Identification and Characterization of an RRM-Containing, RNA Binding Protein in Acinetobacter baumannii
Abstract
:1. Introduction
2. Material and Methods
2.1. Similarity Search for Homologous Proteins to HuR Protein in A. baumannii
2.2. Cloning and Expression of the rAB-Elavl for Biochemical Characterization
2.3. Expression and Purification of rAB-Elavl for X-ray and NMR Analysis
2.4. Crystallization of rAB-Elavl
2.5. X-ray Data Collection and Refinement
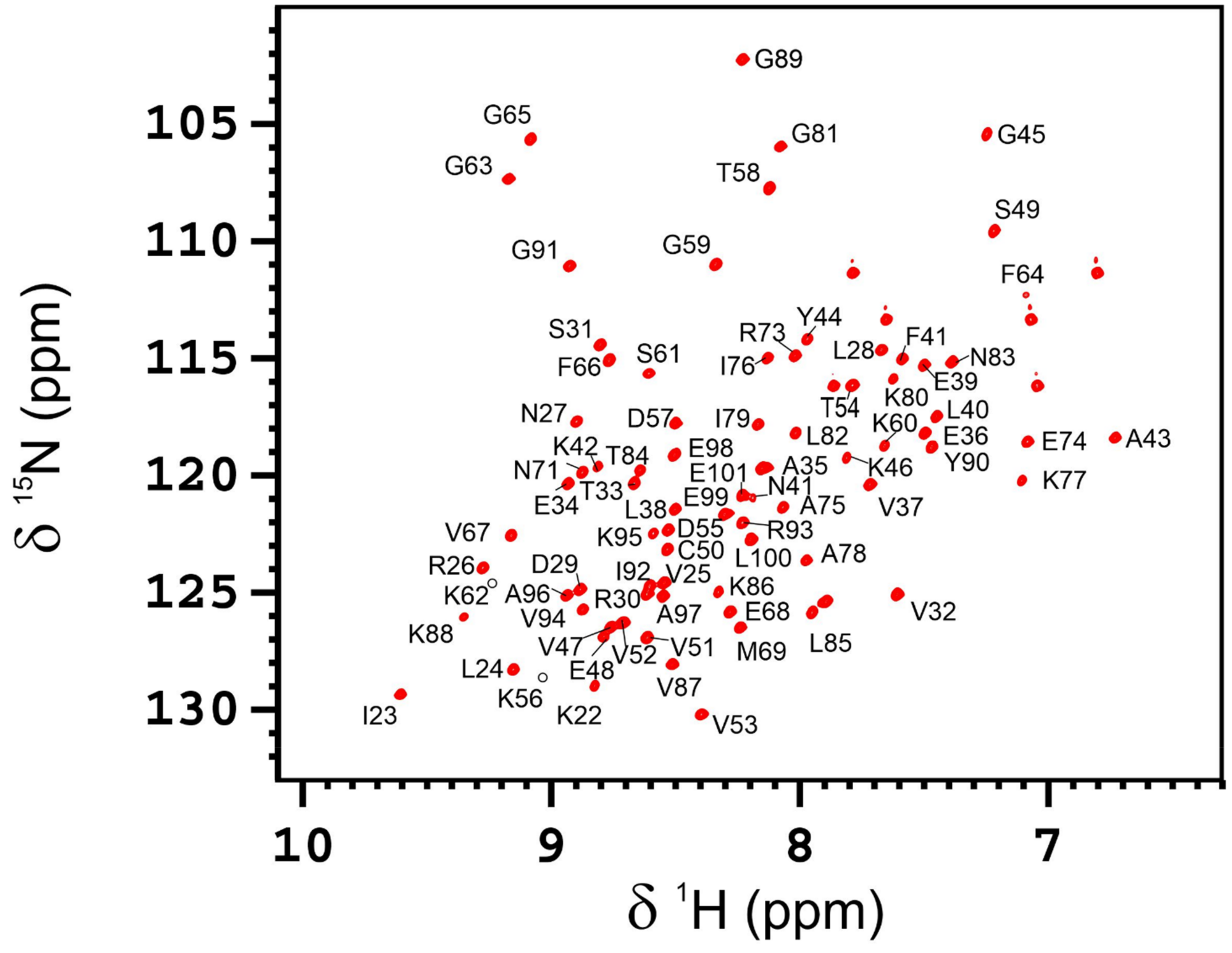
2.6. NMR Measurements and Protein Assignment
2.7. Titration of rAB-Elavl with RNA Probes
2.8. RNA-Electrophoresis Mobility Shift Assay (REMSA)
2.9. Amplified Luminescent Proximity Homogeneous Assay (ALPHA Screen)
2.10. Time Course Experiments Kinetic
2.11. Western Blot from A. baumannii and HEK293 Cells Lysate
2.12. Time Resolved Fluorescence Resonance Energy Transfer (HTRF-FRET)
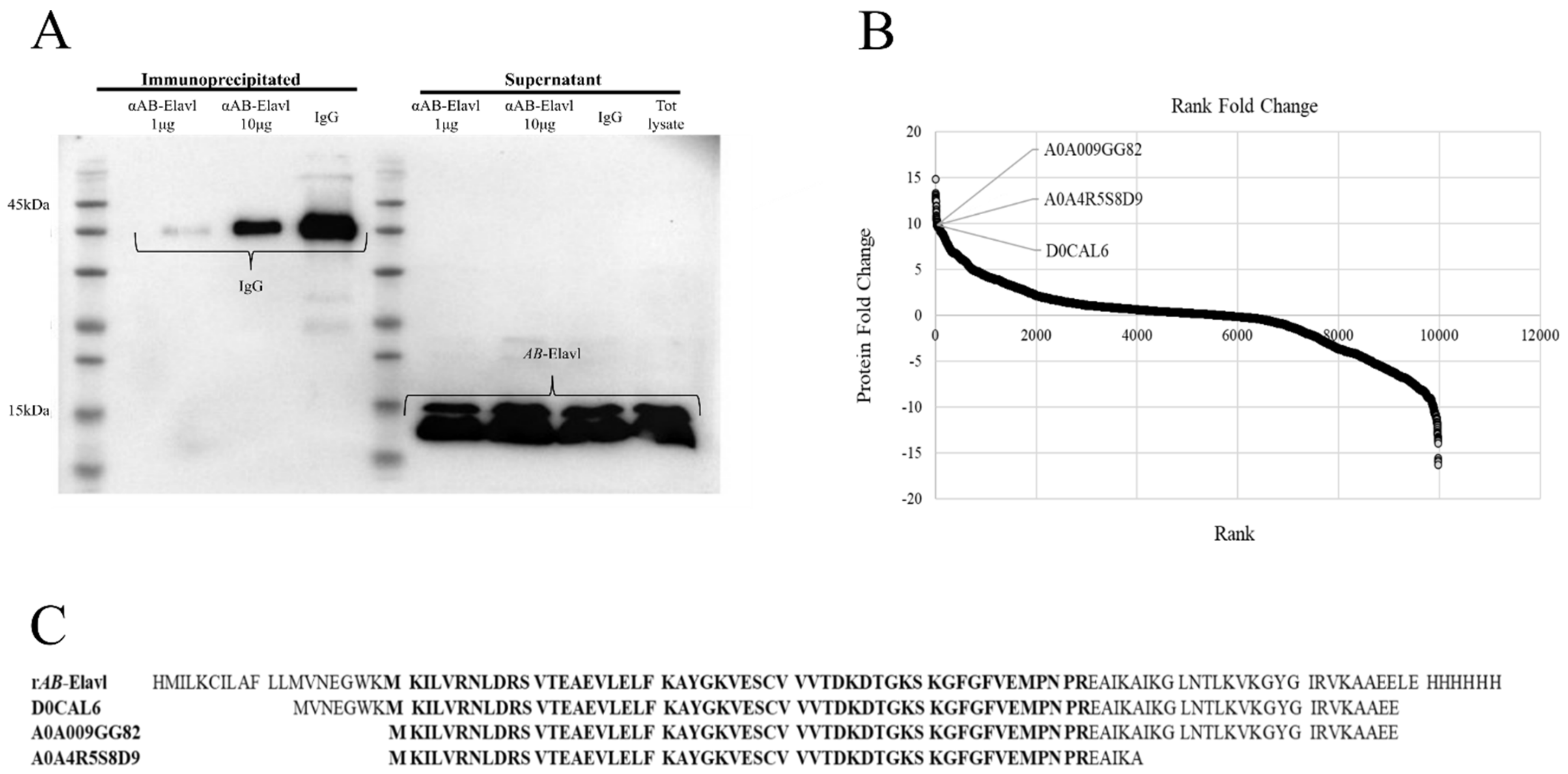
2.13. Immunoprecipitation (IP) Assay of AB-Elavl
2.14. Mass Spectrometry (MS) Analysis
3. Results
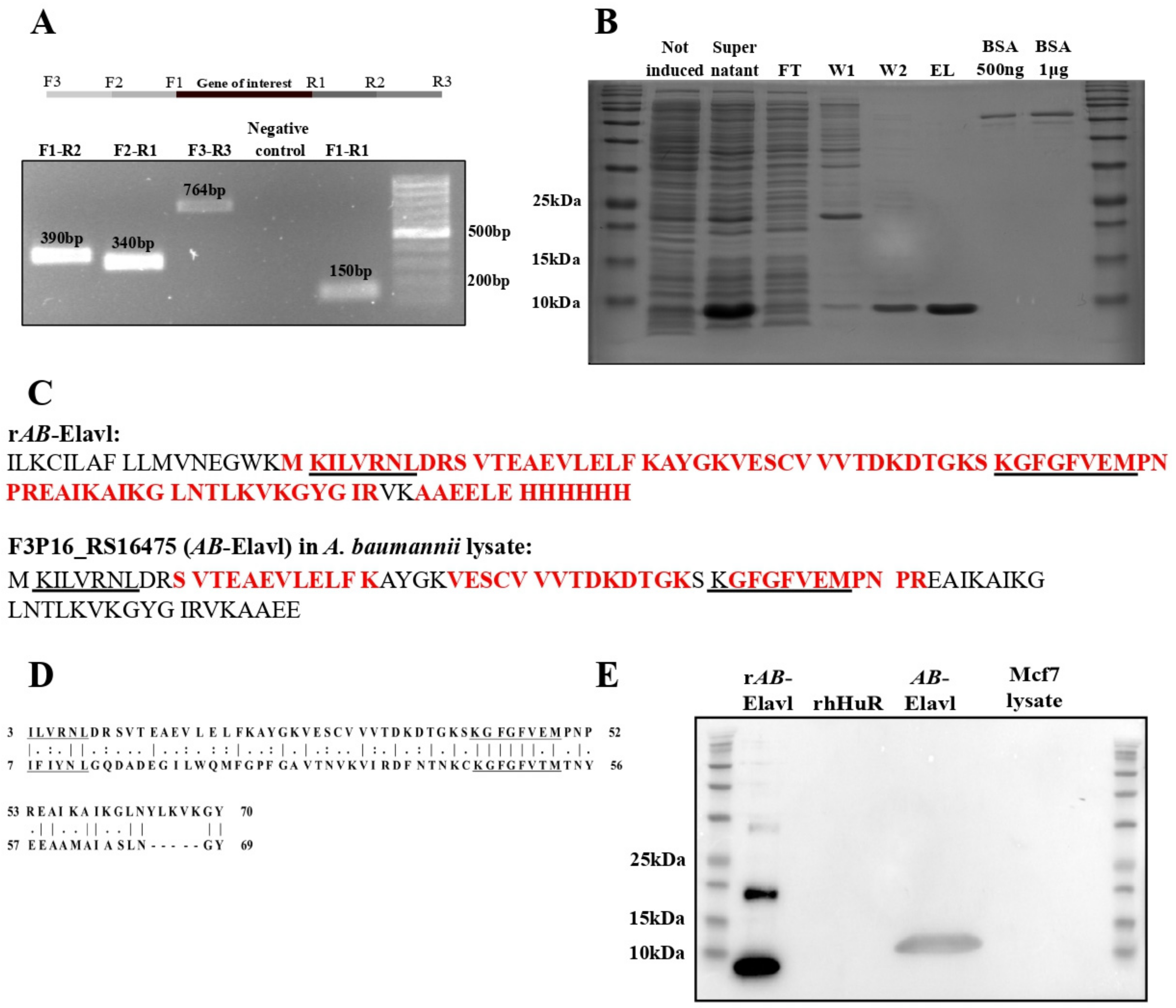
3.1. Identification of a Putative RRM Containing RBP, AB-Elavl
3.2. AB-Elavl Gene Is Expressed and Translated in A. baumannii
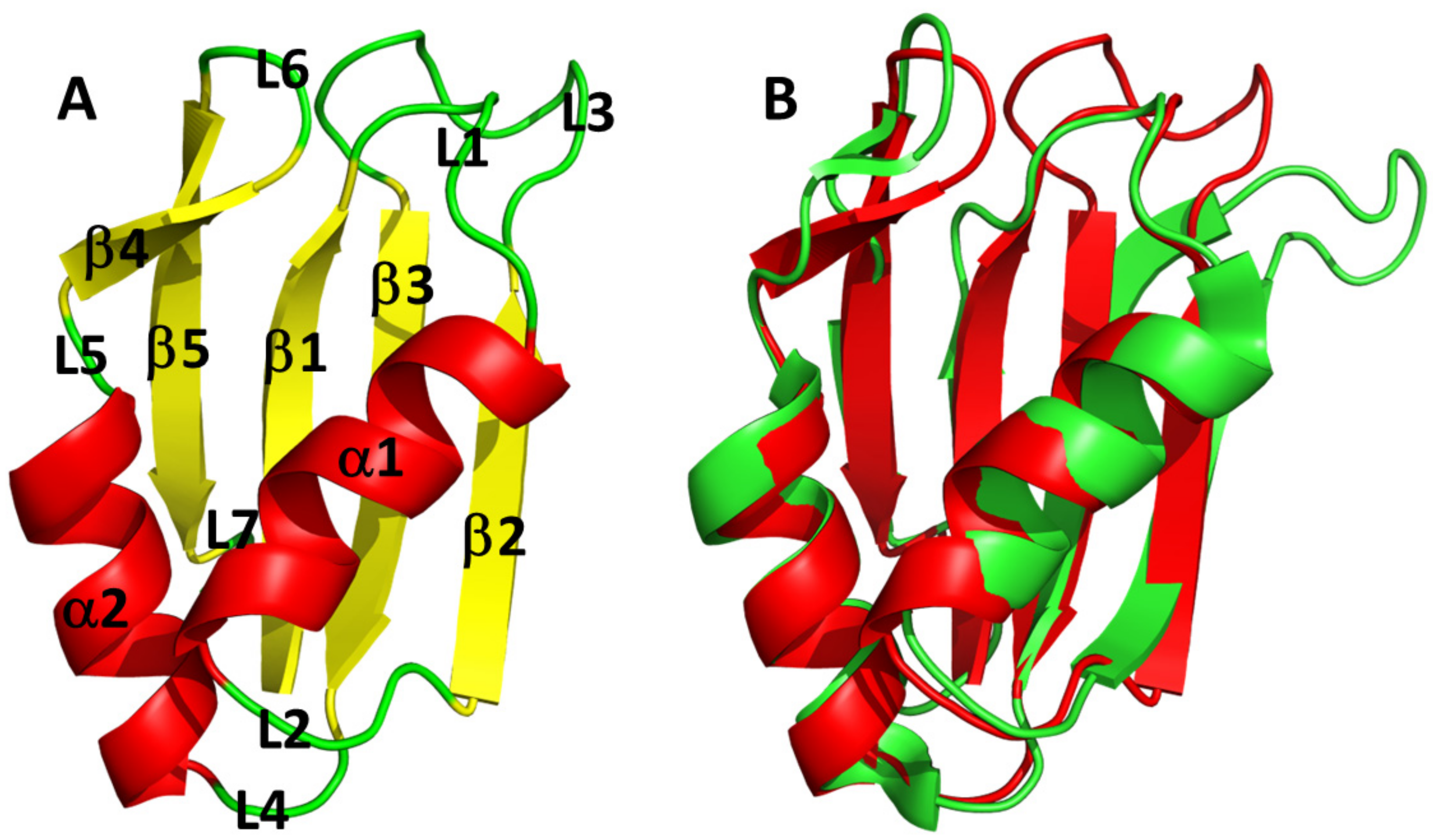
3.3. AB-Elavl Protein Has a Typical Single RRM Domain Structure
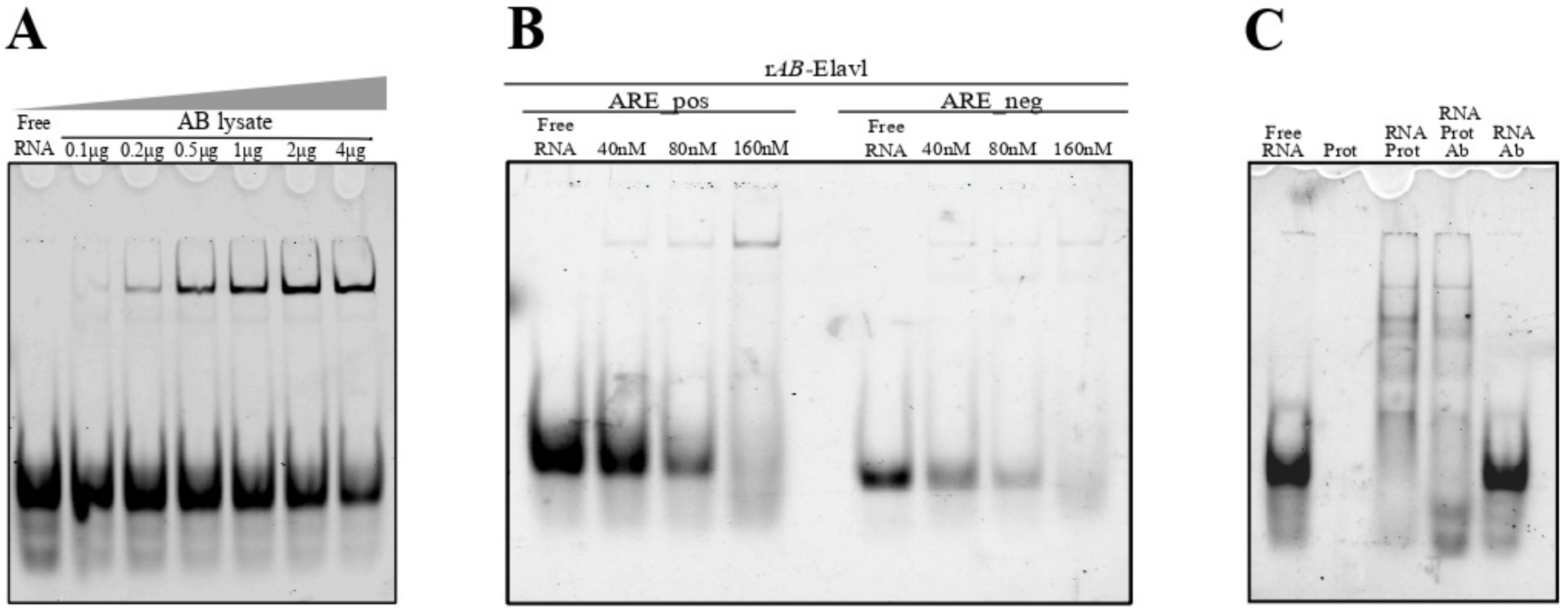
3.4. AB-Elavl Binds AU Rich RNA Probes
3.5. AB-Elavl Binds AU Rich RNA Probes with Nanomolar Affinity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Maris, C.; Dominguez, C.; Allain, F.H.T. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005, 272, 2118–2131. [Google Scholar] [CrossRef] [PubMed]
- Samson, M.L. Rapid functional diversification in the structurally conserved ELAV family of neuronal RNA binding proteins. BMC Genom. 2008, 9, 392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corley, M.; Burns, M.C.; Yeo, G.W. How RNA-Binding Proteins Interact with RNA: Molecules and Mechanisms. Mol. Cell 2020, 78, 9–29. [Google Scholar] [CrossRef] [PubMed]
- Holmqvist, E.; Vogel, J. RNA-binding proteins in bacteria. Nat. Rev. Microbiol. 2018, 16, 601–615. [Google Scholar] [CrossRef]
- Colombrita, C.; Silani, V.; Ratti, A. ELAV proteins along evolution: Back to the nucleus? Mol. Cell Neurosci. 2013, 56, 447–455. [Google Scholar] [CrossRef]
- Gallagher, L.A.; Ramage, E.; Weiss, E.J.; Radey, M.; Hayden, H.S.; Held, K.G.; Huse, H.K.; Zurawski, D.V.; Brittnacher, M.J.; Manoil, C. Resources for Genetic and Genomic Analysis of Emerging Pathogen Acinetobacter baumannii. J. Bacteriol. 2015, 197, 2027–2035. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez-Villoria, A.M.; Valverde-Garduno, V. Antibiotic-Resistant Acinetobacter baumannii Increasing Success Remains a Challenge as a Nosocomial Pathogen. J. Pathog. 2016, 2016, 7318075. [Google Scholar] [CrossRef] [Green Version]
- Farrow, J.M.; Wells, G.; Palethorpe, S.; Adams, M.D.; Pesci, E.C. CsrA Supports both Environmental Persistence and Host-Associated Growth of Acinetobacter baumannii. Infect. Immun. 2020, 88, 58–67. [Google Scholar] [CrossRef]
- Hubloher, J.J.; Schabacker, K.; Müller, V.; Averhoff, B. CsrA Coordinates Compatible Solute Synthesis in Acinetobacter baumannii and Facilitates Growth in Human Urine. Microbiol. Spectr. 2021, 9, e01296-21. [Google Scholar] [CrossRef]
- Moll, I.; Afonyushkin, T.; Vytvytska, O.; Kaberdin, V.R.; Bläsi, U. Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA 2003, 9, 1308–1314. [Google Scholar] [CrossRef] [Green Version]
- Sharma, A.; Dubey, V.; Sharma, R.; Devnath, K.; Gupta, V.K.; Akhter, J.; Bhando, T.; Verma, A.; Ambatipudi, K.; Sarkar, M.; et al. The unusual glycine-rich C terminus of the Acinetobacter baumannii RNA chaperone Hfq plays an important role in bacterial physiology. J. Biol. Chem. 2018, 293, 13377–13388. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Li, R.Q.; Wang, L.; Yang, W.T.; Zou, Q.H.; Xiao, D. Proteomic Analyses of Acinetobacter baumannii Clinical Isolates to Identify Drug Resistant Mechanism. Front. Cell Infect. Microbiol. 2021, 11, 625430. [Google Scholar] [CrossRef]
- Good, P.J. A conserved family of elav-like genes in vertebrates. Proc. Natl. Acad. Sci. USA 1995, 92, 4557–4561. [Google Scholar] [CrossRef] [Green Version]
- Ma, W.J.; Cheng, S.; Campbell, C.; Wright, A.; Furneaux, H. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J. Biol. Chem. 1996, 271, 8144–8151. [Google Scholar] [CrossRef] [Green Version]
- Assoni, G.; La Pietra, V.; Digilio, R.; Ciani, C.; Licata, N.V.; Micaelli, M.; Facen, E.; Tomaszewska, W.; Cerofolini, L.; Pérez-Ràfols, A.; et al. HuR-targeted agents: An insight into medicinal chemistry, biophysical, computational studies and pharmacological effects on cancer models. Adv. Drug Deliv. Rev. 2021, 181, 114088. [Google Scholar] [CrossRef]
- Whiteside, M.D.; Winsor, G.L.; Laird, M.R.; Brinkman, F.S.L. OrtholugeDB: A bacterial and archaeal orthology resource for improved comparative genomic analysis. Nucleic Acids Res. 2013, 4, D366–D376. [Google Scholar] [CrossRef] [Green Version]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, w202–w208. [Google Scholar] [CrossRef]
- Kundu, P.; Fabian, M.R.; Sonenberg, N.; Bhattacharyya, S.N.; Filipowicz, W. HuR protein attenuates miRNA-mediated repression by promoting miRISC dissociation from the target RNA. Nucleic Acids Res. 2012, 40, 5088–5100. [Google Scholar] [CrossRef]
- Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010, 66 Pt 2, 125–132. [Google Scholar] [CrossRef] [Green Version]
- Webb, B.; Sali, A. Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Bioinforma. 2016, 54, 5.6.1–5.6.37. [Google Scholar] [CrossRef] [Green Version]
- Vagin, A.; Teplyakov, A. Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 2010, 66 Pt 1, 22–25. [Google Scholar] [CrossRef]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.-W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010, 66 Pt 2, 213–221. [Google Scholar] [CrossRef] [Green Version]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66 Pt 4, 486–501. [Google Scholar] [CrossRef] [Green Version]
- Chen, V.B.; Arendall, W.B.; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010, 66 Pt 1, 12–21. [Google Scholar] [CrossRef] [Green Version]
- Kay, L.E.; Xu, G.Y.; Yamazaki, T. Enhanced-Sensitivity Triple-Resonance Spectroscopy with Minimal H2O Saturation. J. Magn. Reson. Ser. A 1994, 109, 129–133. [Google Scholar] [CrossRef]
- Shen, Y.; Delaglio, F.; Cornilescu, G.; Bax, A. TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 2009, 44, 213–223. [Google Scholar] [CrossRef]
- Manzoni, L.; Zucal, C.; Di Maio, D.; D’Agostino, V.G.; Thongon, N.; Bonomo, I.; Lal, P.; Miceli, M.; Baj, V.; Brambilla, M.; et al. Interfering with HuR-RNA Interaction: Design, Synthesis and Biological Characterization of Tanshinone Mimics as Novel, Effective HuR Inhibitors. J. Med. Chem. 2018, 61, 1483–1498. [Google Scholar] [CrossRef]
- D’Agostino, V.G.V.G.; Sighel, D.; Zucal, C.; Bonomo, I.; Micaelli, M.; Lolli, G.; Provenzani, A.; Quattrone, A.; Adami, V. Screening Approaches for Targeting Ribonucleoprotein Complexes: A New Dimension for Drug Discovery. SLAS Discov. 2019, 24, 314–331. [Google Scholar] [CrossRef]
- D’Agostino, V.G.V.G.; Adami, V.; Provenzani, A. A Novel High Throughput Biochemical Assay to Evaluate the HuR Protein-RNA Complex Formation. PLoS ONE 2013, 8, e72426. [Google Scholar] [CrossRef] [Green Version]
- Keene, J.D.; Lager, P.J. Post-transcriptional operons and regulons co-ordinating gene expression. Chromosome Res. 2005, 13, 327–337. [Google Scholar] [CrossRef]
- Aguilan, J.T.; Kulej, K.; Sidoli, S. Guide for protein fold change and p-value calculation for non-experts in proteomics. Mol. Omi. 2020, 16, 573–582. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.H.; Han, S.P.; Kassahn, K.S.; Skarshewski, A.; Rothnagel, J.A.; Smith, R. Complex evolutionary relationships among four classes of modular RNA-binding splicing regulators in eukaryotes: The hnRNP, SR, ELAV-like and CELF proteins. J. Mol. Evol. 2012, 75, 214–228. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, K.; Sato, N.; Ohta, N. Conservation of structure and cold-regulation of RNA-binding proteins in cyanobacteria: Probable convergent evolution with eukaryotic glycine-rich RNA-binding proteins. Nucleic Acids Res. 1999, 27, 2029–2036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asensio, N.C.; Rendón, J.M.; Burgas, M.T. Time-Resolved Transcriptional Profiling of Epithelial Cells Infected by Intracellular Acinetobacter baumannii. Microorganisms 2021, 9, 354. [Google Scholar] [CrossRef]
- Kaur, H.; Kalia, M.; Taneja, N. Identification of novel non-homologous drug targets against Acinetobacter baumannii using subtractive genomics and comparative metabolic pathway analysis. Microb. Pathog. 2021, 152, 104608. [Google Scholar] [CrossRef]



| Parameter | Values | Parameter | Values |
|---|---|---|---|
| Wavelength (Å) | 1.541 | R-free | 0.2280 (0.3590) |
| Resolution range | 17.39–1.654 (1.713 –1.654) | CC (work) | 0.953 (0.523) |
| Space group | I 41 | CC (free) | 0.935 (0.489) |
| Unit cell (Å) | 69.56 | Number of non-hydrogen atoms | 642 |
| 69.56 | |||
| 32.46 | |||
| Total reflections | 96620 (2480) | Protein | 601 |
| Unique reflections | 8226 (502) | Solvent | 41 |
| Multiplicity | 11.7 (4.9) | Protein residues | 78 |
| Completeness (%) | 87.01 (53.41) | RMSD (bonds) (Å) | 0.013 |
| Mean I/sigma(I) | 18.74 (1.52) | RMSD (angles) (°) | 1.74 |
| Wilson B-factor | 24.38 | Ramachandran favored (%) | 96.05 |
| R-merge | 0.07861 (0.8945) | Ramachandran allowed (%) | 1.32 |
| R-meas | 0.08199 (0.9341) | Ramachandran outliers (%) | 2.63 |
| R-pim | 0.02281 (0.4356) | Rotamer outliers (%) | 6.15 |
| CC1/2 | 0.999 (0.364) | Clashscore | 12.87 |
| Average B-factor (Å2) | 36.18 | ||
| Reflections used in refinement | 8214 (501) | protein (Å2) | 36.45 |
| Reflections used for R-free | 411 (25) | solvent (Å2) | 32.25 |
| R-work | 0.2049 (0.3354) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ciani, C.; Pérez-Ràfols, A.; Bonomo, I.; Micaelli, M.; Esposito, A.; Zucal, C.; Belli, R.; D’Agostino, V.G.; Bianconi, I.; Calderone, V.; et al. Identification and Characterization of an RRM-Containing, RNA Binding Protein in Acinetobacter baumannii. Biomolecules 2022, 12, 922. https://doi.org/10.3390/biom12070922
Ciani C, Pérez-Ràfols A, Bonomo I, Micaelli M, Esposito A, Zucal C, Belli R, D’Agostino VG, Bianconi I, Calderone V, et al. Identification and Characterization of an RRM-Containing, RNA Binding Protein in Acinetobacter baumannii. Biomolecules. 2022; 12(7):922. https://doi.org/10.3390/biom12070922
Chicago/Turabian StyleCiani, Caterina, Anna Pérez-Ràfols, Isabelle Bonomo, Mariachiara Micaelli, Alfonso Esposito, Chiara Zucal, Romina Belli, Vito Giuseppe D’Agostino, Irene Bianconi, Vito Calderone, and et al. 2022. "Identification and Characterization of an RRM-Containing, RNA Binding Protein in Acinetobacter baumannii" Biomolecules 12, no. 7: 922. https://doi.org/10.3390/biom12070922
APA StyleCiani, C., Pérez-Ràfols, A., Bonomo, I., Micaelli, M., Esposito, A., Zucal, C., Belli, R., D’Agostino, V. G., Bianconi, I., Calderone, V., Cerofolini, L., Massidda, O., Whalen, M. B., Fragai, M., & Provenzani, A. (2022). Identification and Characterization of an RRM-Containing, RNA Binding Protein in Acinetobacter baumannii. Biomolecules, 12(7), 922. https://doi.org/10.3390/biom12070922









