High Energy Channeling and Malleable Transition States: Molecular Dynamics Simulations and Free Energy Landscapes for the Thermal Unfolding of Protein U1A and 13 Mutants
Abstract
1. Introduction
2. Background
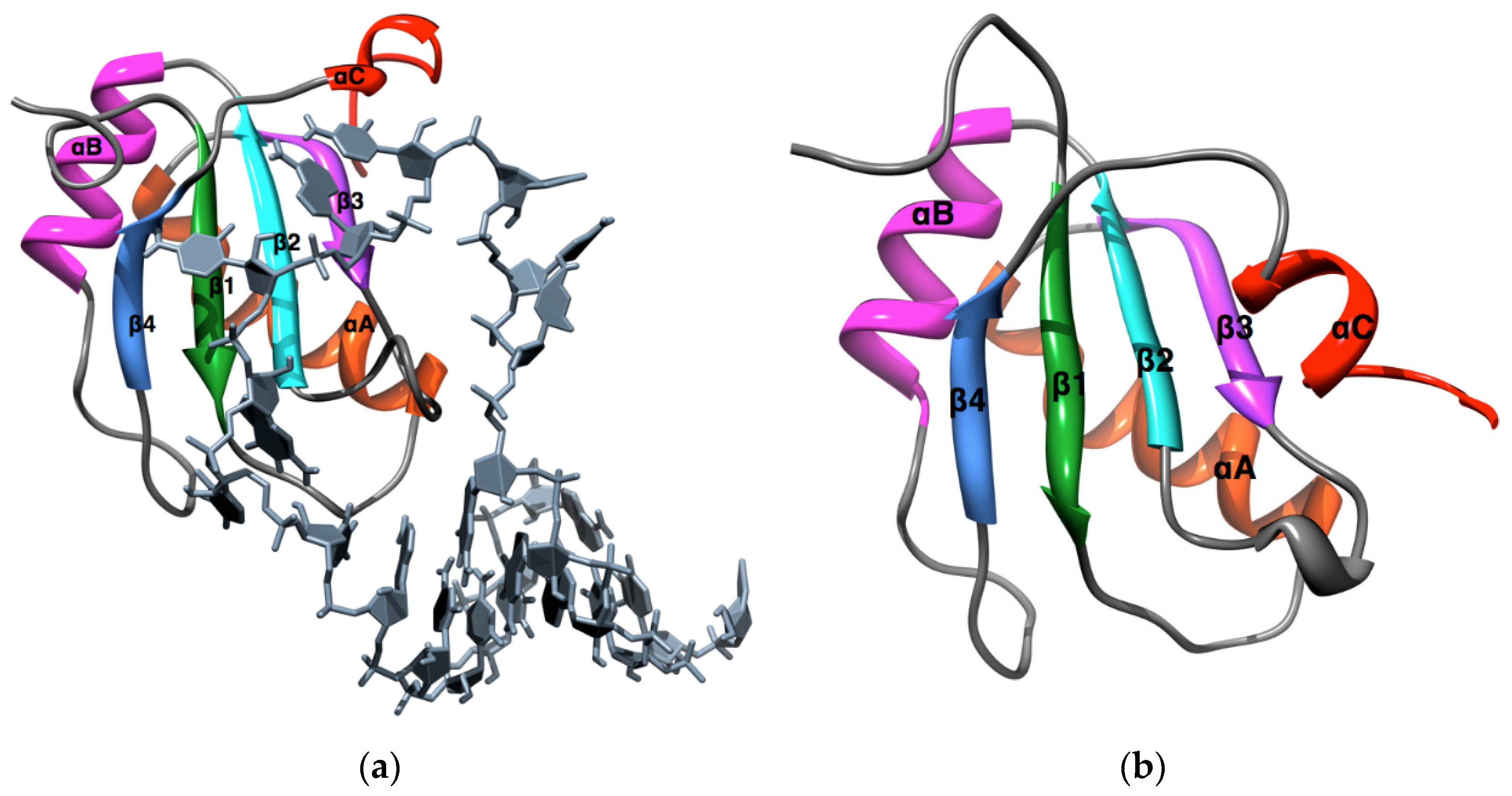
3. Materials and Methods
3.1. Molecular Dynamics Simulations
3.2. Analysis
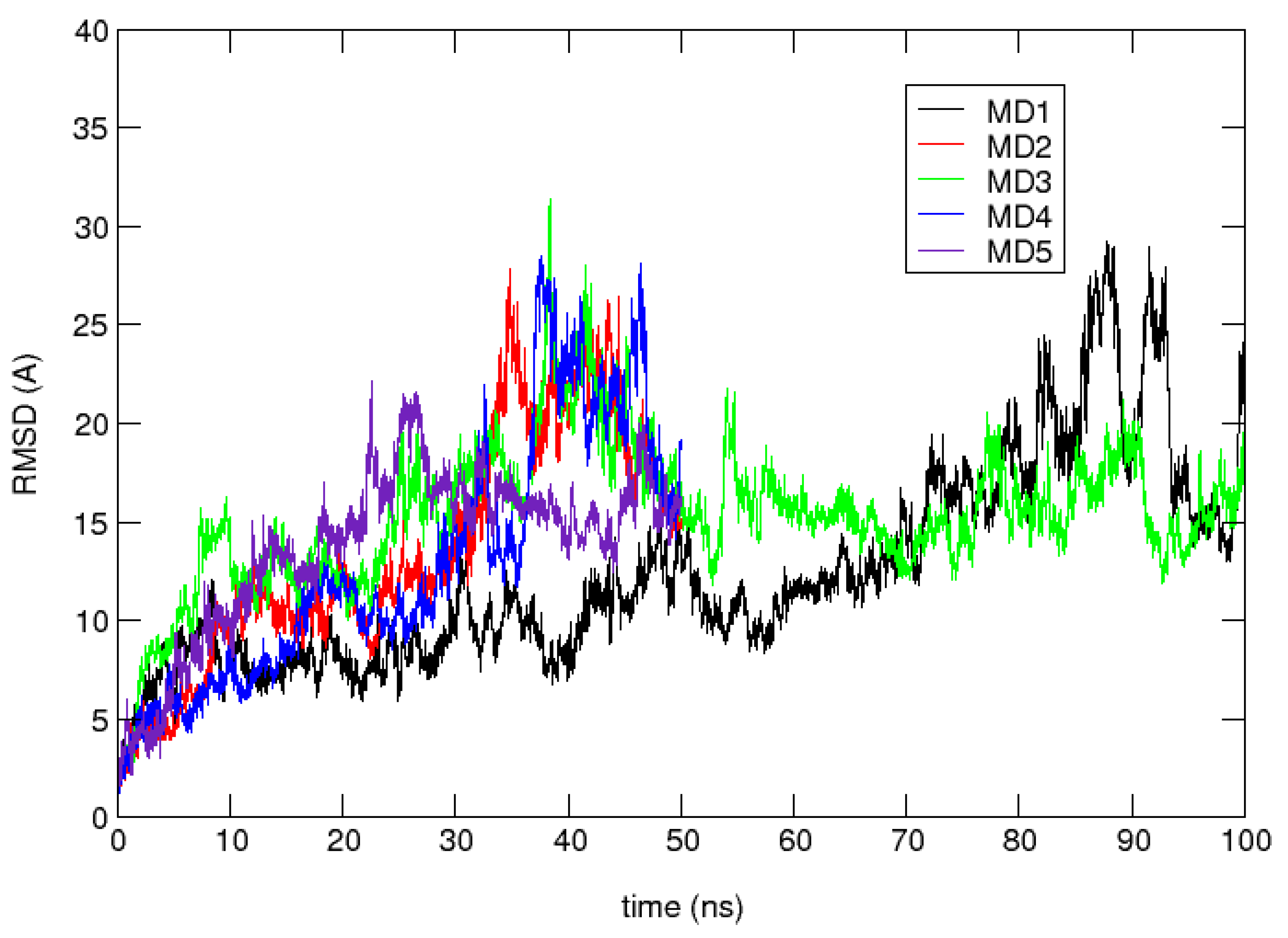
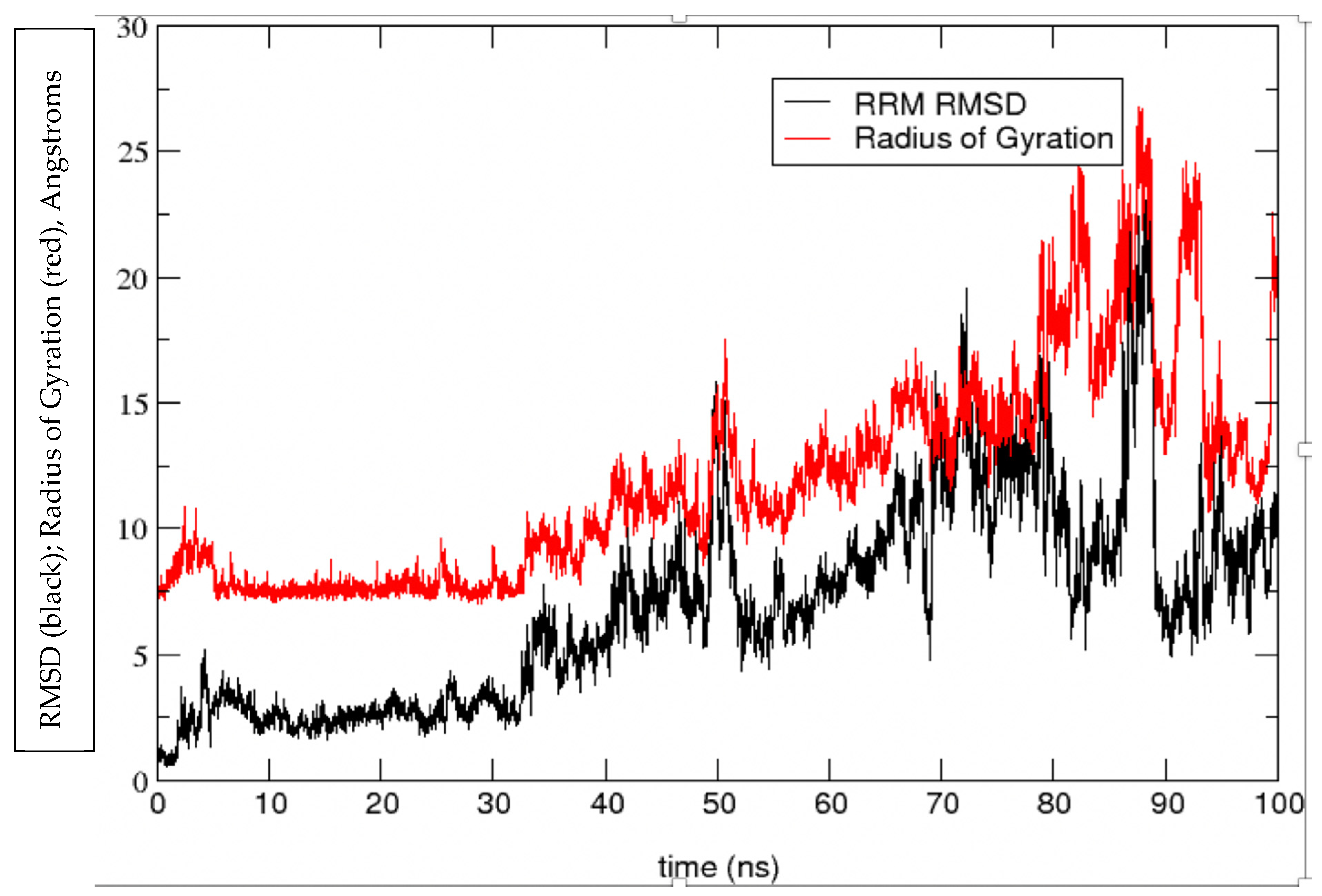
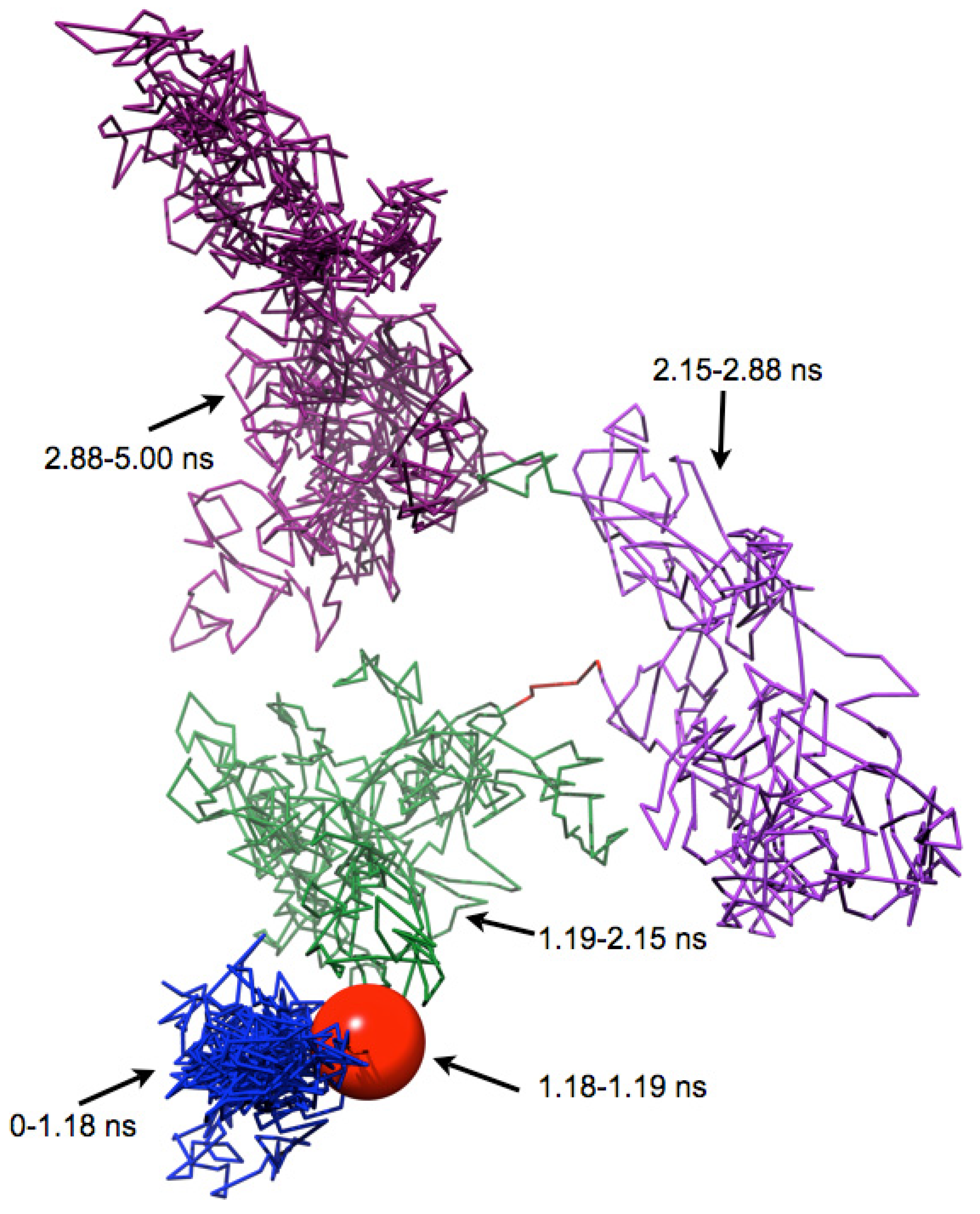
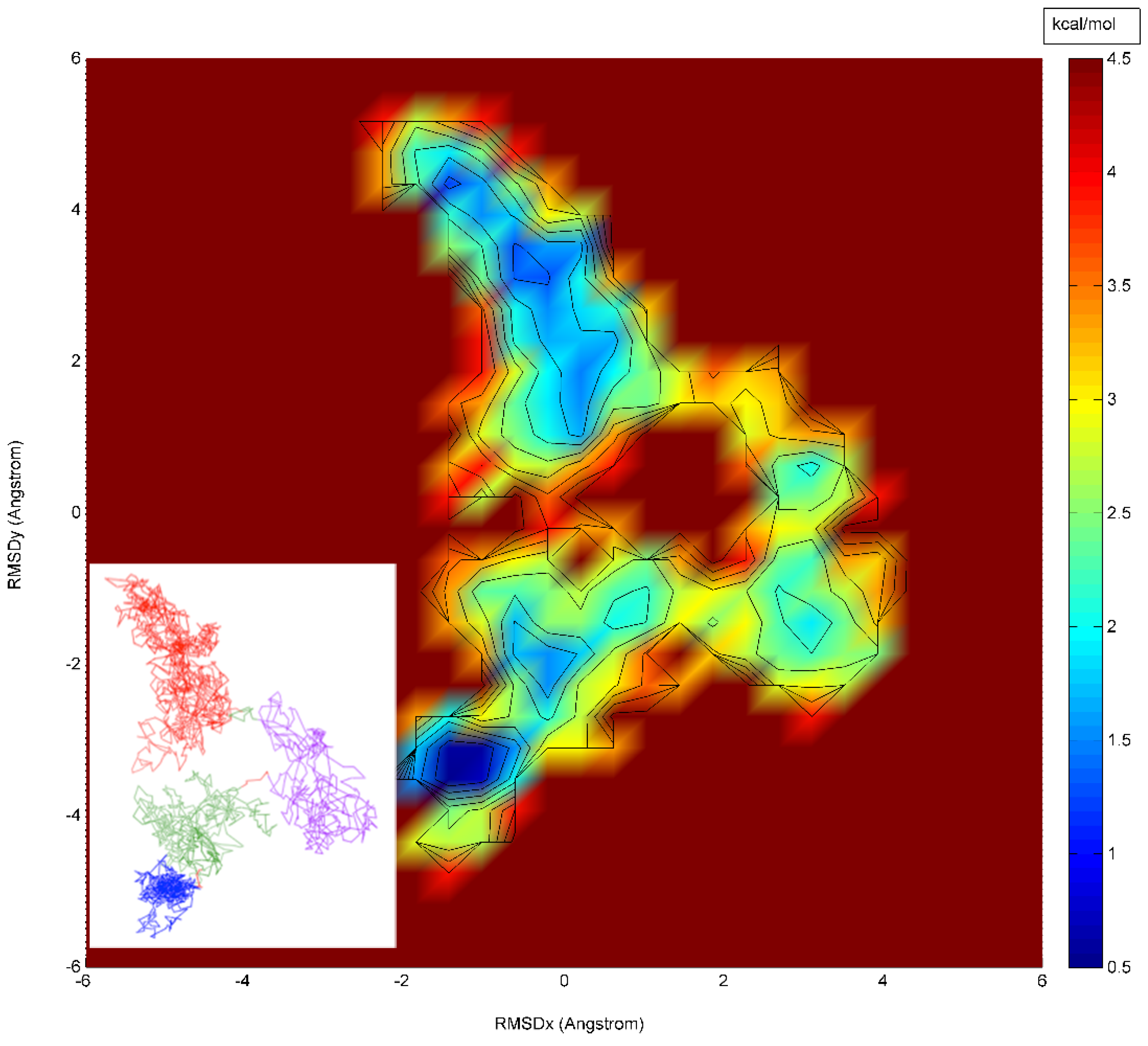
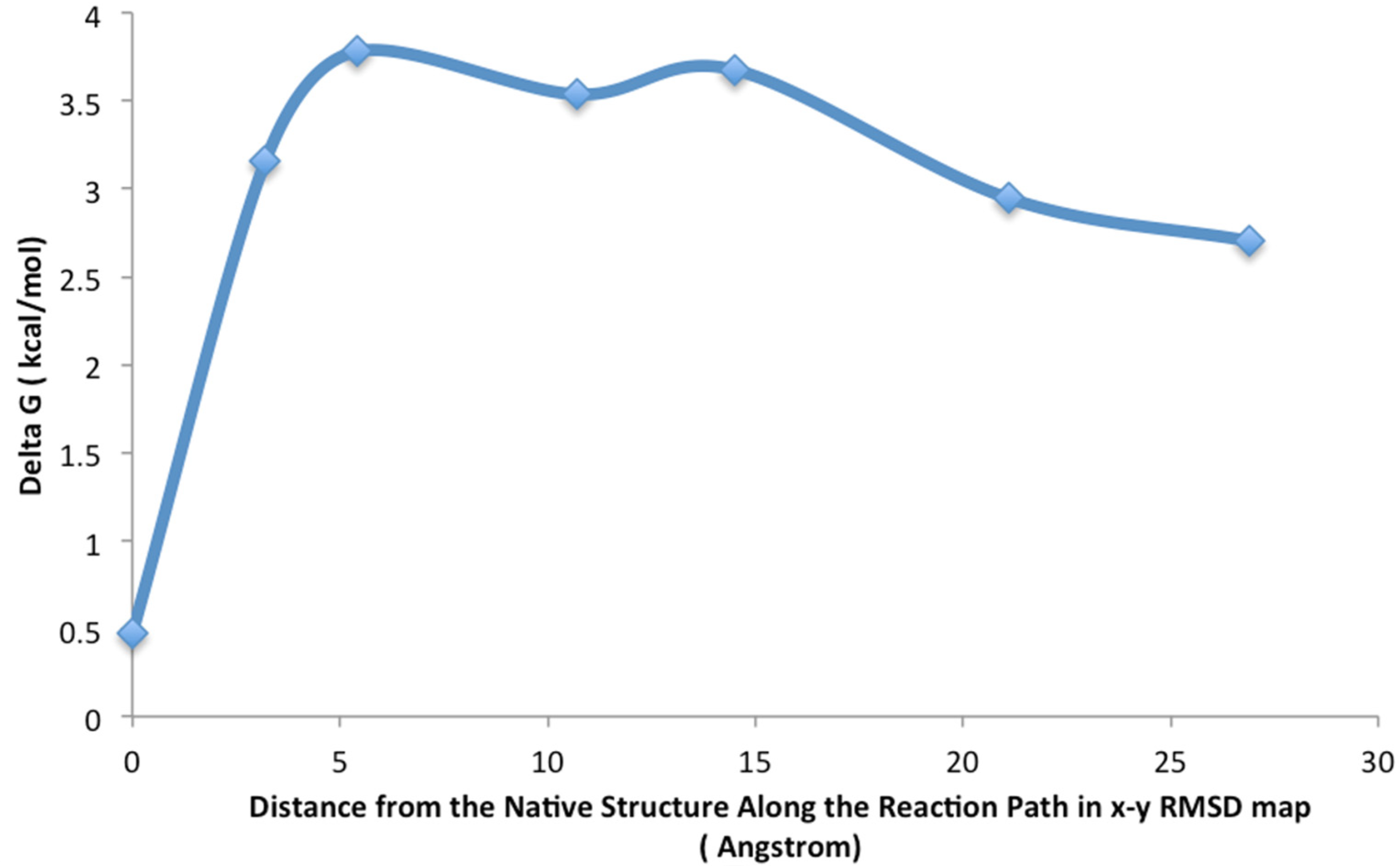
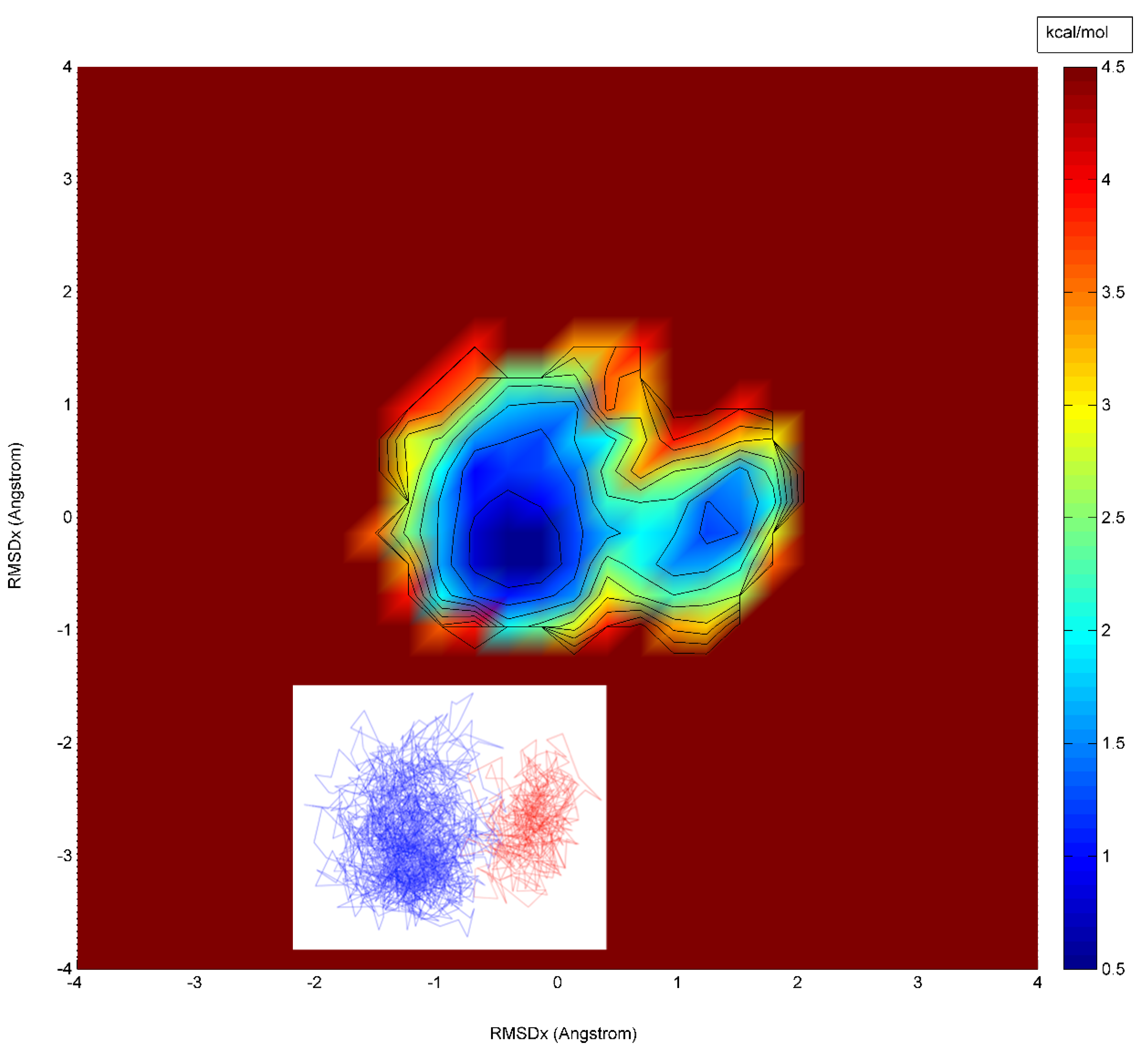
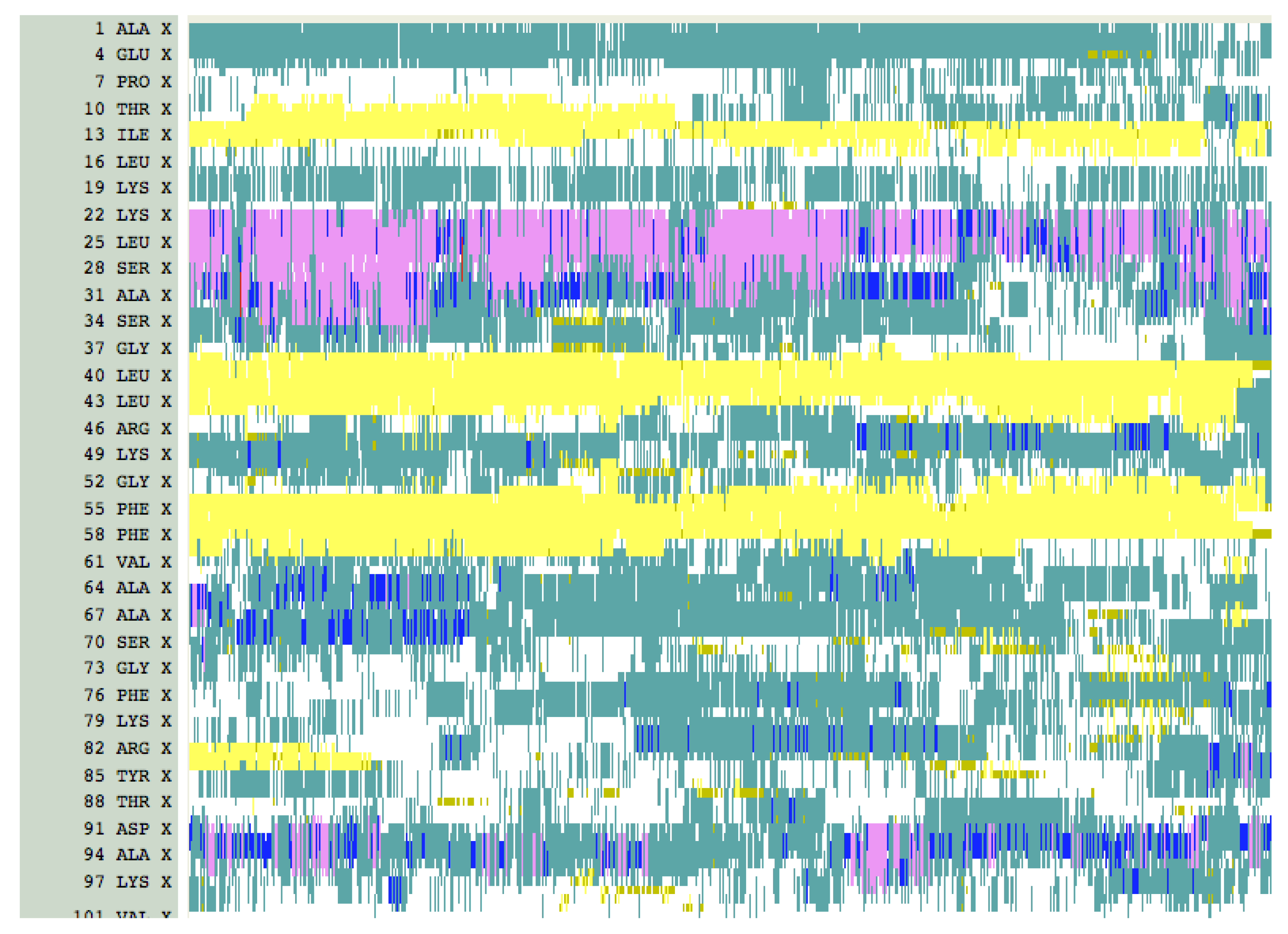
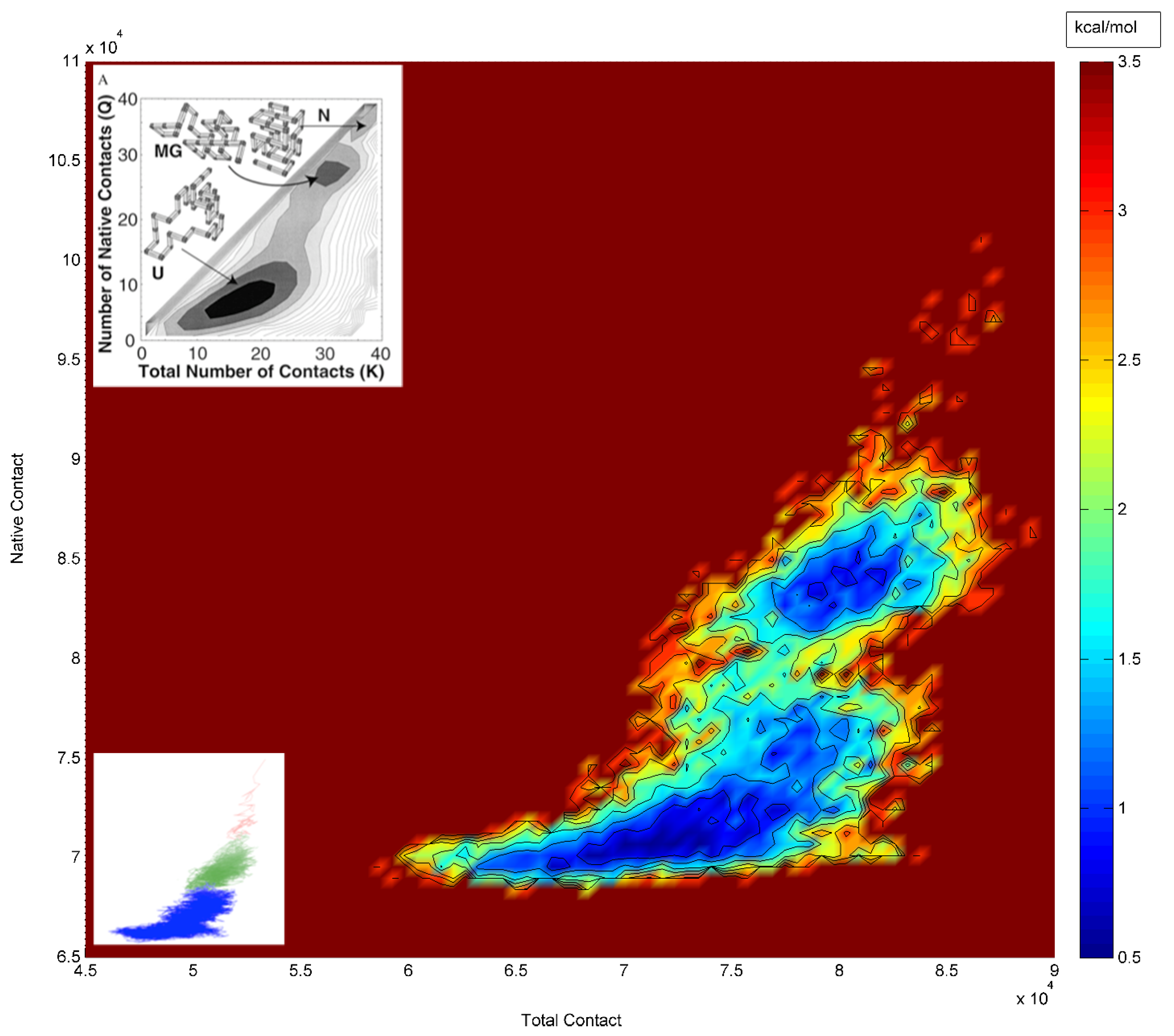
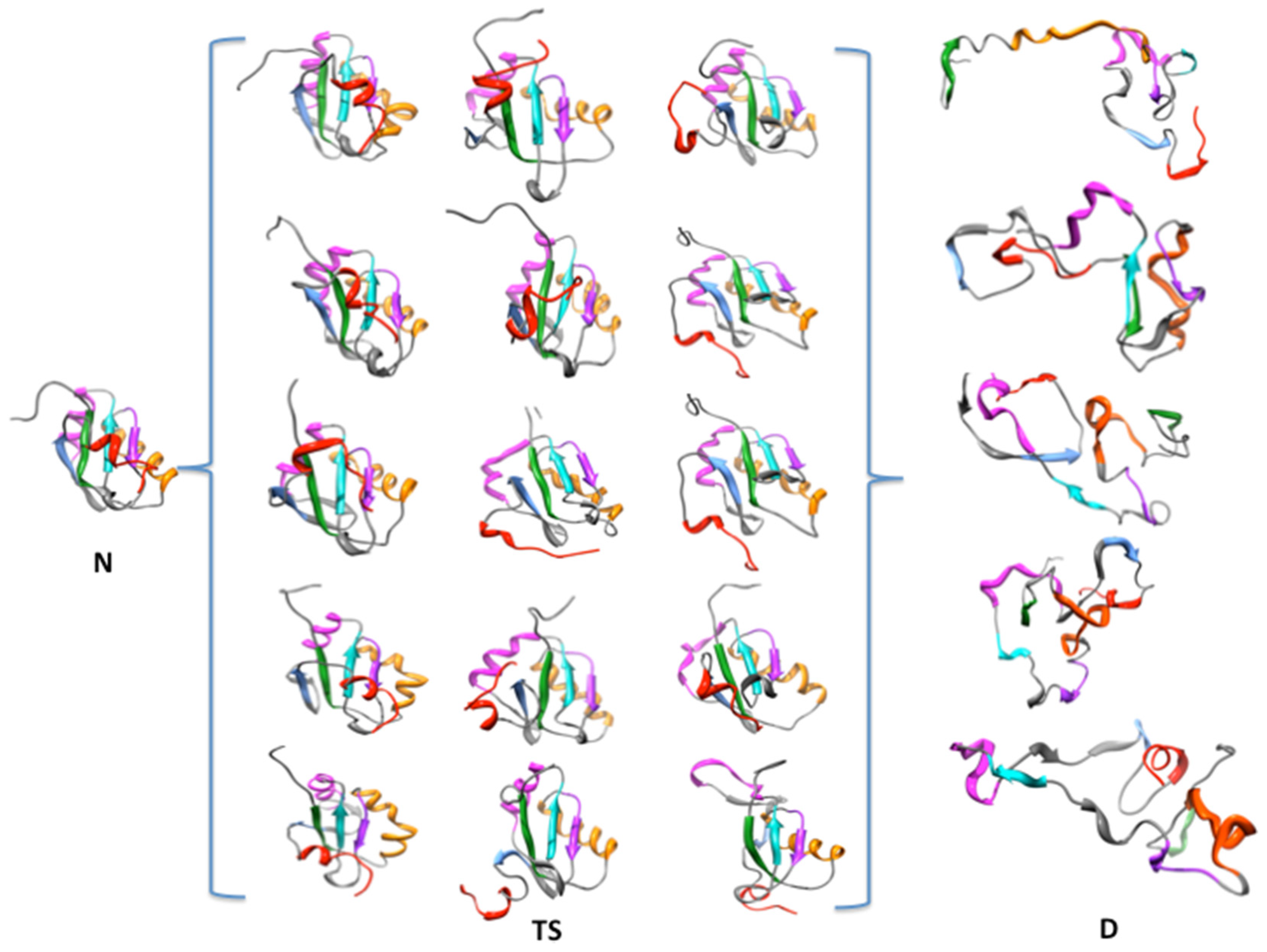
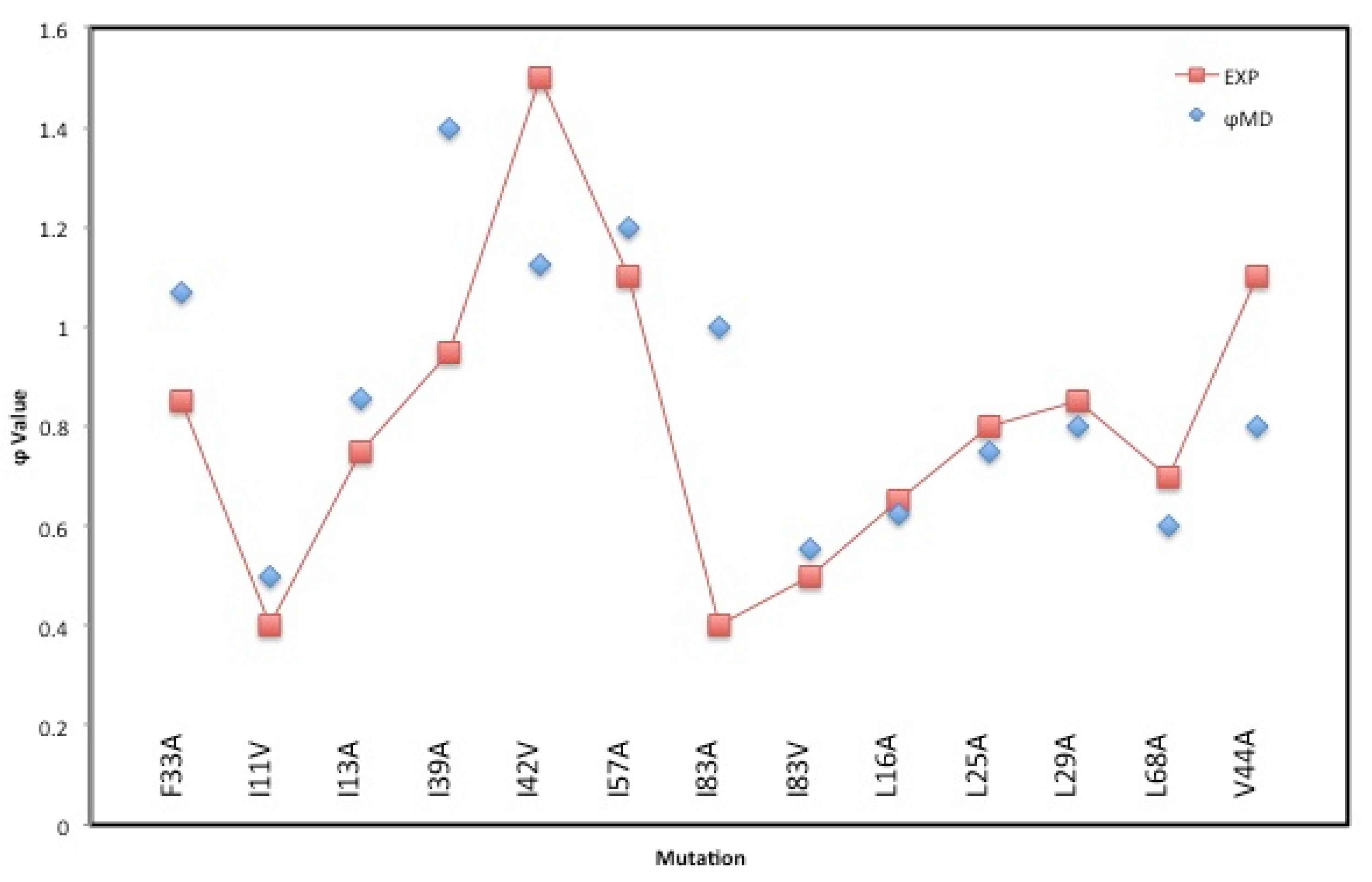
4. Results and Discussion
5. Summary and Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fersht, A.R. Struture and Mechanism in Protein Science; W.H. Freeman & Company: New York, NY, USA, 1998. [Google Scholar]
- Oliveberg, M.; Wolynes, P.G. The experimental survey of protein-folding energy landscapes. Q. Rev. Biophys. 2005, 38, 245–288. [Google Scholar] [CrossRef]
- Ladurner, A.G.; Itzhaki, L.S.; Daggett, V.; Fersht, A.R. Synergy between simulation and experiment in describing the energy landscape of protein folding. Proc. Natl. Acad. Sci. USA 1998, 95, 8473–8478. [Google Scholar] [CrossRef]
- Daggett, V. Long timescale simulations. Curr. Opin. Struct. Biol. 2000, 10, 160–164. [Google Scholar] [CrossRef]
- Mayor, U.; Johnson, C.M.; Daggett, V.; Fersht, A.R. Protein folding and unfolding in microseconds to nanoseconds by experiment and simulation. Proc. Natl. Acad. Sci. USA 2000, 97, 13518–13522. [Google Scholar] [CrossRef] [PubMed]
- Dill, K.A.; Ozkan, S.B.; Shell, M.S.; Weikl, T.R. The protein folding problem. Annu. Rev. Biophys. 2008, 37, 289–316. [Google Scholar] [CrossRef] [PubMed]
- Simmerling, C.; Strockbine, B.; Roitberg, A.E. All-Atom Structure Prediction and Folding Simulations of a Stable Protein. J. Am. Chem. Soc. 2002, 124, 11258–11259. [Google Scholar] [CrossRef]
- Piana, S.; Lindorff-Larsen, K.; Shaw, D.E. How robust are protein folding simulations with respect to force field parameterization? Biophys. J. 2011, 100, L47–L49. [Google Scholar] [CrossRef]
- Li, A.; Daggett, V. Identification and Characterization of the Unfolding Transition State of Chymotrypsin Inhibitor 2 by Molecular Dynamics Simulations. J. Mol. Biol. 1996, 257, 412–429. [Google Scholar] [CrossRef]
- Fersht, A.R.; Daggett, V. Protein Folding and Unfolding at Atomic Resolution. Cell 2002, 108, 573–582. [Google Scholar] [CrossRef]
- Daggett, V. Protein folding-simulation. Chem. Rev. 2006, 106, 1898–1916. [Google Scholar] [CrossRef]
- Li, A.; Daggett, V. Characterization of the transition state of protein unfolding by use of molecular dynamics: Chymotrypsin inhibitor 2. Proc. Natl. Acad. Sci. USA 1994, 91, 10430–10434. [Google Scholar] [CrossRef] [PubMed]
- Beck, D.A.; Daggett, V. A one-dimensional reaction coordinate for identification of transition states from explicit solvent P(fold)-like calculations. Biophys. J. 2007, 93, 3382–3391. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kormos, B.L.; Baranger, A.M.; Beveridge, D.L. Do Collective Atomic Fluctuations Account for Cooperative Effects? Molecular Dynamics Studies of the U1A−RNA Complex. J. Am. Chem. Soc. 2006, 128, 8992–8993. [Google Scholar] [CrossRef]
- Zhao, Y.; Kormos, B.L.; Beveridge, D.L.; Baranger, A.M. Molecular dynamics simulation studies of a protein–RNA complex with a selectively modified binding interface. Biopolymers 2005, 81, 256–269. [Google Scholar] [CrossRef] [PubMed]
- Kormos, B.L.; Benitex, Y.; Baranger, A.M.; Beveridge, D.L. Affinity and Specificity of Protein U1A-RNA Complex Formation Based on an Additive Component Free Energy Model. J. Mol. Biol. 2007, 371, 1405–1419. [Google Scholar] [CrossRef]
- Anunciado, D.; Agumeh, M.; Kormos, B.L.; Beveridge, D.L.; Knee, J.L.; Baranger, A.M. Characterization of the Dynamics of an Essential Helix in the U1A Protein by Time-Resolved Fluorescence Measurements. J. Phys. Chem. B 2008, 112, 6122–6130. [Google Scholar] [CrossRef][Green Version]
- Kormos, B.L.; Pieniazek, S.N.; Beveridge, D.L.; Baranger, A.M. U1A protein-stem loop 2 RNA recognition: Prediction of structural differences from protein mutations. Biopolymers 2011, 95, 591–606. [Google Scholar] [CrossRef]
- Maris, C.; Dominguez, C.; Allain, F.H.-T. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005, 272, 2118–2131. [Google Scholar] [CrossRef]
- Varani, G.; Nagai, K. Rna Recognition By Rnp Proteins During Rna Processing. Annu. Rev. Biophys. Biomol. Struct. 1998, 27, 407–445. [Google Scholar] [CrossRef]
- Cléry, A.; Blatter, M.; Allain, F.H.-T. RNA recognition motifs: Boring? Not quite. Curr. Opin. Struct. Biol. 2008, 18, 290–298. [Google Scholar] [CrossRef]
- Nagai, K.; Oubridge, C.; Ito, N.; Jessen, T.H.; Avis, J.; Evans, P. Crystal structure of the U1A spliceosomal protein complexed with its cognate RNA hairpin. Nucleic Acids Symp. Ser. 1995, 34, 1–2. [Google Scholar]
- Avis, J.M.; Allain, F.H.-T.; Howe, P.W.; Varani, G.; Nagai, K.; Neuhaus, D. Solution Structure of the N-terminal RNP Domain of U1A Protein: The Role of C-terminal Residues in Structure Stability and RNA Binding. J. Mol. Biol. 1996, 257, 398–411. [Google Scholar] [CrossRef] [PubMed]
- Oubridge, C.; Ito, N.; Evans, P.R.; Teo, C.H.; Nagai, K. Crystal structure at 1.92 A resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature 1994, 372, 432–438. [Google Scholar] [CrossRef] [PubMed]
- Silow, M.; Oliveberg, M. High-Energy Channeling in Protein Folding. Biochemistry 1997, 36, 7633–7637. [Google Scholar] [CrossRef]
- Jackson, S.E.; Fersht, A.R. Folding of chymotrypsin inhibitor 2. 1. Evidence for a two-state transition. Biochemistry 1991, 30, 10428–10435. [Google Scholar] [CrossRef]
- Ternstrom, T.; Mayor, U.; Akke, M.; Oliveberg, M. From snapshot to movie: Phi analysis of protein folding transition states taken one step further. Proc. Natl. Acad. Sci. USA 1999, 96, 14854–14859. [Google Scholar] [CrossRef]
- Plotkin, S.S.; Onuchic, J.N. Investigation of routes and funnels in protein folding by free energy functional methods. Proc. Natl. Acad. Sci. USA 2000, 97, 6509–6514. [Google Scholar] [CrossRef]
- Shen, T.; Hofmann, C.P.; Oliveberg, M.; Wolynes, P.G. Scanning Malleable Transition State Ensembles: Comparing Theory and Experiment for Folding Protein U1A. Biochemistry 2005, 44, 6433–6439. [Google Scholar] [CrossRef]
- Lindberg, M.O.; Oliveberg, M. Malleability of protein folding pathways: A simple reason for complex behaviour. Curr. Opin. Struct. Biol. 2007, 17, 21–29. [Google Scholar] [CrossRef]
- Banachewicz, W.; Religa, T.L.; Schaeffer, R.D.; Daggett, V.; Fersht, A.R. Malleability of folding intermediates in the homeodomain superfamily. Proc. Natl. Acad. Sci. USA 2011, 108, 5596–5601. [Google Scholar] [CrossRef]
- Daggett, V.; Levitt, M. Molecular dynamics simulations of helix denaturation. J. Mol. Biol. 1992, 223, 1121–1138. [Google Scholar] [CrossRef]
- Daggett, V.; Levitt, M. Protein Unfolding Pathways Explored Through Molecular Dynamics Simulations. J. Mol. Biol. 1993, 232, 600–619. [Google Scholar] [CrossRef] [PubMed]
- Scheraga, H.A.; Khalili, M.; Liwo, A. Protein-Folding Dynamics: Overview of Molecular Simulation Techniques. Annu. Rev. Phys. Chem. 2007, 58, 57–83. [Google Scholar] [CrossRef] [PubMed]
- van der Kamp, M.W.; Schaeffer, R.D.; Jonsson, A.L.; Scouras, A.D.; Simms, A.M.; Toofanny, R.D.; Benson, N.C.; Anderson, P.C.; Merkley, E.D.; Rysavy, S.; et al. Dynameomics: A comprehensive database of protein dynamics. Structure 2010, 18, 423–435. [Google Scholar] [CrossRef] [PubMed]
- Beck, D.A.; Daggett, V. Methods for molecular dynamics simulations of protein folding/unfolding in solution. Methods 2004, 34, 112–120. [Google Scholar] [CrossRef]
- Bennion, B.J.; Daggett, V. The molecular basis for the chemical denaturation of proteins by urea. Proc. Natl. Acad. Sci. USA 2003, 100, 5142–5147. [Google Scholar] [CrossRef]
- Day, R.; Bennion, B.J.; Ham, S.; Daggett, V. Increasing Temperature Accelerates Protein Unfolding Without Changing the Pathway of Unfolding. J. Mol. Biol. 2002, 322, 189–203. [Google Scholar] [CrossRef]
- Gianni, S.; Guydosh, N.R.; Khan, F.; Caldas, T.D.; Mayor, U.; White, G.W.N.; DeMarco, M.L.; Daggett, V.; Fersht, A.R. Unifying features in protein-folding mechanisms. Proc. Natl. Acad. Sci. USA 2003, 100, 13286–13291. [Google Scholar] [CrossRef]
- Day, R.; Daggett, V. Ensemble versus single-molecule protein unfolding. Proc. Natl. Acad. Sci. USA 2005, 102, 13445–13450. [Google Scholar] [CrossRef]
- Daggett, V.; Li, A.; Itzhaki, L.S.; Otzen, D.; Fersht, A.R. Structure of the Transition State for Folding of a Protein Derived from Experiment and Simulation. J. Mol. Biol. 1996, 257, 430–440. [Google Scholar] [CrossRef]
- Hartigan, J. Clustering Algorithms; John Wiley and Sons: New York, NY, USA, 1975. [Google Scholar]
- Muff, S.; Caflisch, A. Identification of the protein folding transition state from molecular dynamics trajectories. J. Chem. Phys. 2009, 130, 125104. [Google Scholar] [CrossRef] [PubMed]
- DeMarco, M.L.; Alonso, D.O.; Daggett, V. Diffusing and Colliding: The Atomic Level Folding/Unfolding Pathway of a Small Helical Protein. J. Mol. Biol. 2004, 341, 1109–1124. [Google Scholar] [CrossRef] [PubMed]
- Klimov, D.; Thirumalai, D. Progressing from folding trajectories to transition state ensemble in proteins. Chem. Phys. 2004, 307, 251–258. [Google Scholar] [CrossRef]
- Das, P.; Moll, M.; Stamati, H.; Kavraki, L.E.; Clementi, C. Low-dimensional, free-energy landscapes of protein-folding reactions by nonlinear dimensionality reduction. Proc. Natl. Acad. Sci. USA 2006, 103, 9885–9890. [Google Scholar] [CrossRef]
- Rohrdanz, M.A.; Zheng, W.; Maggioni, M.; Clementi, C. Determination of reaction coordinates via locally scaled diffusion map. J. Chem. Phys. 2011, 134, 124116. [Google Scholar] [CrossRef] [PubMed]
- Ceriotti, M.; Tribello, G.A.; Parrinello, M. Simplifying the representation of complex free-energy landscapes using sketch-map. Proc. Natl. Acad. Sci. USA 2011, 108, 13023–13028. [Google Scholar] [CrossRef]
- Krivov, S.V.; Karplus, M. Hidden complexity of free energy surfaces for peptide (protein) folding. Proc. Natl. Acad. Sci. USA 2004, 101, 14766–14770. [Google Scholar] [CrossRef]
- Caflisch, A. Network and graph analyses of folding free energy surfaces. Curr. Opin. Struct. Biol. 2006, 16, 71–78. [Google Scholar] [CrossRef]
- Krivov, S.V.; Muff, S.; Caflisch, A.; Karplus, M. One-dimensional barrier-preserving free-energy projections of a beta-sheet miniprotein: New insights into the folding process. J. Phys. Chem. B 2008, 112, 8701–8714. [Google Scholar] [CrossRef][Green Version]
- Du, R.; Pande, V.S.; Grosberg, A.Y.; Tanaka, T.; Shakhnovich, E.S. On the transition coordinate for protein folding. J. Chem. Phys. 1998, 108, 334–350. [Google Scholar] [CrossRef]
- Hummer, G. From transition paths to transition states and rate coefficients. J. Chem. Phys. 2004, 120, 516–523. [Google Scholar] [CrossRef] [PubMed]
- Best, R.B.; Hummer, G. Reaction coordinates and rates from transition paths. Proc. Natl. Acad. Sci. USA 2005, 102, 6732–6737. [Google Scholar] [CrossRef] [PubMed]
- Becker, O.M.; Karplus, M. The topology of multidimensional potential energy surfaces: Theory and application to peptide structure and kinetics. J. Chem. Phys. 1997, 106, 1495–1517. [Google Scholar] [CrossRef]
- Toofanny, R.D.; Jonsson, A.L.; Daggett, V. A Comprehensive Multidimensional-Embedded, One-Dimensional Reaction Coordinate for Protein Unfolding/Folding. Biophys. J. 2010, 98, 2671–2681. [Google Scholar] [CrossRef][Green Version]
- Case, D.A.; Darden, T.; Cheatham, T.E., III; Simmerling, C.; Wang, J.; Duke, R.E.; Luo, R.; Merz, K.M.; Pearlman, D.A.; Crowley, M.; et al. AMBER 9; The University of California: San Francisco, CA, USA, 2006. [Google Scholar]
- Hornak, V.; Abel, R.; Okur, A.; Strockbine, B.; Roitberg, A.; Simmerling, C. Comparison of multiple AMBER force fields and development of improved protein backbone parameters. Proteins Struct. Funct. Bioinform. 2006, 65, 712–725. [Google Scholar] [CrossRef]
- Jorgensen, W.L. Revised TIPS for simulations of liquid water and aqueous solutions. J. Chem. Phys. 1982, 77, 4156–4163. [Google Scholar] [CrossRef]
- Joung, I.S.; Cheatham, T.E., III. Determination of Alkali and Halide Monovalent Ion Parameters for use in Explicitly Solvated Biomolecular Simulations. J. Phys. Chem. B 2008, 112, 9020–9041. [Google Scholar] [CrossRef]
- Amadei, A.; Linssen, A.B.M.; Berendsen, H.J.C. Essential dynamics of proteins. Proteins 1993, 17, 412–425. [Google Scholar] [CrossRef]
- Daidone, I.; Amadei, A.; Di Nola, A. Thermodynamic and kinetic characterization of a beta-hairpin peptide in solution: An extended phase space sampling by molecular dynamics simulations in explicit water. Proteins 2005, 59, 510–518. [Google Scholar] [CrossRef]
- Prada-Gracia, D.; Gomez-Gardenes, J.; Echenique-Robba, P.; Falo, F. Exploring the Free Energy Landscape: From Dynamics to Networks and Back. PLOS Comput. Biol. 2009, 5, e1000415. [Google Scholar] [CrossRef]
- Bahattacharyya, M.; Ghosg, A.; Hansia, P.; Visveshwara, S. Allostery and conformational free energy changes in human tryptophanyl-t-RNA synthetase from Essential Dynamics and Structure Networks. Proteins 2009, 78, 506–517. [Google Scholar] [CrossRef] [PubMed]
- Pande, V.S.; Rokhsar, D.S. Is the molten globule a third phase of proteins? Proc. Natl. Acad. Sci. USA 1998, 95, 1490–1494. [Google Scholar] [CrossRef] [PubMed]
- Das, P.; Matysiak, S.; Clementi, C. Balancing energy and entropy: A minimalist model for the characterization of protein folding landscapes. Proc. Natl. Acad. Sci. USA 2005, 102, 10141–10146. [Google Scholar] [CrossRef] [PubMed]
- Fersht, A.R. Optimization of rates of protein folding: The nucleation-condensation mechanism and its implications. Proc. Natl. Acad. Sci. USA 1995, 92, 10869–10873. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Ogushi, M.; Wada, A. ‘Molten-globule state’: A compact form of globular proteins with mobile side-chains. FEBS Lett. 1983, 164, 21–24. [Google Scholar] [CrossRef]
- Pande, V.S.; Rokhsar, D.S. Folding pathway of a lattice model for proteins. Proc. Natl. Acad. Sci. USA 1999, 96, 1273–1278. [Google Scholar] [CrossRef]
- Day, R.; Daggett, V. All-Atom Simulations of Protein Folding and Unfolding. Adv. Protein. Chem. 2003, 66, 373–403. [Google Scholar] [CrossRef]
- Mayor, U.; Guydosh, N.R.; Johnson, C.M.; Grossmann, J.G.; Sato, S.; Jas, G.S.; Freund, S.M.V.; Alonso, D.O.V.; Daggett, V.; Fersht, A.R. The complete folding pathway of a protein from nanoseconds to microseconds. Nature 2003, 421, 863–867. [Google Scholar] [CrossRef]
- Seeber, M.; Felline, A.; Raimondi, F.; Muff, S.; Friedman, R.; Rao, F.; Caflisch, A.; Fanelli, F. Wordom: A user-friendly program for the analysis of molecular structures, trajectories, and free energy surfaces. J. Comput. Chem. 2010, 32, 1183–1194. [Google Scholar] [CrossRef]
- Seeber, M.; Cecchini, M.; Rao, F.; Settanni, G.; Caflisch, A. Wordom: A program for efficient analysis of molecular dynamics simulations. Bioinformatics 2007, 23, 2625–2627. [Google Scholar] [CrossRef] [PubMed]
- Rueda, M.; Ferrer-Costa, C.; Meyer, T.; Orozco, M. A Consensus View of Protein Dynmaics. Proc. Natl. Acad. Sci. USA 2007, 104, 796–801. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dang, N.L.; Baranger, A.M.; Beveridge, D.L. High Energy Channeling and Malleable Transition States: Molecular Dynamics Simulations and Free Energy Landscapes for the Thermal Unfolding of Protein U1A and 13 Mutants. Biomolecules 2022, 12, 940. https://doi.org/10.3390/biom12070940
Dang NL, Baranger AM, Beveridge DL. High Energy Channeling and Malleable Transition States: Molecular Dynamics Simulations and Free Energy Landscapes for the Thermal Unfolding of Protein U1A and 13 Mutants. Biomolecules. 2022; 12(7):940. https://doi.org/10.3390/biom12070940
Chicago/Turabian StyleDang, Na Le, Anne M. Baranger, and David L. Beveridge. 2022. "High Energy Channeling and Malleable Transition States: Molecular Dynamics Simulations and Free Energy Landscapes for the Thermal Unfolding of Protein U1A and 13 Mutants" Biomolecules 12, no. 7: 940. https://doi.org/10.3390/biom12070940
APA StyleDang, N. L., Baranger, A. M., & Beveridge, D. L. (2022). High Energy Channeling and Malleable Transition States: Molecular Dynamics Simulations and Free Energy Landscapes for the Thermal Unfolding of Protein U1A and 13 Mutants. Biomolecules, 12(7), 940. https://doi.org/10.3390/biom12070940





