Hu, K.; Johnson, J.; Florens, L.; Fraunholz, M.; Suravajjala, S.; DiLullo, C.; Yates, J.; Roos, D.S.; Murray, J.M. Cytoskeletal components of an invasion machine--the apical complex of Toxoplasma gondii. PLoS Pathog. 2006, 2, e13. [Google Scholar] [CrossRef]
- Lentini, G.; Dubois, D.J.; Maco, B.; Soldati-Favre, D.; Frénal, K. The roles of Centrin 2 and Dynein Light Chain 8a in apical secretory organelles discharge of Toxoplasma gondii. Traffic 2019, 20, 583–600. [Google Scholar] [CrossRef] [PubMed]
- Leung, J.M.; Liu, J.; Wetzel, L.A.; Hu, K. Centrin2 from the human parasite Toxoplasma gondii is required for its invasion and intracellular replication. J. Cell Sci. 2019, 132, jcs228791. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Sandercock, A.M.; Conduit, P.; Robinson, C.V.; Williams, R.L.; Kilmartin, J.V. Structural role of Sfi1p–centrin filaments in budding yeast spindle pole body duplication. J. Cell Biol. 2006, 173, 867–877. [Google Scholar] [CrossRef]
- Kodani, A.; Moyer, T.; Chen, A.; Holland, A.; Walsh, C.A.; Reiter, J.F. SFI1 promotes centriole duplication by recruiting USP9X to stabilize the microcephaly protein STIL. J. Cell Biol. 2019, 218, 2185–2197. [Google Scholar] [CrossRef]
- Suvorova, E.S.; Francia, M.; Striepen, B.; White, M.W. A novel bipartite centrosome coordinates the apicomplexan cell cycle. PLoS Biol. 2015, 13, e1002093. [Google Scholar] [CrossRef]
- Tomasina, R.; González, F.C.; Francia, M.E. Structural and Functional Insights into the Microtubule Organizing Centers of Toxoplasma gondii and Plasmodium spp. Microorganisms 2021, 9, 2503. [Google Scholar] [CrossRef]
- Bombardi, L.; Pedretti, M.; Conter, C.; Dominici, P.; Astegno, A. Distinct Calcium Binding and Structural Properties of Two Centrin Isoforms from Toxoplasma gondii. Biomolecules 2020, 10, 1142. [Google Scholar] [CrossRef] [PubMed]
- Vallone, R.; La Verde, V.; D’Onofrio, M.; Giorgetti, A.; Dominici, P.; Astegno, A. Metal binding affinity and structural properties of calmodulin-like protein 14 from Arabidopsis thaliana. Protein Sci. 2016, 25, 1461–1471. [Google Scholar] [CrossRef] [PubMed]
- La Verde, V.; Dominici, P.; Astegno, A. Determination of Hydrodynamic Radius of Proteins by Size Exclusion Chromatography. Bio-Protocol 2017, 7, e2230. [Google Scholar] [CrossRef] [PubMed]
- Astegno, A.; Bonza, M.C.; Vallone, R.; La Verde, V.; D’Onofrio, M.; Luoni, L.; Molesini, B.; Dominici, P. Arabidopsis calmodulin-like protein CML36 is a calcium (Ca(2+)) sensor that interacts with the plasma membrane Ca(2+)-ATPase isoform ACA8 and stimulates its activity. J. Biol. Chem. 2017, 292, 15049–15061. [Google Scholar] [CrossRef] [PubMed]
- Trande, M.; Pedretti, M.; Bonza, M.C.; Di Matteo, A.; D’Onofrio, M.; Dominici, P.; Astegno, A. Cation and peptide binding properties of CML7, a calmodulin-like protein from Arabidopsis thaliana. J. Inorg. Biochem. 2019, 199, 110796. [Google Scholar] [CrossRef] [PubMed]
- Astegno, A.; La Verde, V.; Marino, V.; Dell’Orco, D.; Dominici, P. Biochemical and biophysical characterization of a plant calmodulin: Role of the N- and C-lobes in calcium binding, conformational change, and target interaction. Biochim. Biophys. Acta (BBA)—Proteins Proteom. 2016, 1864, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef]
- Pérez Santero, S.; Favretto, F.; Zanzoni, S.; Chignola, R.; Assfalg, M.; D’Onofrio, M. Effects of macromolecular crowding on a small lipid binding protein probed at the single-amino acid level. Arch. Biochem. Biophys. 2016, 606, 99–110. [Google Scholar] [CrossRef]
- Vranken, W.F.; Boucher, W.; Stevens, T.J.; Fogh, R.H.; Pajon, A.; Llinas, M.; Ulrich, E.L.; Markley, J.L.; Ionides, J.; Laue, E.D. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins 2005, 59, 687–696. [Google Scholar] [CrossRef]
- Martinez-Sanz, J.; Assairi, L. New insights into the interaction of centrin with Sfi1. Biochim. Biophys. Acta 2016, 1864, 319–330. [Google Scholar] [CrossRef]
- Schneider, T.D.; Stephens, R.M. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 1990, 18, 6097–6100. [Google Scholar] [CrossRef]
- Favretto, F.; Ceccon, A.; Zanzoni, S.; D’Onofrio, M.; Ragona, L.; Molinari, H.; Assfalg, M. The unique ligand binding features of subfamily-II iLBPs with respect to bile salts and related drugs. Prostaglandins Leukot. Essent. Fat. Acids 2015, 95, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Latham, M.P.; Zimmermann, G.R.; Pardi, A. NMR chemical exchange as a probe for ligand-binding kinetics in a theophylline-binding RNA aptamer. J. Am. Chem. Soc. 2009, 131, 5052–5053. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Favretto, F.; Flores, D.; Baker, J.D.; Strohaker, T.; Andreas, L.B.; Blair, L.J.; Becker, S.; Zweckstetter, M. Catalysis of proline isomerization and molecular chaperone activity in a tug-of-war. Nat. Commun. 2020, 11, 6046. [Google Scholar] [CrossRef] [PubMed]
- Breeze, A.L. Isotope-filtered NMR methods for the study of biomolecular structure and interactions. Prog. Nucl. Magn. Reson. Spectrosc. 2000, 36, 323–372. [Google Scholar] [CrossRef]
- Konrat, R. NMR contributions to structural dynamics studies of intrinsically disordered proteins. J. Magn. Reson. 2014, 241, 74–85. [Google Scholar] [CrossRef]
- La Verde, V.; Dominici, P.; Astegno, A. Towards Understanding Plant Calcium Signaling through Calmodulin-Like Proteins: A Biochemical and Structural Perspective. Int. J. Mol. Sci. 2018, 19, 1331. [Google Scholar] [CrossRef]
- Grecu, D.; Assairi, L. CK2 phosphorylation of human centrins 1 and 2 regulates their binding to the DNA repair protein XPC, the centrosomal protein Sfi1 and the phototransduction protein transducin β. FEBS Open Bio 2014, 4, 407–419. [Google Scholar] [CrossRef]
- Thompson, J.R.; Ryan, Z.C.; Salisbury, J.L.; Kumar, R. The Structure of the Human Centrin 2-Xeroderma Pigmentosum Group C Protein Complex. J. Biol. Chem. 2006, 281, 18746–18752. [Google Scholar] [CrossRef]
- Azimzadeh, J.; Hergert, P.; Delouvée, A.; Euteneuer, U.; Formstecher, E.; Khodjakov, A.; Bornens, M. hPOC5 is a centrin-binding protein required for assembly of full-length centrioles. J. Cell Biol. 2009, 185, 101–114. [Google Scholar] [CrossRef]
- Charbonnier, J.B.; Renaud, E.; Miron, S.; Le Du, M.H.; Blouquit, Y.; Duchambon, P.; Christova, P.; Shosheva, A.; Rose, T.; Angulo, J.F.; et al. Structural, thermodynamic, and cellular characterization of human centrin 2 interaction with xeroderma pigmentosum group C protein. J. Mol. Biol. 2007, 373, 1032–1046. [Google Scholar] [CrossRef] [PubMed]
- Matei, E.; Miron, S.; Blouquit, Y.; Duchambon, P.; Durussel, I.; Cox, J.A.; Craescu, C.T. C-terminal half of human centrin 2 behaves like a regulatory EF-hand domain. Biochemistry 2003, 42, 1439–1450. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.; Miron, S.; Mouawad, L.; Duchambon, P.; Blouquit, Y.; Craescu, C.T. Flexibility and Plasticity of Human Centrin 2 Binding to the Xeroderma Pigmentosum Group C Protein (XPC) from Nuclear Excision Repair. Biochemistry 2006, 45, 3653–3663. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Sanz, J.; Kateb, F.; Assairi, L.; Blouquit, Y.; Bodenhausen, G.; Abergel, D.; Mouawad, L.; Craescu, C.T. Structure, dynamics and thermodynamics of the human centrin 2/hSfi1 complex. J. Mol. Biol. 2010, 395, 191–204. [Google Scholar] [CrossRef] [PubMed]



| Buffer | n | Kd (µM) | ΔH (kcal mol−1) | TΔS (kcal mol−1) | |
|---|---|---|---|---|---|
| TgCEN1 + R10 | CaCl2 | n1 = 0.7 ± 0.1 | 0.014 ± 0.002 | −19.1 ± 0.4 | −6.3 ± 0.4 |
| n2 = 0.9 ± 0.1 | 2.5 ± 0.3 | −12.1 ± 0.3 | −4.4 ± 0.3 | ||
| EGTA | 0.8 ± 0.1 | 0.8 ± 0.1 | −24.7 ± 0.4 | −16.4 ± 0.4 | |
| N-TgCEN1 + R10 | CaCl2 | 0.7 ± 0.1 | 1.2 ± 0.4 | −12.6 ± 0.9 | −4.4 ± 1.0 |
| EGTA | - | - | - | - | |
| C-TgCEN1 + R10 | CaCl2 | 0.7 ± 0.1 | 0.027 ± 0.005 | −15.8 ± 0.1 | −5.5 ± 0.5 |
| EGTA | 0.7 ± 0.1 | 1.8 ± 0.1 | −19.3 ± 0.1 | −11.5 ± 0.7 | |
| TgCEN1 + R16 | CaCl2 | n1 = 0.7 ± 0.1 | 0.009 ± 0.001 | −20.9 ± 2.5 | −9.9 ± 2.4 |
| n2 = 0.8 ± 0.1 | 0.5 ± 0.1 | −11.8 ± 2.7 | −3.3 ± 1.7 | ||
| EGTA | 0.9 ± 0.2 | 3.4 ± 0.3 | −26.2 ± 3.4 | −18.7 ± 3.3 | |
| N-TgCEN1 + R16 | CaCl2 | 0.7 ± 0.1 | 0.4 ± 0.1 | −11.6 ± 0.6 | −2.8 ± 0.7 |
| EGTA | - | - | - | - | |
| C-TgCEN1 + R16 | CaCl2 | 0.7 ± 0.1 | 0.006 ± 0.001 | −18.8 ± 1.5 | −8.9 ± 1.6 |
| EGTA | 0.8 ± 0.1 | 9.1 ± 0.8 | −22.4 ± 0.6 | −15.7 ± 2.2 | |
| TgCEN1 + R17 | CaCl2 | 0.7 ± 0.1 | 0.8 ± 0.2 | −18.1 ± 0.5 | −9.7 ± 0.7 |
| EGTA | - | - | - | - | |
| N-TgCEN1 + R17 | CaCl2 | - | - | - | - |
| EGTA | - | - | - | - | |
| C-TgCEN1 + R17 | CaCl2 | 0.9 ± 0.1 | 2.7 ± 0.1 | −16.9 ± 0.2 | −9.3 ± 0.3 |
| EGTA | - | - | - | - | |
| TgCEN1 + R24 | CaCl2 | 0.9 ± 0.1 | 3.4 ± 0.6 | −8.9 ± 0.9 | −2.1 ± 0.7 |
| EGTA | - | - | - | - | |
| N-TgCEN1 + R24 | CaCl2 | - | - | - | - |
| EGTA | - | - | - | - | |
| C-TgCEN1 + R24 | CaCl2 | 0.8 ± 0.1 | 2.6 ± 0.3 | −8.6 ± 0.2 | −0.9 ± 0.3 |
| EGTA | - | - | - | - | |
| TgCEN1 + R31 | CaCl2 | 0.8 ± 0.1 | 2.9 ± 0.1 | −24.2 ± 1.4 | −16.6 ± 1.5 |
| EGTA | - | - | - | - | |
| N-TgCEN1 + R31 | CaCl2 | - | - | - | - |
| EGTA | - | - | - | - | |
| C-TgCEN1 + R31 | CaCl2 | 0.8 ± 0.1 | 1.9 ± 0.1 | −22.2 ± 0.3 | −14.4 ± 0.3 |
| EGTA | - | - | - | - | |
| TgCEN1 + R5 | CaCl2 | 0.7 ± 0.1 | 0.5 ± 0.1 | −26.4 ± 2.4 | −22.8 ± 1.8 |
| EGTA | 0.6 ± 0.1 | 36.9 ± 6.6 | −24.2 ± 2.1 | −10.6 ± 2.3 | |
| N-TgCEN1 + R5 | CaCl2 | - | - | - | - |
| EGTA | - | - | - | - | |
| C-TgCEN1 + R5 | CaCl2 | 0.7 ±0.1 | 0.9 ± 0.1 | −23.3 ± 0.1 | −15.1 ± 0.3 |
| EGTA | 0.9 ± 0.3 | 47.8 ± 4.9 | −10.2 ± 3.1 | −14.4 ± 3.2 | |
| TgCEN1 + R12 | CaCl2 | 0.9 ± 0.1 | 0.5 ± 0.1 | −21.5 ± 1.3 | −12.7 ± 1.2 |
| EGTA | 1.1 ± 0.1 | 29.8 ± 3.4 | −20.4 ± 2.4 | −14.2 ± 2.5 | |
| N-TgCEN1 + R12 | CaCl2 | - | - | - | - |
| EGTA | - | - | - | - | |
| C-TgCEN1 + R12 | CaCl2 | 0.8 ± 0.1 | 0.4 ± 0.1 | −18.5 ± 0.1 | −9.8 ± 0.5 |
| EGTA | 1.2 ± 0.5 | 69.9 ± 7.0 | −13.3 ± 1.9 | −7.6 ± 2.9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bombardi, L.; Favretto, F.; Pedretti, M.; Conter, C.; Dominici, P.; Astegno, A. Conformational Plasticity of Centrin 1 from Toxoplasma gondii in Binding to the Centrosomal Protein SFI1. Biomolecules 2022, 12, 1115. https://doi.org/10.3390/biom12081115
Bombardi L, Favretto F, Pedretti M, Conter C, Dominici P, Astegno A. Conformational Plasticity of Centrin 1 from Toxoplasma gondii in Binding to the Centrosomal Protein SFI1. Biomolecules. 2022; 12(8):1115. https://doi.org/10.3390/biom12081115
Chicago/Turabian StyleBombardi, Luca, Filippo Favretto, Marco Pedretti, Carolina Conter, Paola Dominici, and Alessandra Astegno. 2022. "Conformational Plasticity of Centrin 1 from Toxoplasma gondii in Binding to the Centrosomal Protein SFI1" Biomolecules 12, no. 8: 1115. https://doi.org/10.3390/biom12081115
APA StyleBombardi, L., Favretto, F., Pedretti, M., Conter, C., Dominici, P., & Astegno, A. (2022). Conformational Plasticity of Centrin 1 from Toxoplasma gondii in Binding to the Centrosomal Protein SFI1. Biomolecules, 12(8), 1115. https://doi.org/10.3390/biom12081115






