Glioblastoma Stem-Like Cells (GSCs) with Mesenchymal Signature: Lipid Profiles of Mobile Lipids Obtained with MRS before and after Radio/Chemical Treatments
Abstract
1. Introduction
2. Materials and Methods
2.1. GSCs Isolation, Cell Culture
2.2. 1H MRS Cell Sample Preparation
2.3. Etomoxir Treatment
2.4. Irradiation Treatments
2.5. 1H MRS Measurements
2.6. Statistical Analysis
3. Results
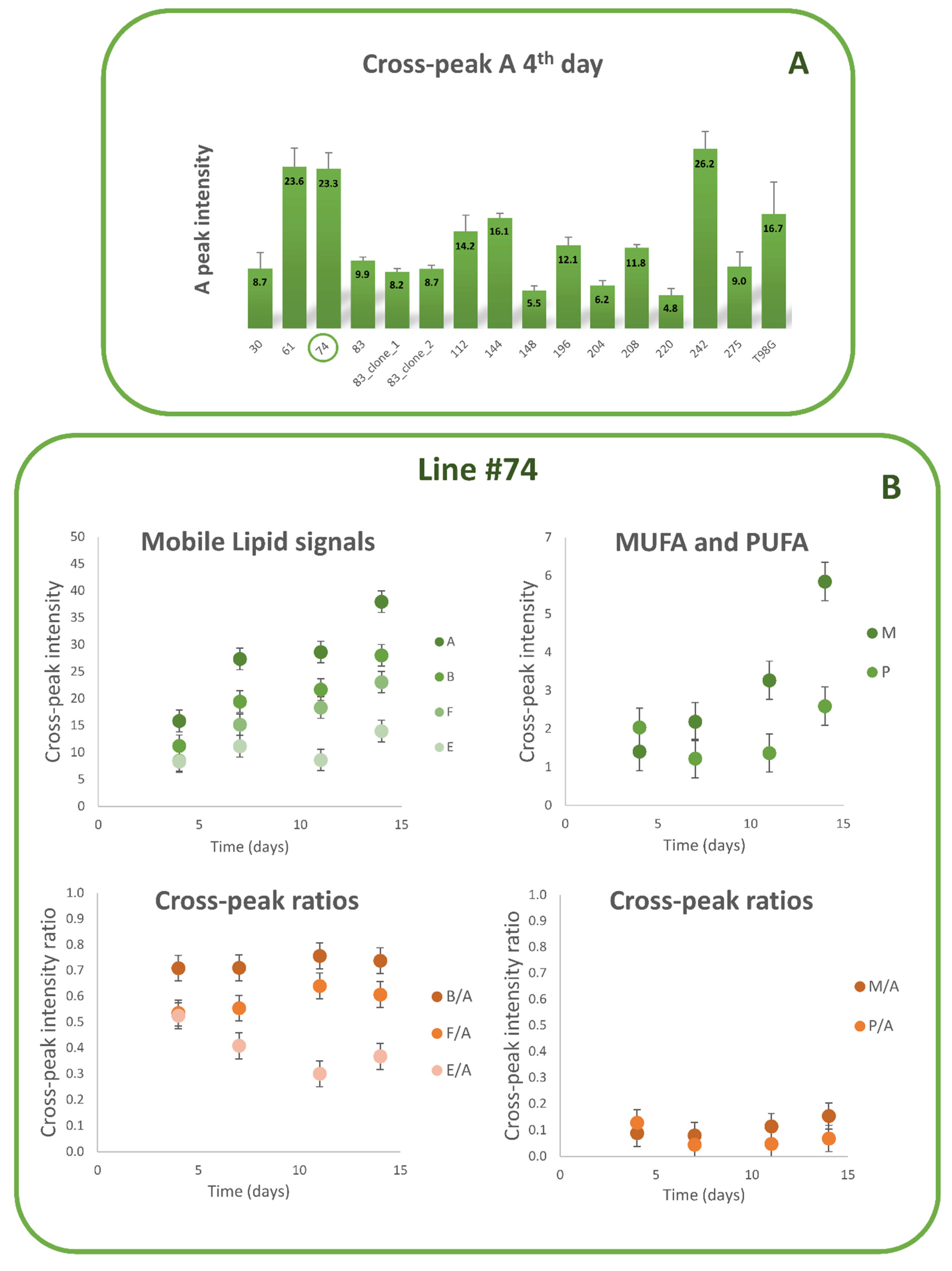
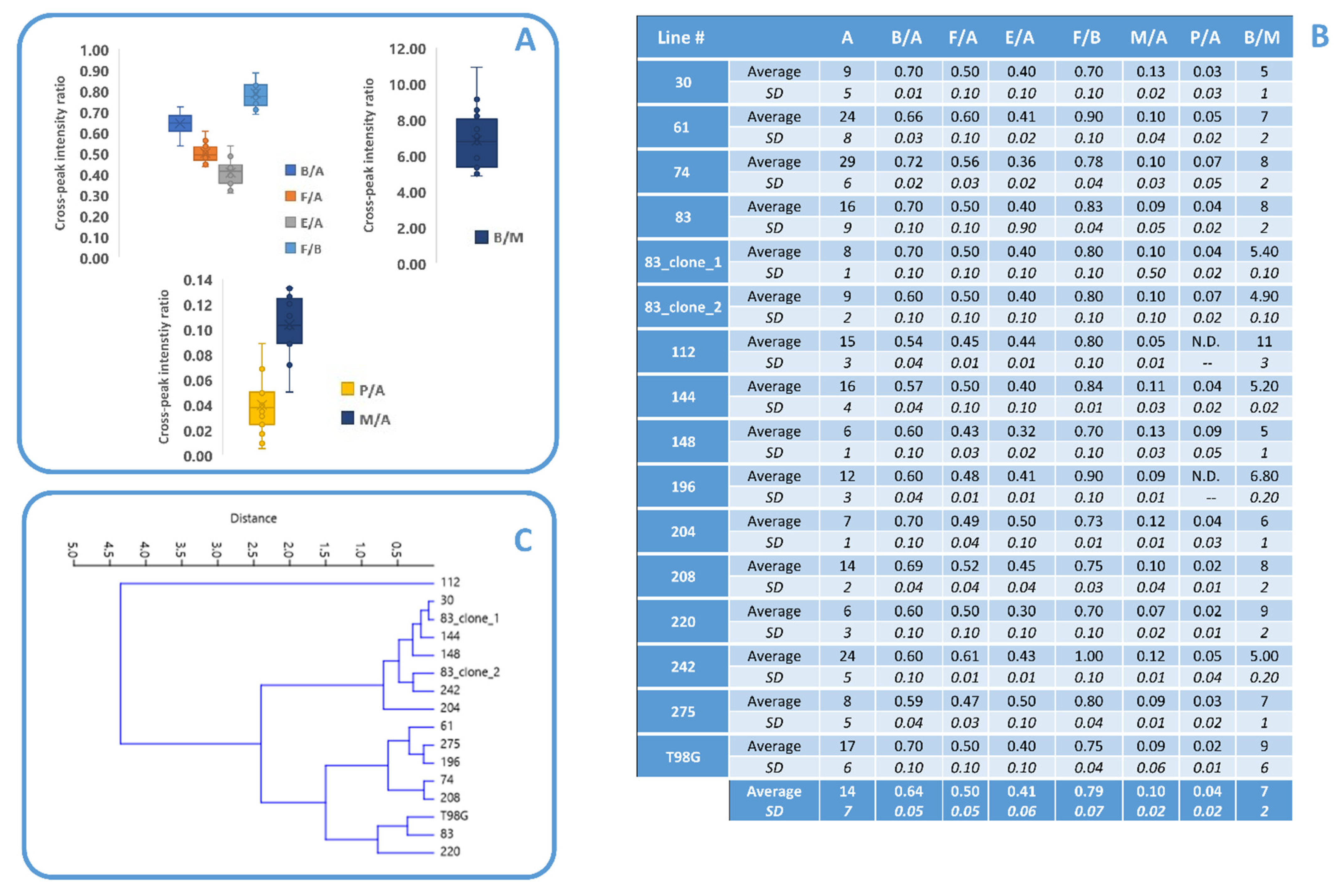
3.1. Lipid Metabolism in MES GSCs Lines
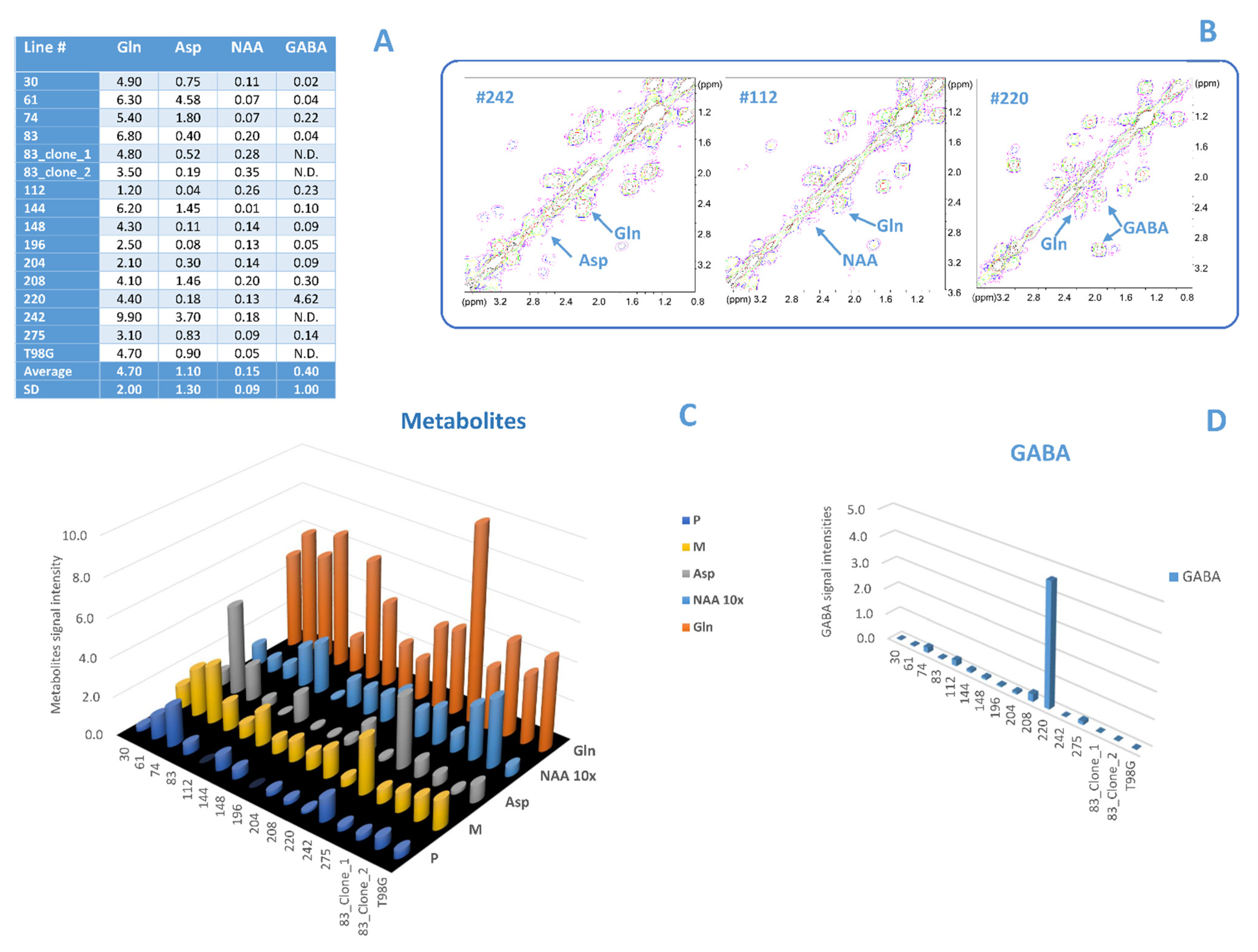
3.2. Comparison between Lipid and No Lipid Metabolism
3.3. Etomoxir and Irradiation Treatment
4. Discussion
4.1. Comparison between Lipid and No Lipid Metabolism
4.2. Irradiation and Etomoxir Treatment
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Omuro, A. Glioblastoma and Other Malignant Gliomas: A Clinical Review. JAMA 2013, 310, 1842–1850. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Hu, B.; Hu, X.; Kim, H.; Squatrito, M.; Scarpace, L.; de Carvalho, A.C.; Lyu, S.; Li, P.; Li, Y.; et al. Tumor evolution of glioma intrinsic gene expression subtype associates with immunological changes in the microenvironment. Cancer Cell. 2017, 32, 42–56. [Google Scholar] [CrossRef] [PubMed]
- Molenaar, R.J.; Wilmink, J.W. IDH1/2 Mutations in Cancer Stem Cells and Their Implications for Differentiation Therapy. J. Histochem. Cytochem. 2022, 70, 83–97. [Google Scholar] [CrossRef]
- Prager, B.C.; Bhargava, S.; Mahadev, V.; Hubert, C.G.; Rich, J.N. Glioblastoma Stem Cells: Driving Resilience through Chaos. Trends Cancer 2020, 6, 223–235. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.; Hasan, M.R. Cancer Metabolism and Drug Resistance. Lipid metabolism in cancer. FEBS J. 2015, 279, 2610–2623. [Google Scholar]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Lathia, J.D.; Mack, S.C.; Mulkearns-Hubert, E.E.; Valentim, C.C.L.; Rich, J.N. Cancer stem cells in glioblastoma. Genes Dev. 2015, 29, 1203–1217. [Google Scholar] [CrossRef] [PubMed]
- Hira, V.V.; Van Noorden, C.J.; Molenaar, R.J. CXCR4 Antagonists as Stem Cell Mobilizers and Therapy Sensitizers for Acute Myeloid Leukemia and Glioblastoma? Biology 2020, 9, 31. [Google Scholar] [CrossRef]
- Hira, V.V.; Molenaar, R.J.; Breznik, B.; Lah, T.; Aronica, E.; Van Noorden, C.J. Immunohistochemical Detection of Neural Stem Cells and Glioblastoma Stem Cells in the Subventricular Zone of Glioblastoma Patients. J. Histochem. Cytochem. 2021, 69, 349–364. [Google Scholar] [CrossRef]
- Singh, S.K.; Hawkins, C.; Clarke, I.D.; Squire, J.A.; Bayani, J.; Hide, T.; Henkelman, R.M.; Michael, D.; Cusimano, M.D.; Dirk, P.B. Identifi-cation of human brain tumour initiating cells. Nature 2004, 432, 396–401. [Google Scholar] [CrossRef]
- Bao, S.; Wu, Q.; McLendon, R.E.; Hao, Y.; Shi, Q.; Hjelmeland, A.B.; Dewhirst, M.W.; Bigner, D.D.; Rich, J.N. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 2006, 444, 756–760. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, Y.; Yu, T.-S.; McKay, R.M.; Burns, D.K.; Kernie, S.G.; Parada, L.F. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature 2012, 488, 522–526. [Google Scholar] [CrossRef] [PubMed]
- Chena, X.; Chen, L.; Chen, J.; Hu, W.; Gao, H.; Xie, B.; Wang, X.; Yin, Z.; Li, S.; Wang, X. ADAM17 promotes U87 glio-blastoma stem cell migration and invasion. Brain Res. 2013, 1538, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Wub, Q.; Guryanova, O.A.; Huang, Z.; Huanga, Q.; Rich, J.N.; Bao, S. Elevated invasive potential of glioblastoma stem cells. BBRC 2011, 4, 643–648. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Li, C.; Guo, L.; Garland, M. Experimental evidence for a significant homometallic catalytic binuclear elimination reaction: Linear-quadratic kinetics in the rhodium catalyzed hydroformylation of cyclooctene. J. Catal. 2006, 237, 67–78. [Google Scholar] [CrossRef]
- Grande, S.; Palma, A.; Ricci-Vitiani, L.; Luciani, A.M.; Buccarelli, M.; Biffoni, M.; Molinari, A.; Calcabrini, A.; D’Amore, E.; Guidoni, L.; et al. Metabolic Heterogeneity Evidenced by MRS among Patient-Derived Glioblastoma Multiforme Stem-Like Cells Accounts for Cell Clustering and Different Responses to Drugs. Stem Cells Int. 2018, 2018, 3292704. [Google Scholar] [CrossRef]
- Marziali, G.; Signore, M.; Buccarelli, M.; Grande, S.; Palma, A.; Biffoni, M.; Rosi, A.; D’Alessandris, Q.G.; Martini, M.; Larocca, L.M.; et al. Metabolic/Proteomic Signature Defines Two Glioblastoma Subtypes With Different Clinical Outcome. Sci. Rep. 2016, 6, 21557. [Google Scholar] [CrossRef]
- Marziali, G.; Buccarelli, M.; Giuliani, A.; Ilari, R.; Grande, S.; Palma, A.; D’Alessandris, Q.G.; Martini, M.; Biffoni, M.; Pallini, R.; et al. A three-microRNA signature identifies two subtypes of glioblastoma patients with different clinical outcomes. Mol. Oncol. 2017, 11, 1115–1129. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, P.; Gong, Z.; Shultz, L.D.; Ren, G. Mesenchymal Stem Cells: From Regeneration to Cancer. Pharmacol. Ther. 2019, 200, 42–54. [Google Scholar] [CrossRef]
- Ono, M.; Kosaka, N.; Tominaga, N.; Yoshioka, Y.; Takeshita, F.; Takahashi, R.U.; Yoshida, M.; Tsuda, H.; Tamura, K.; Ochiya, T.; et al. Exosomes from Bone Marrow Mesenchymal Stem Cells Contain a MicroRNA that Promotes Dormancy in Metastatic Breast Cancer Cells. Sci. Signal. 2014, 7, ra63. [Google Scholar] [CrossRef]
- Zhang, L.; Pan, Y.; Qin, G.; Chen, L.; Chatterjee, T.K.; Weintraub, N.L.; Tang, Y. Inhibition of Stearoyl-coA Desaturase Se-lectively Eliminates Tumorigenic Nanog-Positive Cells: Improving the Safety of iPS Cell Transplantation to Myocardium. Cell Cycle 2014, 13, 762–771. [Google Scholar] [CrossRef] [PubMed]
- Santos, C.R.; Schulze, A. Lipid metabolism in cancer. FEBS J. 2012, 279, 2610–2623. [Google Scholar] [CrossRef]
- Beloribi-Djefaflia, S.; Vasseur, S.; Guillaumond, F. Lipid metabolic reprogramming in cancer cells. Oncogenesis 2016, 5, e189. [Google Scholar] [CrossRef] [PubMed]
- Broadfield, L.A.; Pane, A.A.; Talebi, A.; Swinnen, J.V.; Fendt, S.-M. Lipid metabolism in cancer: New perspectives and emerging mechanisms. Dev. Cell 2021, 56, 1363–1393. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Geng, F.; Cheng, X.; Guo, Q.; Zhong, Y.; Cloughesy, T.F.; Yong, W.H.; Chakravarti, A.; Guo, D. Lipid Droplets Maintain Energy Homeostasis and Glioblastoma Growth via Autophagic Release of Stored Fatty Acids. iScience 2020, 23, 101569. [Google Scholar] [CrossRef]
- Peck, B.; Schulze, A. Lipid desaturation—The next step in targeting lipogenesis in cancer? FEBS J. 2016, 283, 2767–2778. [Google Scholar] [CrossRef] [PubMed]
- Petan, T. Lipid Droplets in Cancer. In Reviews of Physiology, Biochemistry and Pharmacology; Springer: Berlin/Heidelberg, Germany, 2020; pp. 1–34. [Google Scholar] [CrossRef]
- Guo, D.; Bell, E.H.; Chakravarti, A. Lipid metabolism emerges as a promising target for malignant glioma therapy. CNS Oncol. 2013, 2, 289–299. [Google Scholar] [CrossRef]
- Delikatny, E.J.; Chawla, S.; Leung, D.-J.; Poptani, H. MR-visible lipids and the tumor microenvironment. NMR Biomed. 2011, 24, 592–611. [Google Scholar] [CrossRef]
- Alexandri, E.; Ahmed, R.; Siddiqui, H.; Choudhary, M.I.; Constantinos, G.; Tsiafoulis, C.G.; Gerothanassis, I.P. High Resolution NMR Spectroscopy as a Structural and Analytical Tool for Unsaturated Lipids in Solution. Molecules 2017, 22, 1663. [Google Scholar] [CrossRef] [PubMed]
- Prescot, A.P.; Renshaw, P.F. Two-dimensionalJ-resolved proton MR spectroscopy and prior knowledge fitting (ProFit) in the frontal and parietal lobes of healthy volunteers: Assessment of metabolite discrimination and general reproducibility. J. Magn. Reson. Imaging 2013, 37, 642–651. [Google Scholar] [CrossRef]
- Lauretti, L.; Cenci, T.; Montano, N.; Offi, M.; Giordano, M.; Caccavella, V.M.; Mangraviti, A.; Agostini, L.; Olivi, A.; Gabriele, L.; et al. Molecular Analysis in a Glioblastoma Cohort—Results of a Prospective Analysis. J. Pers. Med. 2022, 12, 685. [Google Scholar] [CrossRef] [PubMed]
- Kleihues, P.; Cavenee, W. Pathology and genetics of tumours of the nervous system. In World Health Organization Classification of Tumours, 2nd ed.; France IARC Press: Lyon, France, 2000. [Google Scholar]
- Pallini, R.; Ricci-Vitiani, L.; Banna, G.L.; Signore, M.; Lombardi, D.; Todaro, M.; Stassi, G.; Martini, M.; Maira, G.; Larocca, L.M.; et al. Cancer Stem Cell Analysis and Clinical Outcome in Patients with Glioblastoma Multiforme. Clin. Cancer Res. 2008, 14, 8205–8212. [Google Scholar] [CrossRef] [PubMed]
- Guidoni, L.; Ricci-Vitiani, L.; Rosi, A.; Palma, A.; Grande, S.; Luciani, A.M.; Pelacchi, F.; di Martino, S.; Colosimo, C.; Biffoni, M.; et al. 1H NMR detects different metabolic profiles in glioblastoma stem-like cells. NMR Biomed. 2014, 27, 129–145. [Google Scholar] [CrossRef] [PubMed]
- Dahshan, B.A.; Weir, J.S.; Bice, R.P.; Renz, P.; Cifarelli, D.T.; Poplawski, L.; Hack, J.; Vargo, J.A.; Cifarelli, C.P. Dose homogeneity analysis of adjuvant radiation treatment in surgically resected brain metastases: Comparison of IORT, SRS, and IMRT indices. Brachy-Therapy 2021, 20, 426–432. [Google Scholar] [CrossRef] [PubMed]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological Statistic software package for education and data analysis. Paleontol. Electron. 2001, 4, 76. [Google Scholar]
- Peterson, P.; Trinh, L.; Mansson, S. Quantitative 1 HMRI and MRS of fatty acid composition. Magn. Reson. Med. 2021, 85, 49–67. [Google Scholar] [CrossRef]
- Ramadan, S.; Ratai, E.-M.; Wald, L.L.; Mountford, C.E. In vivo 1D and 2D correlation MR spectroscopy of the soleus muscle at 7T. J. Magn. Reson. 2010, 204, 91–98. [Google Scholar] [CrossRef]
- Palma, A.; Grande, S.; Ricci-Vitiani, L.; Luciani, A.M.; Buccarelli, M.; Biffoni, M.; Dini, V.; Cirrone, G.A.P.; Ciocca, M.; Guidoni, L.; et al. Different Mechanisms underlie the Metabolic Response of GBM Stem-Like Cells to Ionizing Radiation: Biological and MRS Studies on Effects of Photons and Carbon Ions. Int. J. Mol. Sci. 2020, 21, 5167. [Google Scholar] [CrossRef] [PubMed]
- Palma, A.; Grande, S.; Luciani, A.M.; Ricci-Vitiani, L.; Buccarelli, M.; Pallini, R.; Triveri, A.; Pirota, V.; Doria, F.; D’Alessandris, Q.G.; et al. Effects of the Combined Treatment with a G-Quadruplex-Stabilizing Ligand and Photon Beams on Glioblastoma Stem-like Cells: A Magnetic Resonance Study. Int. J. Mol. Sci. 2021, 22, 12709. [Google Scholar] [CrossRef] [PubMed]
- Luciani, A.M.; Grande, S.; Palma, A.; Rosi, A.; Giovannini, C.; Sapora, O.; Viti, V.; Guidoni, L. Characterization of 1H NMR detectable mobile lipids in cells from human adenocarcinomas. FEBS J. 2009, 276, 1333–1346. [Google Scholar] [CrossRef] [PubMed]
- Alkan, H.F.; Walter, K.E.; Luengo, A.; Madreiter-Sokolowski, C.T.; Stryeck, S.; Lau, A.N.; Al-Zoughbi, W.; Lewis, C.A.; Thomas, C.J.; Hoefler, G.; et al. Cytosolic Aspartate Availability Determines Cell Survival When Glutamine Is Limiting. Cell Metab. 2018, 28, 706–720.e6. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Bermudez, J.; Baudrier, L.; La, K.; Zhu, X.G.; Fidelin, J.; Sviderskiy, V.O.; Papagiannakopoulos, T.; Molina, H.; Snuderl, M.; Lewis, C.A.; et al. Aspartate is a limiting metabolite for cancer cell proliferation under hypoxia and in tumours. Nat. Cell Biol. 2018, 20, 775–781. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.C.; Gonzalez, M.E.; Burman, B.; Zhao, X.; Anwar, T.; Tran, M.; Medhora, N.; Hiziroglu, A.B.; Lee, W.; Cheng, Y.-H.; et al. Mesenchymal Stem/Stromal Cell Engulfment Reveals Metastatic Advantage in Breast Cancer. Cell Rep. 2019, 27, 3916–3926. [Google Scholar] [CrossRef]
- Liu, Y.; Ren, H.; Zhou, Y.; Shang, L.; Zhang, Y.; Yang, F.; Shi, X. The Hypoxia Conditioned Mesenchymal Stem Cells Promote Hepatocellular Carcinoma Progression Through YAP Mediated Lipogenesis Reprogramming. J. Exp. Clin. Cancer Res. 2019, 38, 228. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Liang, B.; Wang, C.; Li, S.; Zhao, Y.; Huang, Q.; Liu, Z.; Yao, Z.; Wu, Q.; Liao, W.; et al. MSC-regulated lncRNA MACC1-AS1 Promotes Stemness and Chemoresistance Through Fatty Acid Oxidation in Gastric Cancer. Oncogene 2019, 38, 4637–4654. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Liu, B.; Chen, Z.; Li, G.; Zhang, Z. MSC-induced lncRNA HCP5 Drove Fatty Acid Oxidation Through miR-3619-5p/AMPK/PGC1?/CEBPB Axis to Promote Stemness and Chemo-Resistance of Gastric Cancer. Cell Death Dis. 2020, 11, 233. [Google Scholar] [CrossRef]
- Han, J.; Qu, H.; Han, M.; Ding, Y.; Xie, M.; Hu, J.; Chen, Y.; Dong, H. MSC-induced lncRNA AGAP2-AS1 Promotes Stemness and Trastuzumab Resistance Through Regulating CPT1 Expression and Fatty Acid Oxidation in Breast Cancer. Oncogene 2021, 40, 833–847. [Google Scholar] [CrossRef] [PubMed]
- Libby, C.J.; Tran, A.N.; Scott, S.E.; Griguer, C.; Hjelmeland, A.B. The pro-tumorigenic effects of metabolic alterations in glioblastoma including brain tumor initiating cells. Biochim. Biophys. Acta Rev. Cancer 2018, 1869, 175–188. [Google Scholar] [CrossRef]
- Pan, T.; Liu, J.; Xu, S.; Yu, Q.; Wang, H.; Sun, H.; Wu, J.; Zhu, Y.; Zhou, J.; Zhu, Y. A Novel Tumor Microenvironment-Induced Mitochondrial Protein Pro-motes Metabolic Reprogramming of Colorectal Cancer Cells. Theranostics 2020, 10, 516–536. [Google Scholar] [CrossRef]
- Batlle, E.; Clevers, H. Cancer stem cells revisited. Nat. Med. 2017, 23, 1124–1134. [Google Scholar] [CrossRef]
- Li, H.; Feng, Z.; He, M.-L. Lipid metabolism alteration contributes to and maintains the properties of cancer stem cells. Theranostics 2020, 10, 7053–7069. [Google Scholar] [CrossRef]
- Daniel, Y.; Lelou, E.; Aninat, C.; Corlu, A.; Cabillic, F. Interplay Between Metabolism Reprogramming and Epitheli-al-To-Mesenchymal Transition in Cancer Stem Cells. Cancers 2021, 13, 197313. [Google Scholar] [CrossRef]
- Vazquez-Martin, A.; Corominas-Faja, B.; Cufi, S.; Vellon, L.; Oliveras-Ferraros, C.; Menendez, O.J.; Joven, J.; Lupu, R.; Menendez, J.A. The Mitochondrial H(+)-ATP Synthase and the Lipogenic Switch: New Core Components of Metabolic Reprogramming in Induced Pluripotent Stem (iPS) Cells. Cell Cycle 2013, 12, 207–218. [Google Scholar] [CrossRef]
- Rysman, E.; Brusselmans, K.; Scheys, K.; Timmermans, L.; Derua, R.; Munck, S.; Van Veldhoven, P.P.; Waltregny, D.; Daniëls, V.W.; Machiels, J.; et al. De novo Lipogenesis Protects Cancer Cells from Free Radicals and Chemotherapeutics by Promoting Membrane Lipid Saturation. Cancer Res. 2010, 70, 8117–8126. [Google Scholar] [CrossRef]
- Antalis, C.J.; Arnold, T.; Rasool, T.; Lee, B.; Buhman, K.K.; Siddiqui, R.A. High ACAT1 expression in estrogen receptor negative basal-like breast cancer cells is associated with LDL-induced proliferation. Breast Cancer Res. Treat. 2010, 122, 661–670. [Google Scholar] [CrossRef] [PubMed]
- Chamras, H.; Ardashian, A.; Heber, D.; Glaspy, J.A. Fatty acid modulation of MCF-7 human breast cancer cell proliferation, apoptosis and differentiation. J. Nutr. Biochem. 2002, 13, 711–716. [Google Scholar] [CrossRef]
- Przybytkowski, E.; Joly, É.; Nolan, C.; Hardy, S.; Francoeur, A.-M.; Langelier, Y.; Prentki, M. Upregulation of cellular triacylglycerol—Free fatty acid cycling by oleate is associated with long-term serum-free survival of human breast cancer cells. Biochem. Cell Biol. 2007, 85, 301–310. [Google Scholar] [CrossRef]
- Shiu, R.P.; Paterson, J.A. Alteration of cell shape, adhesion, and lipid accumulation in human breast cancer cells (T-47D) by human prolactin and growth hormone. Cancer Res. 1984, 44, 1178–1186. [Google Scholar]
- Mohammadzadeh, F.; Mosayebi, G.; Montazeri, V.; Darabi, M.; Fayezi, S.; Shaaker, M.; Rahmati, M.; Baradaran, B.; Mehdizadeh, A.; Darabi, M. Fatty Acid Composition of Tissue Cultured Breast Carcinoma and the Effect of Stearoyl-CoA Desaturase 1 Inhibition. J. Breast Cancer 2014, 17, 136–142. [Google Scholar] [CrossRef]
- Montecillo-Aguado, M.; Tirado-Rodriguez, B.; Tong, Z.; Vega, O.M.; Morales-Martínez, M.; Abkenari, S.; Pedraza-Chaverri, J.; Huerta-Yepez, S. Importance of the Role of ω-3 and ω-6 Polyunsaturated Fatty Acids in the Progression of Brain Cancer. Brain Sci. 2020, 10, 381. [Google Scholar] [CrossRef]
- Yi, M.; Li, J.; Chen, S.; Cai, J.; Ban, Y.; Peng, Q.; Zhou, Y.; Zeng, Z.; Peng, S.; Li, X.; et al. Emerging role of lipid metabolism alterations in Cancer stem cells. J. Exp. Clin. Cancer Res. 2018, 37, 1–18. [Google Scholar] [CrossRef]
- Fu, Y.; Zou, T.; Shen, X.; Nelson, P.J.; Li, J.; Wu, C.; Yang, J.; Zheng, Y.; Bruns, C.; Zhao, Y.; et al. Lipid metabolism in cancer progression and therapeutic strategies. Med. Commun. 2021, 2, 27–59. [Google Scholar] [CrossRef]
- Li, J.; Condello, S.; Thomes-Pepin, J.; Ma, X.; Xia, Y.; Thomas, T.D.; Matei, D.; Cheng, J.X. Lipid Desaturation Is a Metabolic Marker and Therapeutic Target of Ovarian Cancer Stem Cells. Cell Stem Cell 2017, 20, 303–314.e5. [Google Scholar] [CrossRef]
- Qin, X.Y.; Su, T.; Yu, W.; Kojima, S. Lipid desaturation-associated endoplasmic reticulum stress regulates MYCN gene expression in hepa-tocellular carcinoma cells. Cell Death Dis. 2020, 11, 66. [Google Scholar] [CrossRef]
- Noto, A.; Raffa, S.; De Vitis, C.; Roscilli, G.; Malpicci, D.; Coluccia, P.; Di Napoli, A.; Ricci, A.; Giovagnoli, M.R.; Aurisicchio, L.; et al. Stearoyl-CoA desaturase-1 is a key factor for lung cancer-initiating cells. Cell Death Dis. 2013, 4, e947. [Google Scholar] [CrossRef]
- Pinkham, K.; Park, D.J.; Hashemiaghdam, A.; Kirov, A.B.; Adam, I.; Rosiak, K.; da Hora, C.C.; Teng, J.; Cheah, P.S.; Carvalho, L.; et al. Stearoyl CoA Desaturase Is Essential for Regulation of Endoplasmic Reticulum Homeostasis and Tumor Growth in Glioblastoma Cancer Stem Cells. Stem Cell Rep. 2019, 12, 712–727. [Google Scholar] [CrossRef] [PubMed]
- Hosios, A.M.; Hecht, V.C.; Danai, L.V.; Johnson, M.O.; Rathmell, J.C.; Steinhauser, M.L.; Manalis, S.R.; Vander Heiden, M.G. Amino Acids Rather than Glucose Account for the Majority of Cell Mass in Proliferating Mammalian Cells. Dev. Cell 2016, 36, 540–549. [Google Scholar] [CrossRef] [PubMed]
- Catalina-Rodriguez, O.; Kolukula, V.K.; Tomita, Y.; Preet, A.; Palmieri, F.; Wellstein, A.; Byers, S.; Giaccia, A.J.; Glasgow, E.; Albanese, C.; et al. The mitochondrial citrate transporter, CIC, is essential for mitochondrial homeostasis. Oncotarget 2012, 3, 1220–1235. [Google Scholar] [CrossRef]
- Jiang, L.; Boufersaoui, A.; Yang, C.; Ko, B.; Rakheja, D.; Guevara, G.; Hu, Z.; DeBerardinis, R.J. Quantitative metabolic flux analysis reveals an unconventional pathway of fatty acid synthesis in cancer cells deficient for the mitochondrial citrate transport protein. Metab. Eng. 2016, 43, 198–207. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Xie, B.; Xiao, W.; Fan, M.; Xu, S.; Duan, Y.; Hamsafar, Y.; Evans, A.C.; Huang, J.; Zhou, W.; et al. Fatty acid oxidation fuels glioblastoma radioresistance with CD47-mediated immune evasion. Nat. Commun. 2022, 13, 1511. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grande, S.; Palma, A.; Luciani, A.M.; Anello, P.; Ricci-Vitiani, L.; Buccarelli, M.; D’Alessandris, Q.G.; Pallini, R.; Guidoni, L.; Viti, V.; et al. Glioblastoma Stem-Like Cells (GSCs) with Mesenchymal Signature: Lipid Profiles of Mobile Lipids Obtained with MRS before and after Radio/Chemical Treatments. Biomolecules 2022, 12, 1051. https://doi.org/10.3390/biom12081051
Grande S, Palma A, Luciani AM, Anello P, Ricci-Vitiani L, Buccarelli M, D’Alessandris QG, Pallini R, Guidoni L, Viti V, et al. Glioblastoma Stem-Like Cells (GSCs) with Mesenchymal Signature: Lipid Profiles of Mobile Lipids Obtained with MRS before and after Radio/Chemical Treatments. Biomolecules. 2022; 12(8):1051. https://doi.org/10.3390/biom12081051
Chicago/Turabian StyleGrande, Sveva, Alessandra Palma, Anna Maria Luciani, Pasqualino Anello, Lucia Ricci-Vitiani, Mariachiara Buccarelli, Quintino Giorgio D’Alessandris, Roberto Pallini, Laura Guidoni, Vincenza Viti, and et al. 2022. "Glioblastoma Stem-Like Cells (GSCs) with Mesenchymal Signature: Lipid Profiles of Mobile Lipids Obtained with MRS before and after Radio/Chemical Treatments" Biomolecules 12, no. 8: 1051. https://doi.org/10.3390/biom12081051
APA StyleGrande, S., Palma, A., Luciani, A. M., Anello, P., Ricci-Vitiani, L., Buccarelli, M., D’Alessandris, Q. G., Pallini, R., Guidoni, L., Viti, V., & Rosi, A. (2022). Glioblastoma Stem-Like Cells (GSCs) with Mesenchymal Signature: Lipid Profiles of Mobile Lipids Obtained with MRS before and after Radio/Chemical Treatments. Biomolecules, 12(8), 1051. https://doi.org/10.3390/biom12081051





