l-Ornithine-N5-monooxygenase (PvdA) Substrate Analogue Inhibitors for Pseudomonas aeruginosa Infections Treatment: Drug Repurposing Computational Studies
Abstract
1. Introduction
2. Experimental Section
2.1. Collection of Substrate Analogues
2.2. Computational Details
2.3. Preparation of Protein and Ligand Structures
2.4. Molecular Docking Studies
2.5. Prediction of Drug Properties
2.6. Drug-Likeness
2.7. Molecular Dynamics Simulation
3. Results
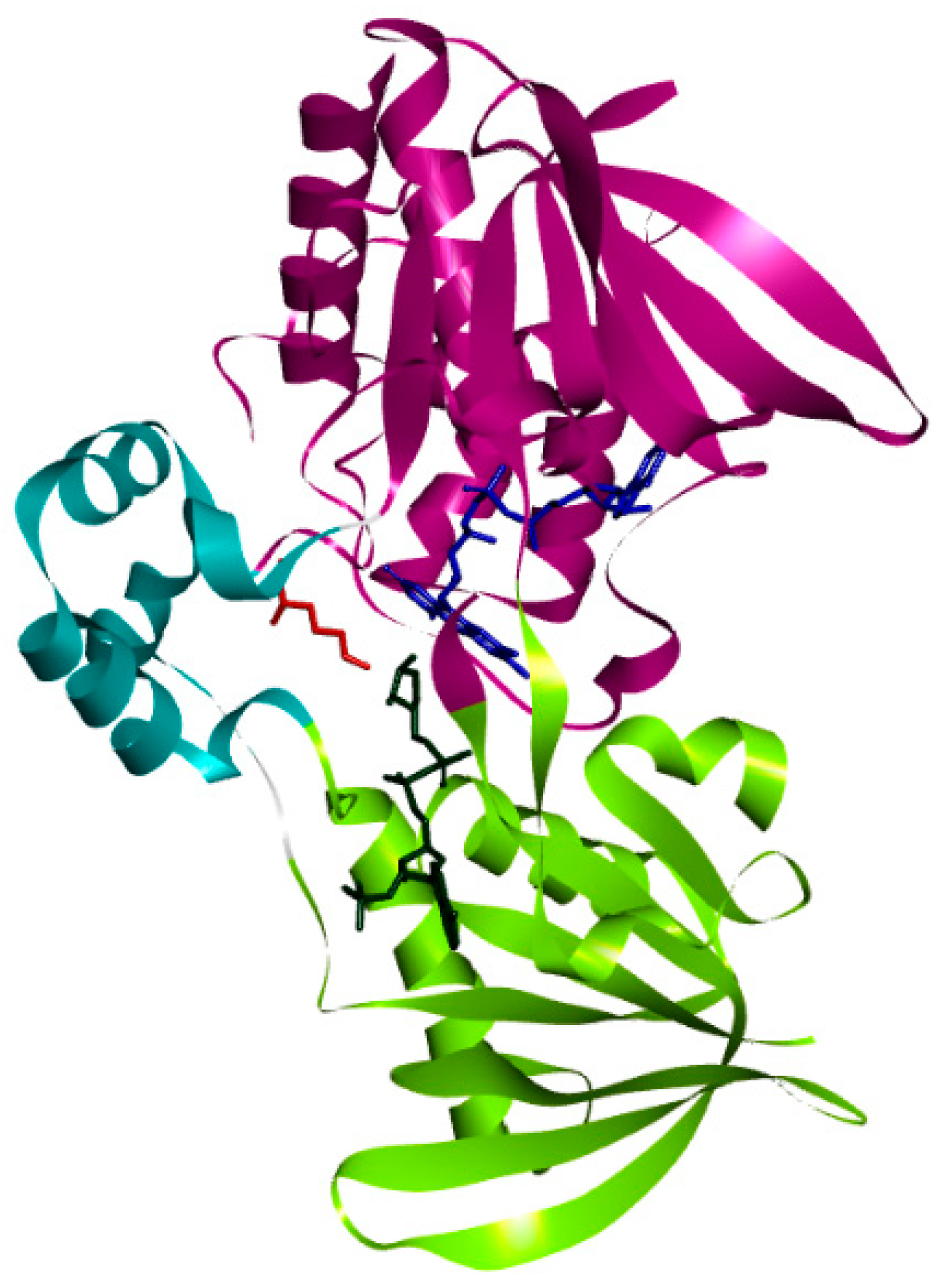
3.1. Structure of PvdA
3.2. Top Binders of PvdA
3.3. ADME Properties and Drug-Likeness
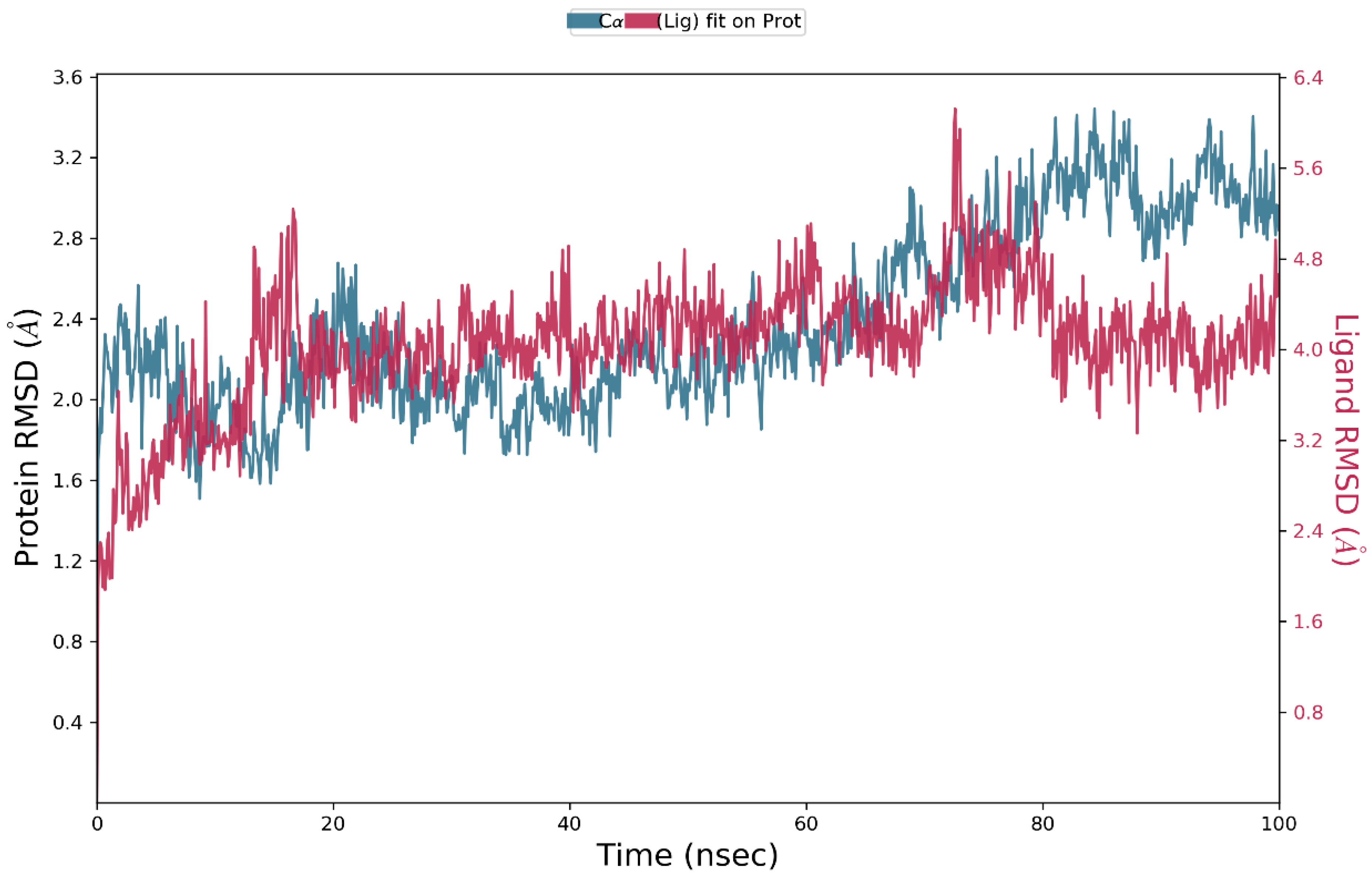
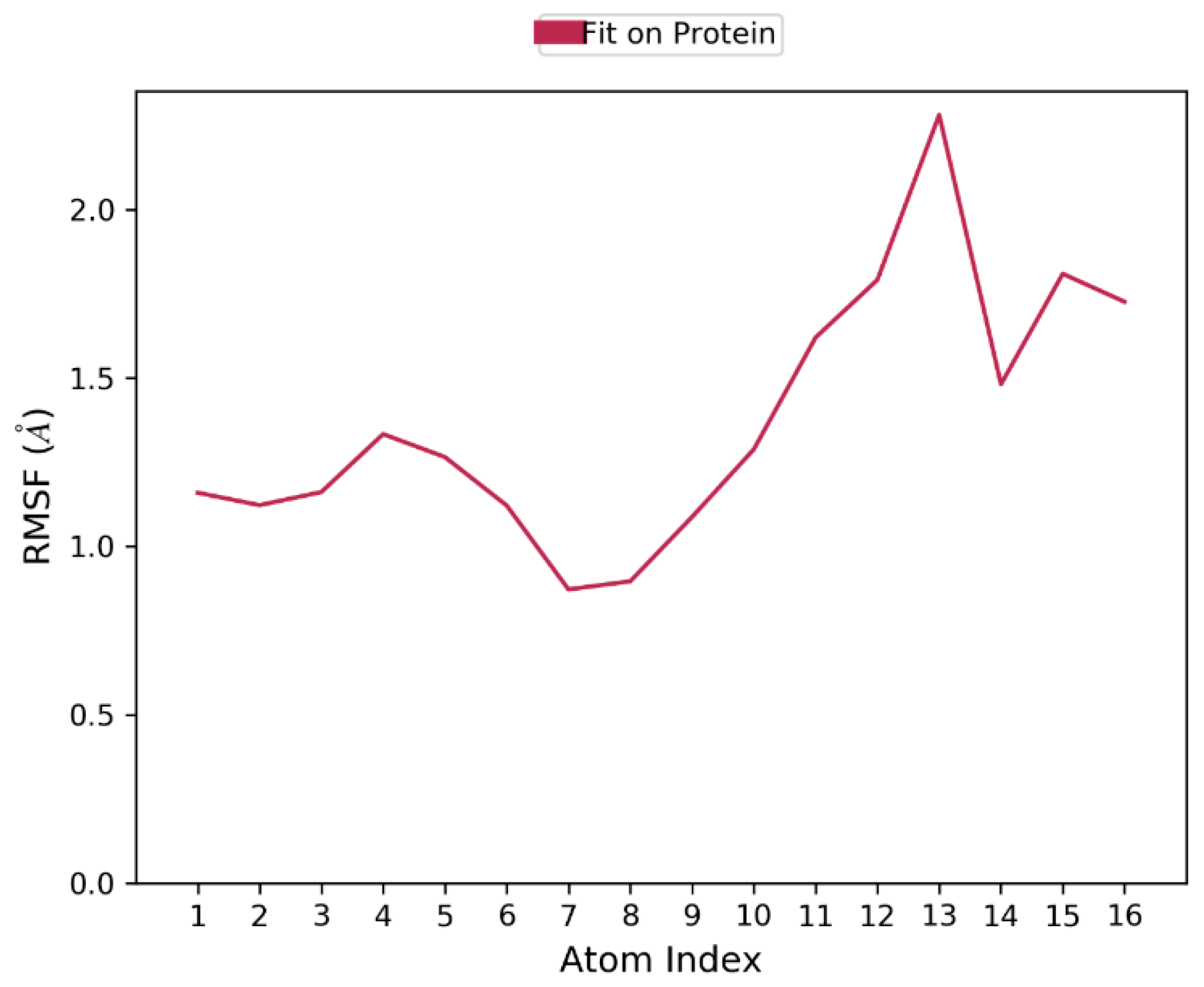
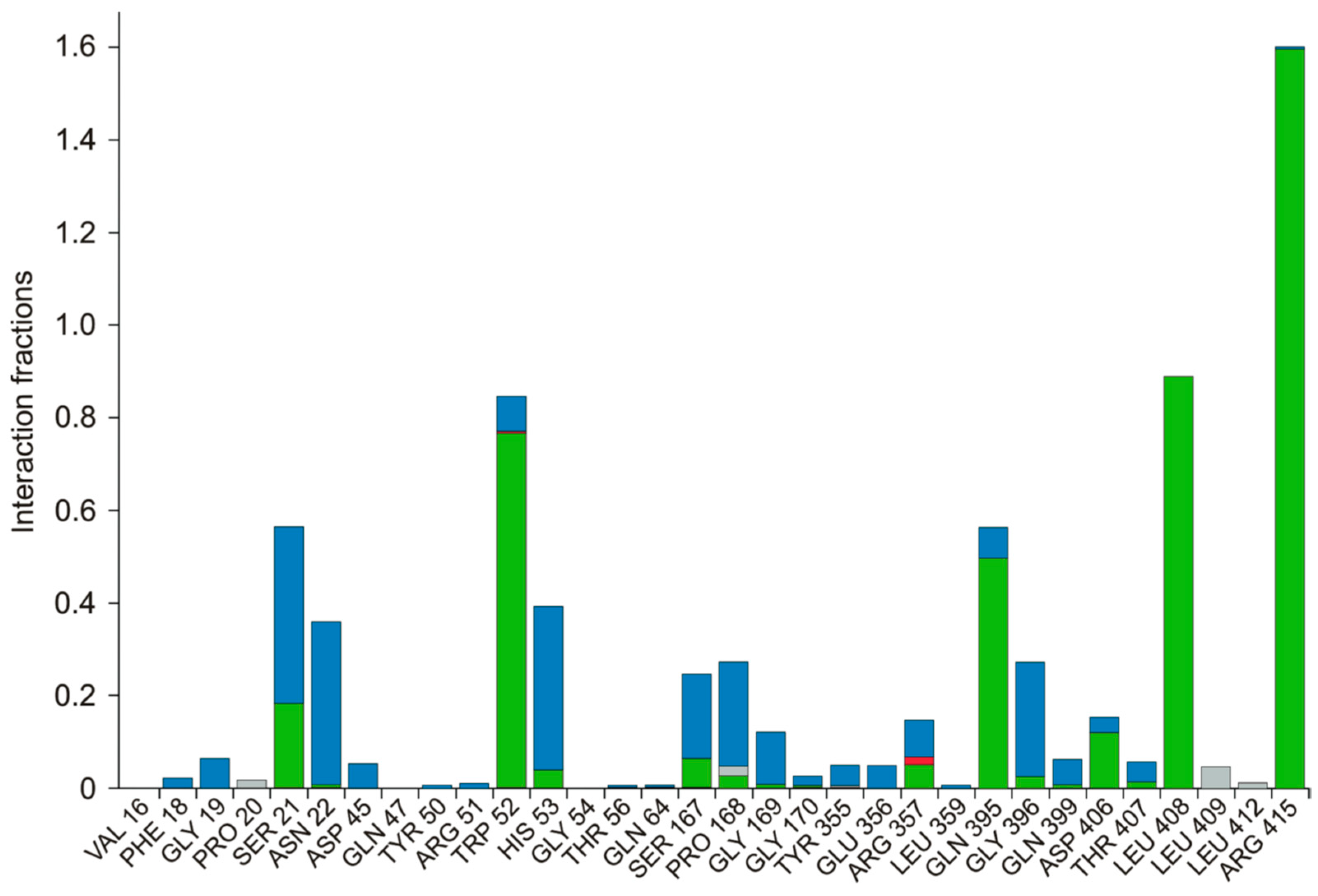
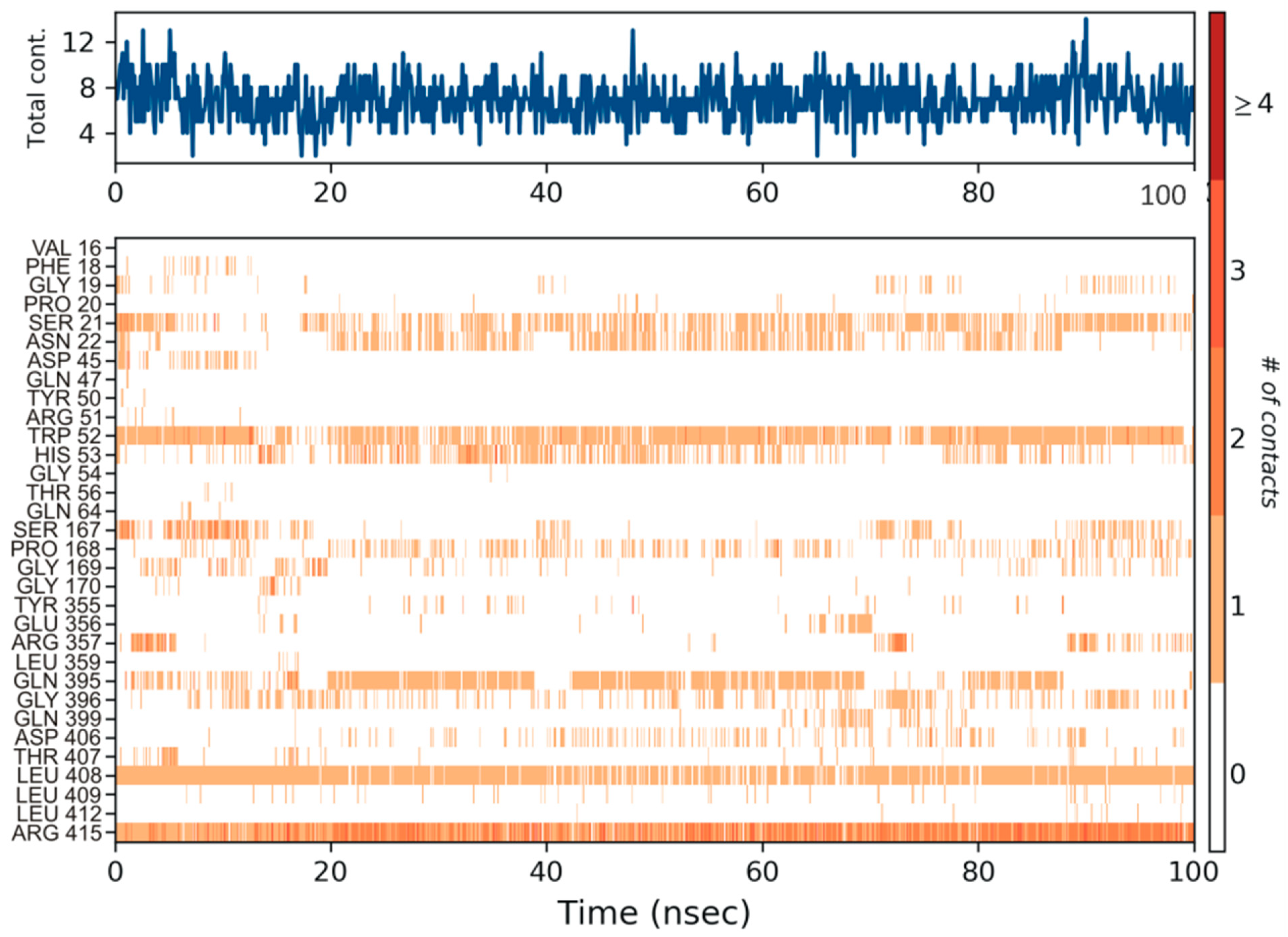
3.4. Molecular Dynamics Simulation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baron, C. Antivirulence drugs to target bacterial secretion systems. Curr. Opin. Microbiol. 2010, 13, 100–105. [Google Scholar] [CrossRef] [PubMed]
- Calvert, M.B.; Jumde, V.R.; Titz, A. Pathoblockers or antivirulence drugs as a new option for the treatment of bacterial infections. Beilstein J. Org. Chem. 2018, 14, 2607–2617. [Google Scholar] [CrossRef] [PubMed]
- D’Angelo, F.; Baldelli, V.; Halliday, N.; Pantalone, P.; Polticelli, F.; Fiscarelli, E.; Williams, P.; Visca, P.; Leoni, L.; Rampioni, G. Identification of FDA-approved drugs as antivirulence agents targeting the pqs quorum-sensing system of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2018, 62, e01296-18. [Google Scholar] [CrossRef] [PubMed]
- Foley, T.L.; Simeonov, A. Targeting iron assimilation to develop new antibacterials. Expert Opin. Drug Discov. 2012, 7, 831–847. [Google Scholar] [CrossRef]
- Smith, D.J.; Lamont, I.L.; Anderson, G.J.; Reid, D.W. Targeting iron uptake to control Pseudomonas aeruginosa infections in cystic fibrosis. Eur. Respir. J. 2013, 42, 1723–1736. [Google Scholar] [CrossRef]
- Litton, E.; Lim, J. Iron metabolism: An emerging therapeutic target in critical illness. Annu. Update Intensive Care Emerg. Med. 2019, 2019, 573–584. [Google Scholar]
- Breidenstein, E.B.; de la Fuente-Núñez, C.; Hancock, R.E. Pseudomonas aeruginosa: All roads lead to resistance. Trends Microbiol. 2011, 19, 419–426. [Google Scholar] [CrossRef]
- Cox, C.D.; Adams, P. Siderophore activity of pyoverdin for Pseudomonas aeruginosa. Infect. Immun. 1985, 48, 130–138. [Google Scholar] [CrossRef]
- Cox, C.D.; Rinehart, K.L.; Moore, M.L.; Cook, J.C. Pyochelin: Novel structure of an iron-chelating growth promoter for Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 1981, 78, 4256–4260. [Google Scholar] [CrossRef]
- Meyer, J.M.; Neely, A.; Stintzi, A.; Georges, C.; Holder, I.A. Pyoverdin is essential for virulence of Pseudomonas aeruginosa. Infect. Immun. 1996, 64, 518–523. [Google Scholar] [CrossRef]
- Takase, H.; Nitanai, H.; Hoshino, K.; Otani, T. Impact of Siderophore Production on Pseudomonas aeruginosa Infections in Immunosuppressed Mice. Infect. Immun. 2000, 68, 1834–1839. [Google Scholar] [CrossRef] [PubMed]
- Kirienko, N.V.; Kirienko, D.R.; Larkins-Ford, J.; Wählby, C.; Ruvkun, G.; Ausubel, F.M. Pseudomonas aeruginosa disrupts Caenorhabditis elegans iron homeostasis, causing a hypoxic response and death. Cell Host Microbe 2013, 13, 406–416. [Google Scholar] [CrossRef]
- Minandri, F.; Imperi, F.; Frangipani, E.; Bonchi, C.; Visaggio, D.; Facchini, M.; Pasquali, P.; Bragonzi, A.; Visca, P. Role of iron uptake systems in Pseudomonas aeruginosa virulence and airway infection. Infect. Immun. 2016, 84, 2324–2335. [Google Scholar] [CrossRef] [PubMed]
- Kirienko, N.V.; Ausubel, F.M.; Ruvkun, G. Mitophagy confers resistance to siderophore-mediated killing by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2015, 112, 1821–1826. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.; Kirienko, D.R.; Webster, P.; Fisher, A.L.; Kirienko, N.V. Pyoverdine, a siderophore from Pseudomonas aeruginosa, translocates into C. elegans, removes iron, and activates a distinct host response. Virulence 2018, 9, 804–817. [Google Scholar] [CrossRef] [PubMed]
- Kirienko, D.R.; Kang, D.; Kirienko, N.V. Novel pyoverdine inhibitors mitigate Pseudomonas aeruginosa pathogenesis. Front. Microbiol. 2019, 9, 3317. [Google Scholar] [CrossRef]
- Caspi, R.; Altman, T.; Billington, R.; Dreher, K.; Foerster, H.; Fulcher, C.A.; Holland, T.A.; Keseler, I.M.; Kothari, A.; Kubo, A.; et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res. 2014, 42, D459–D471. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Olucha, J.; Meneely, K.M.; Chilton, A.S.; Lamb, A.L. Two structures of an N-hydroxylating flavoprotein monooxygenase: Ornithine hydroxylase from Pseudomonas aeruginosa. J. Biol. Chem. 2011, 286, 31789–31798. [Google Scholar] [CrossRef] [PubMed]
- Al-Masoudi, N.A.; Elias, R.S.; Saeed, B. Molecular docking studies of some antiviral and antimalarial drugs via bindings to 3cl-protease and polymerase enzymes of the novel coronavirus (Sars-cov-2). Biointerface Res. Appl. Chem. 2020, 10, 6444–6459. [Google Scholar]
- Yedidi, R.S.; Liu, Z.; Kovari, I.A.; Woster, P.M.; Kovari, L.C. P1 and P1′ para-fluoro phenyl groups show enhanced binding and favorable predicted pharmacological properties: Structure-based virtual screening of extended lopinavir analogs against multi-drug resistant HIV-1 protease. J. Mol. Graph. Model. 2014, 47, 18–24. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Meneely, K.M.; Lamb, A.L. Biochemical characterization of a flavin adenine dinucleotide-dependent monooxygenase, ornithine hydroxylase from Pseudomonas aeruginosa, suggests a novel reaction mechanism. Biochemistry 2007, 46, 11930–11937. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997, 23, 3–25. [Google Scholar] [CrossRef]
- Ghose, A.K.; Viswanadhan, V.N.; Wendoloski, J.J. A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. A qualitative and quantitative characterization of known drug databases. J. Comb. Chem. 1999, 1, 55–68. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.-Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef]
- Egan, W.J.; Merz, K.M.; Baldwin, J.J. Prediction of drug absorption using multivariate statistics. J. Med. Chem. 2000, 43, 3867–3877. [Google Scholar] [CrossRef]
- Muegge, I.; Heald, S.L.; Brittelli, D. Simple selection criteria for drug-like chemical matter. J. Med. Chem. 2001, 44, 1841–1846. [Google Scholar] [CrossRef]
- Chinnamanyakar, R.; Ramanathan, E.M. Anti-cancer and antimicrobial activity, in-silico ADME and docking studies of biphenyl pyrazoline derivatives. Biointerface Res. Appl. Chem. 2021, 11, 8266–8282. [Google Scholar]
- Mark, P.; Nilsson, L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J. Phys. Chem. A 2001, 105, 9954–9960. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Maxwell, D.S.; Tirado-Rives, J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 1996, 118, 11225–11236. [Google Scholar] [CrossRef]
- Tuckerman, M.; Berne, B.J.; Martyna, G.J. Reversible multiple time scale molecular dynamics. J. Chem. Phys. 1992, 97, 1990–2001. [Google Scholar] [CrossRef]
- Cheng, A.; Merz, K.M. Application of the nosé−hoover chain algorithm to the study of protein dynamics. J. Phys. Chem. 1996, 100, 1927–1937. [Google Scholar] [CrossRef]
- Kalibaeva, G.; Ferrario, M.; Ciccotti, G. Constant pressure-constant temperature molecular dynamics: A correct constrained NPT ensemble using the molecular virial. Mol. Phys. 2003, 101, 765–778. [Google Scholar] [CrossRef]
- Kumar, B.K.; Faheem, N.; Sekhar, K.V.G.C.; Ojha, R.; Prajapati, V.K.; Pai, A.; Murugesan, S. Pharmacophore based virtual screening, molecular docking, molecular dynamics and MM-GBSA approach for identification of prospective SARS-CoV-2 inhibitor from natural product databases. J. Biomol. Struct. Dyn. 2022, 40, 1363–1386. [Google Scholar] [CrossRef]
- Dreyer, G.B.; Metcalf, B.W.; ATomaszek, T.; Carr, T.J.; Chandler, A.C.; Hyland, L.; AFakhoury, S.; Magaard, V.W.; Moore, M.L.; EStrickler, J. Inhibition of human immunodeficiency virus 1 protease in vitro: Rational design of substrate analogue inhibitors. Proc. Natl. Acad. Sci. USA 1989, 86, 9752–9756. [Google Scholar] [CrossRef]
- Tomasselli, A.G.; Olsen, M.K.; Hui, J.O.; Staples, D.J.; Sawyer, T.K.; Heinrikson, R.L.; Tomich, C.S.C. Substrate analog inhibition and active site titration of purified recombinant HIV-1 protease. Biochemistry 1990, 29, 264–269. [Google Scholar] [CrossRef]
- Coggins, B.E.; Li, X.; McClerren, A.L.; Hindsgaul, O.; Raetz, C.R.H.; Zhou, P. Structure of the LpxC deacetylase with a bound substrate-analog inhibitor. Nat. Struct. Mol. Biol. 2003, 10, 645–651. [Google Scholar] [CrossRef]
- Drews, J. Drug discovery: A historical perspective. Science 2000, 287, 1960–1964. [Google Scholar] [CrossRef] [PubMed]
- Jann, A.; Stalon, V.; Wauven, C.V.; Leisinger, T.; Haas, D. N-Succinylated intermediates in an arginine catabolic pathway of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 1986, 83, 4937–4941. [Google Scholar] [CrossRef] [PubMed]
- Szucs, T. Cilazapril. A review. Drugs 1991, 41, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Murakami, T.; Horibata, Y.; Morimoto, Y.; Tateno, S.; Kawasoe, Y.; Niwa, K. Syndrome of inappropriate secretion of antidiuretic hormone associated with angiotensin-converting enzyme inhibitor administration. Pediatr. Cardiol. 2013, 34, 1261–1263. [Google Scholar] [CrossRef] [PubMed]

| S. No. | Name of the Drug | Structure | Binding Energy (kcal mol−1) |
|---|---|---|---|
| 1. | (2s)-2,8-Diaminooctanoic Acid |  | −5.2 |
| 2. | 2,6-Diaminopimelic Acid |  | −5.1 |
| 3. | 2-Methylleucine |  | −4.9 |
| 4. | Allo-Isoleucine |  | −4.8 |
| 5. | 4-Carboxy-4-Aminobutanal |  | −4.6 |
| 6. | d-Leucine |  | −4.6 |
| 7. | 2-Aminopimelic Acid |  | −4.6 |
| 8. | d-Lysine |  | −4.6 |
| 9. | 2-Amino-6-Oxo-Hexanoic Acid | 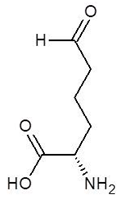 | −4.5 |
| 10. | d-Glutamine | 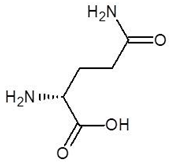 | −4.5 |
| 11. | 6-hydroxy-l-norleucine |  | −4.4 |
| 12. | Norvaline |  | −4.4 |
| 13. | 5-Hydroxy Norvaline | 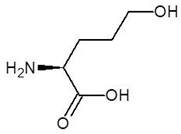 | −4.3 |
| 14. | Alpha-Aminobutyric Acid |  | −4.1 |
| 15. | Delta-Amino Valeric Acid | 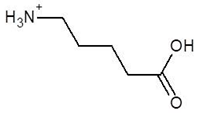 | −3.9 |
| S. No. | Name of the Drug | Structure | Binding Energy (kcal mol−1) |
|---|---|---|---|
| 1. | N-2-Succinyl ornithine | 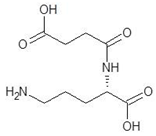 | −12.8 |
| 2. | Trypanothione | 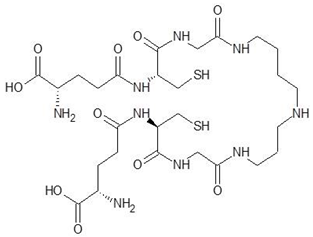 | −11.7 |
| 3. | Talotrexin | 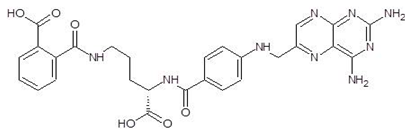 | −11.0 |
| 4. | Davunetide | 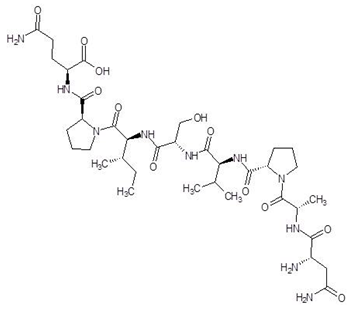 | −11.0 |
| 5. | CTT-1057 |  | −9.4 |
| 6. | Cilazapril | 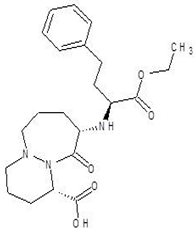 | −9.1 |
| 7. | S-P-Nitrobenzyloxycarbonylglutathione | 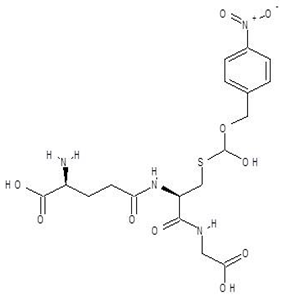 | −8.3 |
| 8. | Glutathione disulfide |  | −8.1 |
| 9. | S-Hydroxymethyl Glutathione | 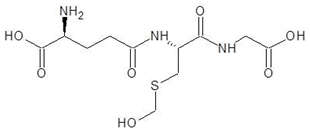 | −7.6 |
| 10. | Glutathione |  | −7.2 |
| 11. | Argininosuccinate | 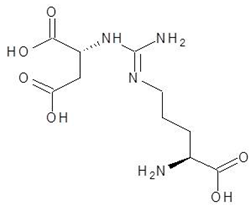 | −6.9 |
| 12. | Glutathione Sulfinate | 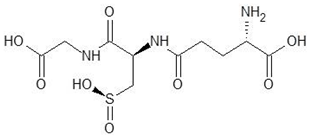 | −6.6 |
| 13. | N-(Phosphonoacetyl)-l-Ornithine |  | −6.3 |
| 14. | N-Alpha-l-Acetyl-Arginine | 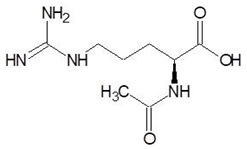 | −6.0 |
| 15. | Gamma-Glutamylcysteine | 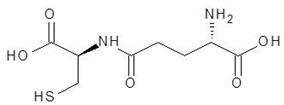 | −6.0 |
| 16. | N-omega-nitro-l-arginine methyl ester |  | −6.0 |
| 17. | S-methyl-glutathione | 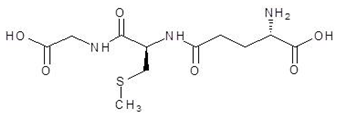 | −5.9 |
| 18. | N-Acetyl-l-Citrulline | 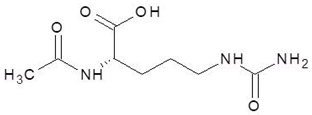 | −5.8 |
| 19. | Nitroarginine | 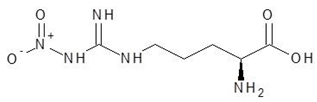 | −5.8 |
| 20. | N3, N4-Dimethylarginine | 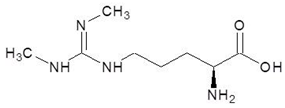 | −5.6 |
| 21. | l-Eflornithine | 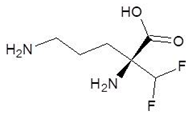 | −5.5 |
| 22. | 5-N-Allyl-Arginine | 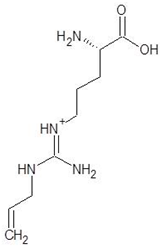 | −5.5 |
| 23. | Glutamine t-butyl ester | 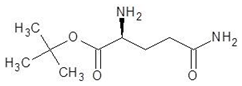 | −5.5 |
| 24. | N, N-dimethylarginine | 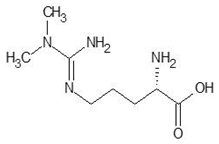 | −5.4 |
| 25. | Aceglutamide | 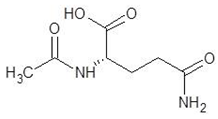 | −5.4 |
| 26. | Tilarginine |  | −5.4 |
| 27. | l-Citrulline | 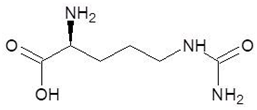 | −5.3 |
| 28. | Glutamine hydroxamate | 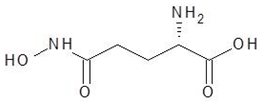 | −5.2 |
| 29. | N5-Methylglutamine | 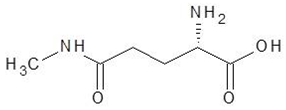 | −4.9 |
| 30. | l-Thiocitrulline | 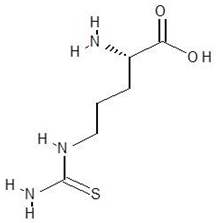 | −4.9 |
| Name of the Protein | l-Ornithine-N5-Monooxygenase (or) Ornithine Hydroxylase |
|---|---|
| Number of amino acids | 463 |
| PDB Id | 3S5W |
| Resolution | 1.9 Å |
| Number of chains | 2 (Homodimer) |
| Number of Domains | Three FAD Binding Domain NADPH Binding Domain Ornithine Binding Domain |
| Amino acids interacting with natural ligand (N5- hydroxyl ornithine) | Lys69, Asn254, Phe257, Asn284, Ser410 |
| Lipinski (Lipinski et al., 1997) | Ghose Ghose, (Viswanadhan, Wendoloski, 1999) | Veber (Veber et al., 2002) | Egan (Egan, Merz, Baldwin, 2000) | Muegge (Muegge, Heald, Brittelli, 2001) |
|---|---|---|---|---|
| MW ≤ 500 MLOGP ≤ 4.15 N or O ≤ 10 NH or OH ≤ 5 | 160 ≤ MW ≤ 480 −0.4 ≤ WLOGP ≤ 5.6 40 ≤ MR ≤ 130 20 ≤ atoms ≤ 70 | Rotatable bonds ≤ 10 TPS ≤ 140 | WLOGP ≤ 5.88 TPSA ≤ 131.6 | 200 ≤ MW ≤ 600 −2 ≤ XLOGP ≤ 5 TPSA ≤ 150 Num. rings ≤ 7 Num. carbon > 4 Num. heteroatoms > 1 Num. rotatable bonds ≤ 15 H-bond acc. ≤ 10 H-bond don. ≤ 5 |
| Name of the Compound | Molecular Weight (g × mol−1) | H-Bond Acceptors | H-Bond Donors | Rotatable Bonds | LogPo/w | GI Absorption | BBB Permeation | Bioavailability Score |
|---|---|---|---|---|---|---|---|---|
| N2- Succinyl-l-ornithine | 232.23 | 6 | 4 | 9 | −1.18 | High | No | 0.56 |
| Trypanothione | 723.86 | 13 | 11 | 33 | −4.20 | Low | No | 0.17 |
| Talotrexin | 573.56 | 10 | 7 | 14 | 0.18 | Low | No | 0.11 |
| Davunetide | 824.92 | 13 | 10 | 29 | −3.07 | Low | No | 0.17 |
| CTT-1057 | 706.61 | 16 | 9 | 28 | −0.12 | Low | No | 0.11 |
| Cilazapril | 417.50 | 7 | 2 | 9 | 1.37 | High | No | 0.55 |
| Name of the Compound | Lipinski | Ghose | Veber | Egan | Muegge |
|---|---|---|---|---|---|
| N-2-Succinyl ornithine | Yes; 0 violation | No; 1 violation: WLOGP < −0.4 | Yes; 0 violation | Yes; 0 violation | No; 1 violation: XLOGP3 < −2 |
| Trypanothione | No; 3 violations: MW > 500, N or O > 10, NH or OH > 5 | No; 4 violations: MW > 480, WLOGP < −0.4, MR > 130, #atoms > 70 | No; 2 violations: Rotors > 10, TPSA > 140 | No; 1 violation: TPSA > 131.6 | No; 6 violations: MW > 600, XLOGP3 < −2, TPSA > 150, Rotors > 15, H-acc > 10, H-don > 5 |
| Talotrexin | No; 3 violations: MW > 500, N or O > 10, NH or OH > 5 | No; 2 violations: MW > 480, MR > 130 | No; 2 violations: Rotors > 10, TPSA > 140 | No; 1 violation: TPSA > 131.6 | No; 2 violations: TPSA > 150, H-don > 5 |
| Davunetide | No; 3 violations: MW > 500, N or O > 10, NH or OH > 5 | No; 4 violations: MW > 480, WLOGP < −0.4, MR > 130, #atoms > 70 | No; 2 violations: Rotors > 10, TPSA > 140 | No; 1 violation: TPSA > 131.6 | No; 6 violations: MW > 600, XLOGP3 < −2, TPSA > 150, Rotors > 15, H-acc > 10, H-don > 5 |
| CTT-1057 | No; 3 violations: MW > 500, N or O > 10, NH or OH > 5 | No; 4 violations: MW > 480, WLOGP < −0.4, MR > 130, #atoms > 70 | No; 2 violations: Rotors > 10, TPSA > 140 | No; 1 violation: TPSA > 131.6 | No; 6 violations : MW > 600, XLOGP3 < −2, TPSA > 150, Rotors > 15, H-acc > 10, H-don > 5 |
| Cilazapril | Yes; 0 violation | Yes; 0 violation | Yes; 0 violation | Yes; 0 violation | Yes; 0 violation |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rosy, J.C.; Babkiewicz, E.; Maszczyk, P.; Mrówka, P.; Kumar, B.K.; Murugesan, S.; Kunjiappan, S.; Sundar, K. l-Ornithine-N5-monooxygenase (PvdA) Substrate Analogue Inhibitors for Pseudomonas aeruginosa Infections Treatment: Drug Repurposing Computational Studies. Biomolecules 2022, 12, 887. https://doi.org/10.3390/biom12070887
Rosy JC, Babkiewicz E, Maszczyk P, Mrówka P, Kumar BK, Murugesan S, Kunjiappan S, Sundar K. l-Ornithine-N5-monooxygenase (PvdA) Substrate Analogue Inhibitors for Pseudomonas aeruginosa Infections Treatment: Drug Repurposing Computational Studies. Biomolecules. 2022; 12(7):887. https://doi.org/10.3390/biom12070887
Chicago/Turabian StyleRosy, Joseph Christina, Ewa Babkiewicz, Piotr Maszczyk, Piotr Mrówka, Banoth Karan Kumar, Sankaranarayanan Murugesan, Selvaraj Kunjiappan, and Krishnan Sundar. 2022. "l-Ornithine-N5-monooxygenase (PvdA) Substrate Analogue Inhibitors for Pseudomonas aeruginosa Infections Treatment: Drug Repurposing Computational Studies" Biomolecules 12, no. 7: 887. https://doi.org/10.3390/biom12070887
APA StyleRosy, J. C., Babkiewicz, E., Maszczyk, P., Mrówka, P., Kumar, B. K., Murugesan, S., Kunjiappan, S., & Sundar, K. (2022). l-Ornithine-N5-monooxygenase (PvdA) Substrate Analogue Inhibitors for Pseudomonas aeruginosa Infections Treatment: Drug Repurposing Computational Studies. Biomolecules, 12(7), 887. https://doi.org/10.3390/biom12070887








