Djhsp60 Is Required for Planarian Regeneration and Homeostasis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Identification of Djhsp60 and RNAi Experiments
2.3. QRT-PCR
2.4. In Situ Hybridization
2.5. Paraffin Section, HE Staining and Immunochemistry
2.6. Immunostaining on Dissociated Cells
2.7. Whole-Mount Immunostaining
2.8. Whole-Mount TUNEL
2.9. RNA Sequencing and Gene Expression Analysis
2.10. Assessment of Mitochondrial Respiration
2.11. Transmission Electron Microscopy (TEM)
2.12. Statistical Analysis
3. Results
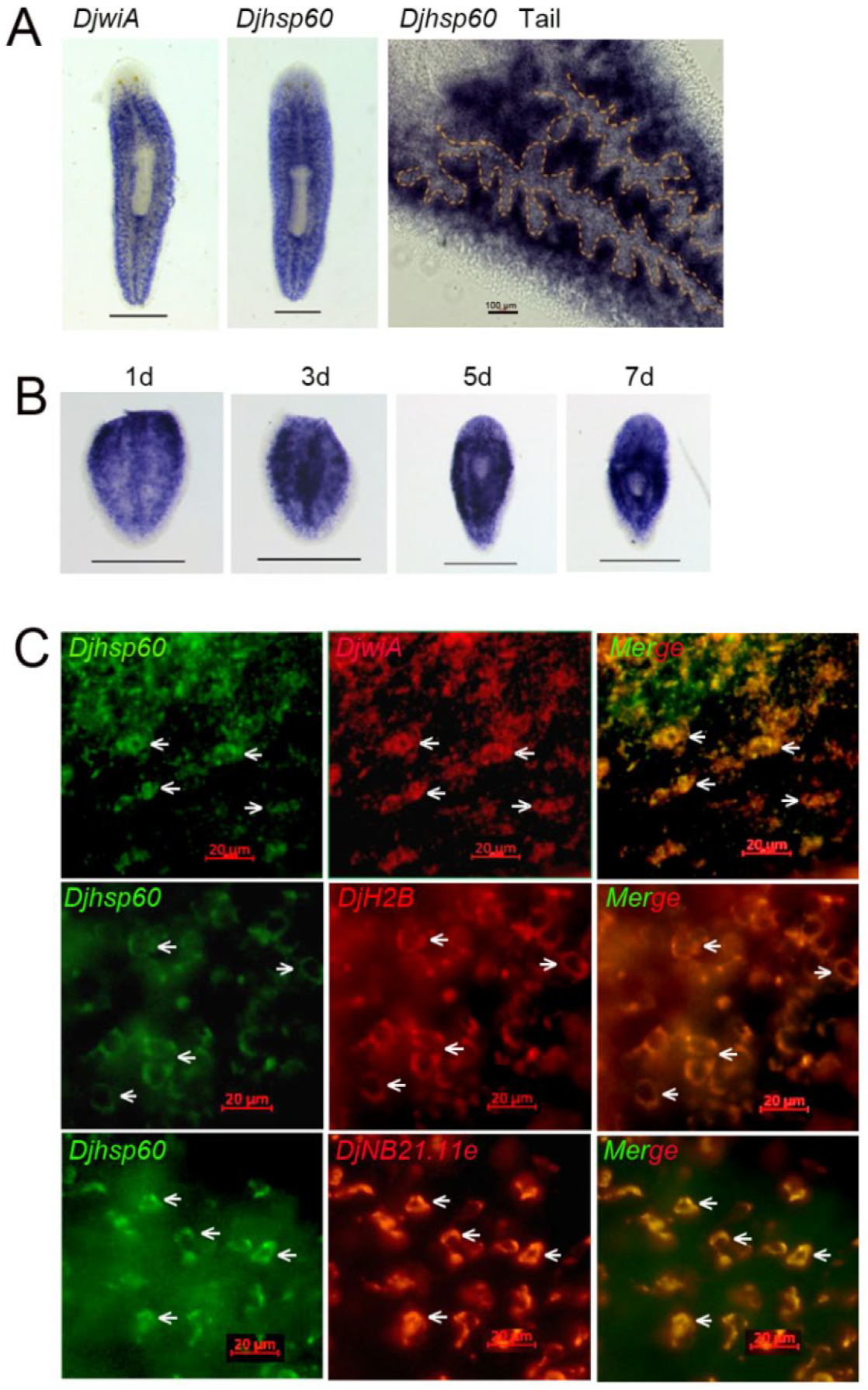
3.1. Expression Patterns and Protein Localization of HSP60 in the Planarian Dugesia japonica
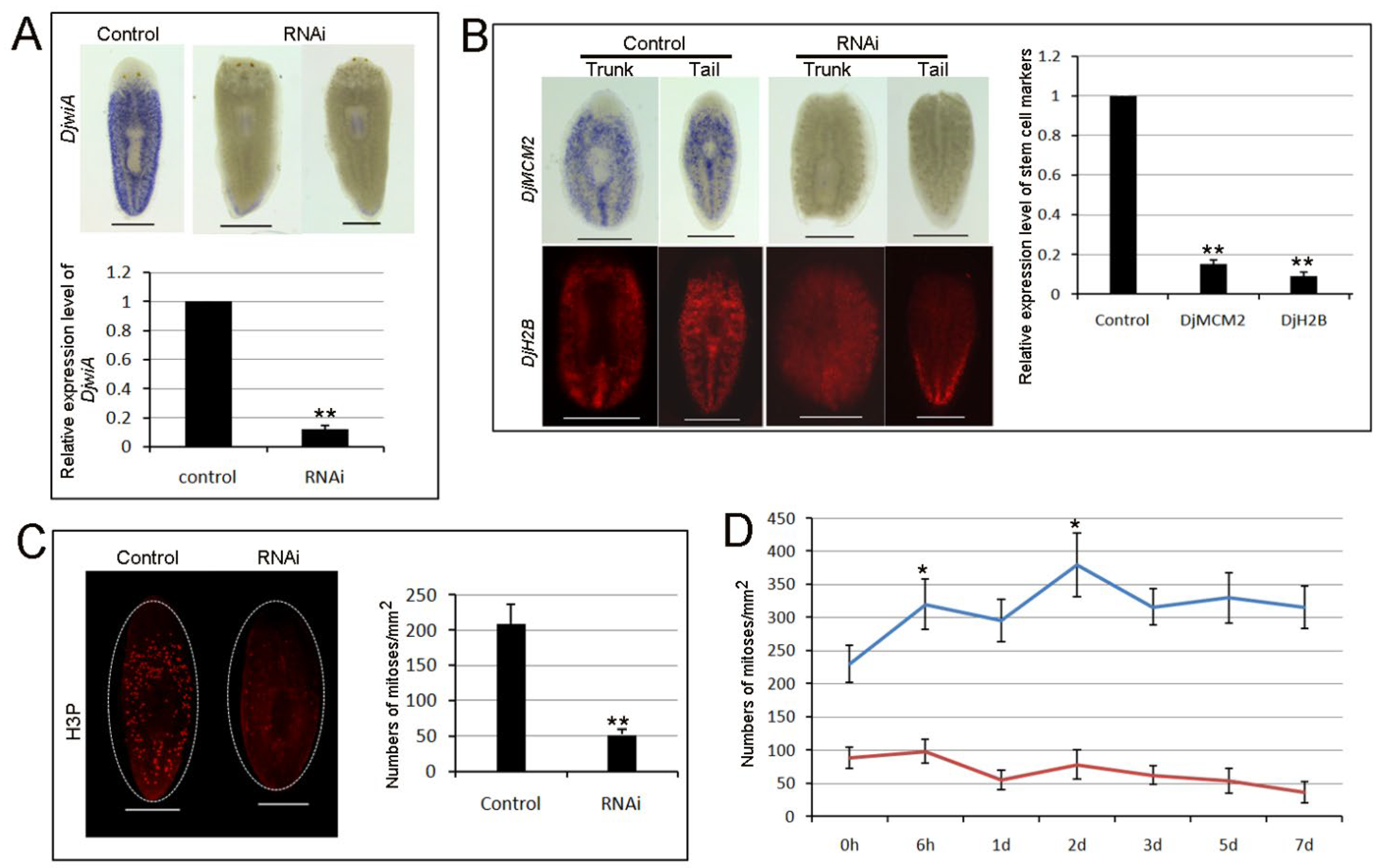
3.2. RNAi-Djhsp60 Affects Planarian Regeneration and Tissue Homeostasis
3.3. Effects of RNAi-Djhsp60 on Stem Cell Viability
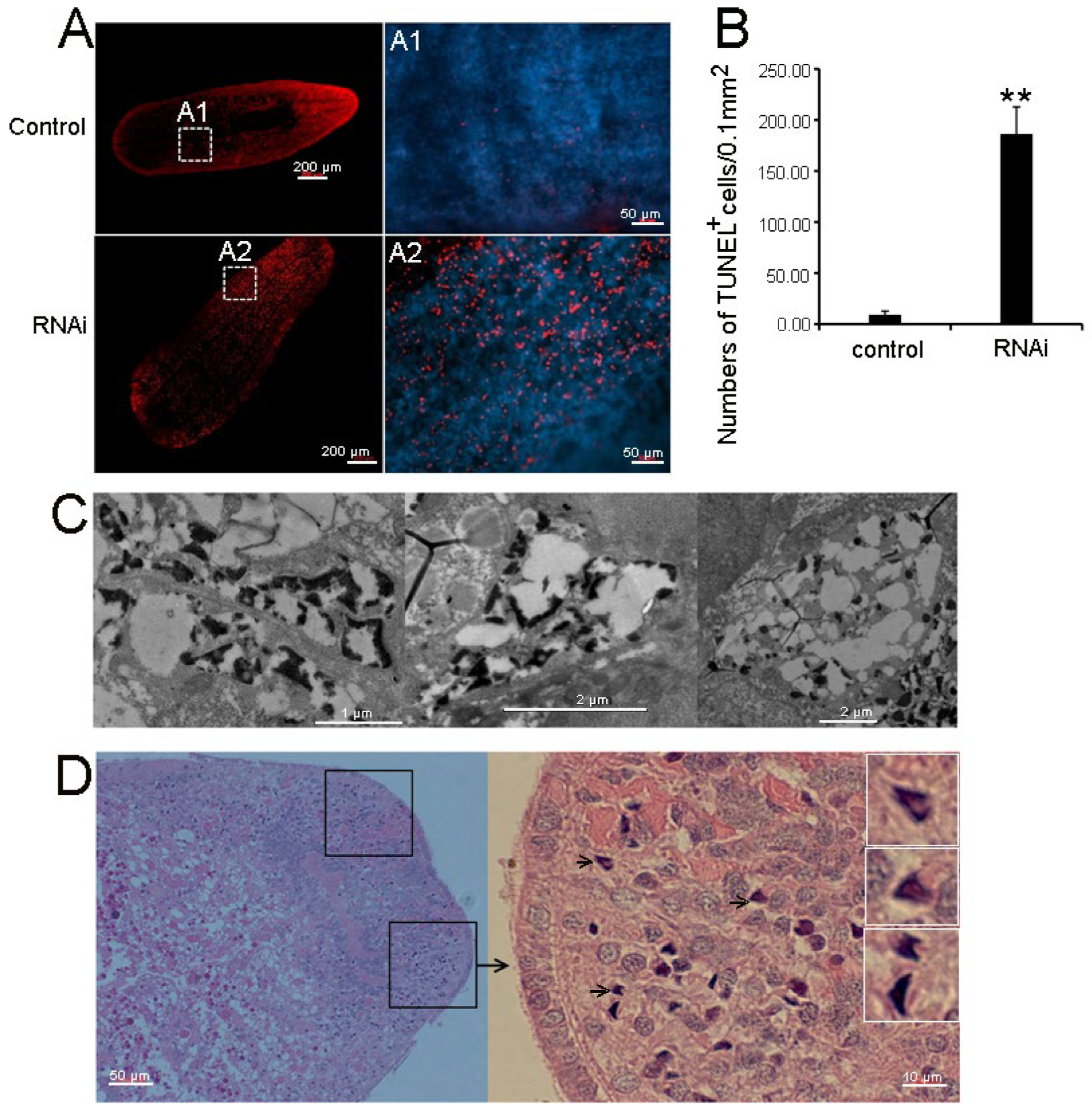
3.4. RNAi-Djhsp60 Caused Necrotic Cell Death
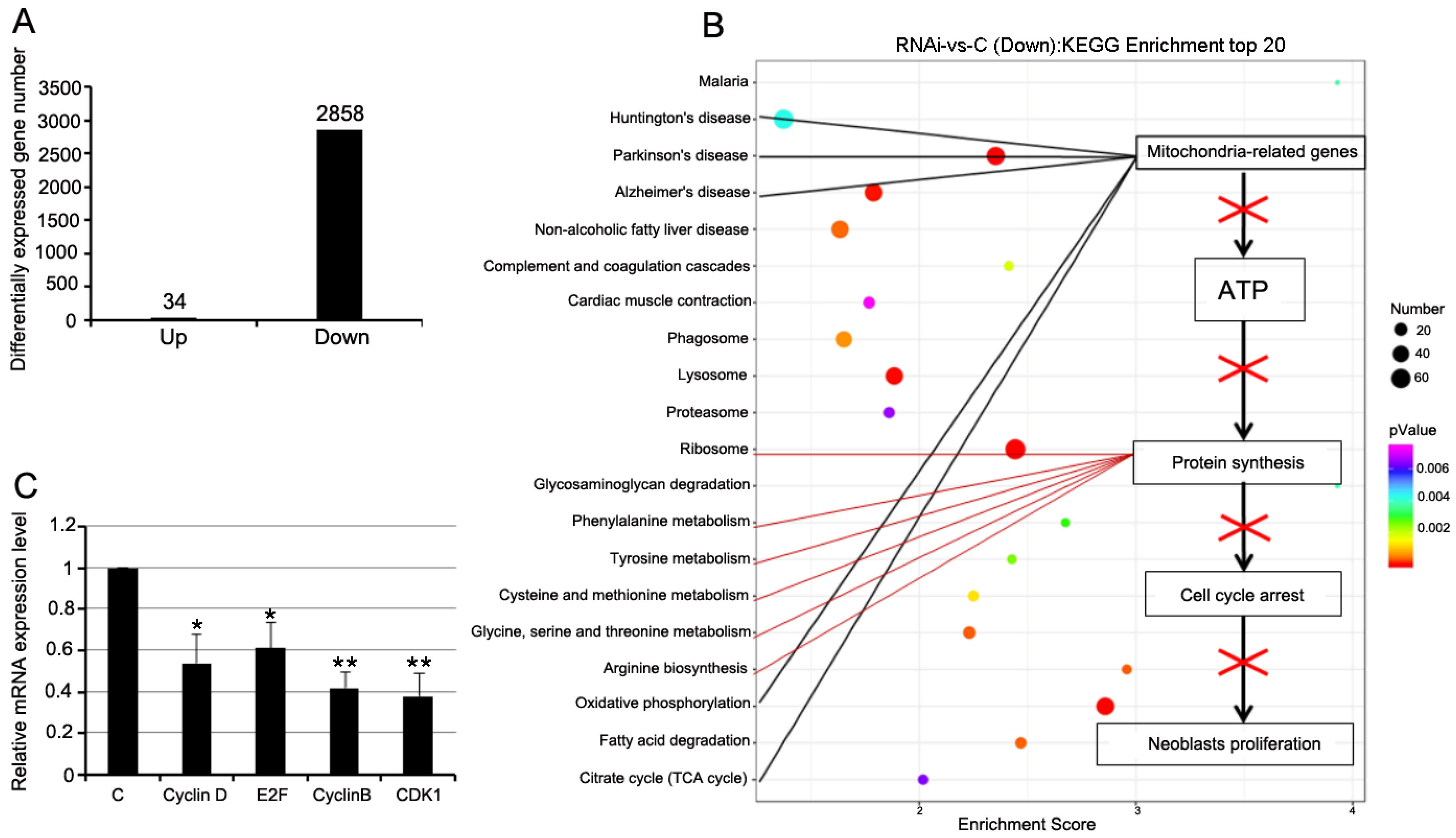
3.5. RNAi-Djhsp60 Globally Inhibits Gene Expression
3.6. RNAi-Djhsp60 Downregulates the Genes Related to the Glycolysis and Pentose Phosphate Pathway
3.7. RNAi-Djhsp60 Severely Affects Mitochondrial Function
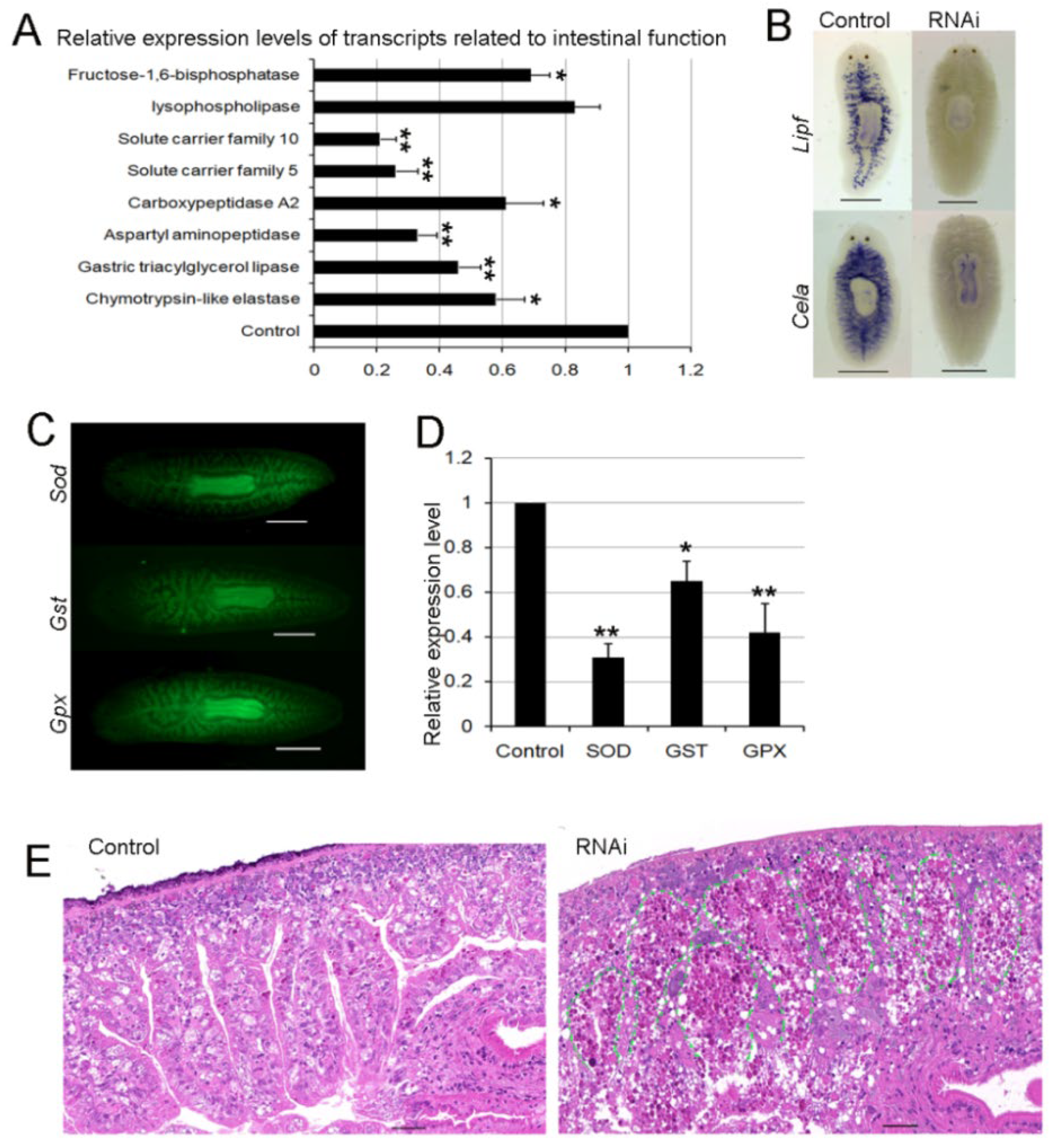
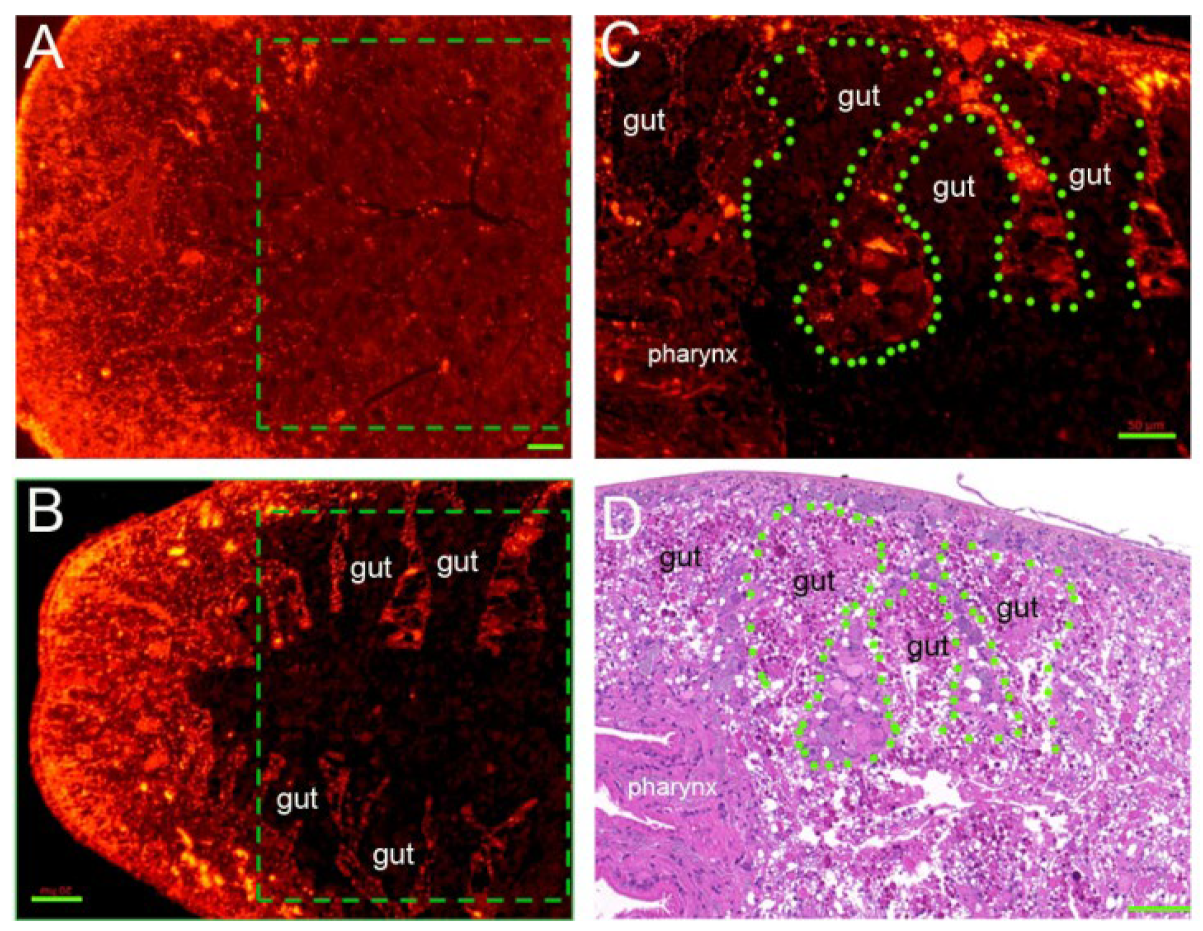
3.8. RNAi-Djhsp60 Severely Damages the Structure and Function of the Intestinal System
3.9. RNAi-Djhsp60 Activates Cathepsin-Mediated Cell Death
4. Discussion
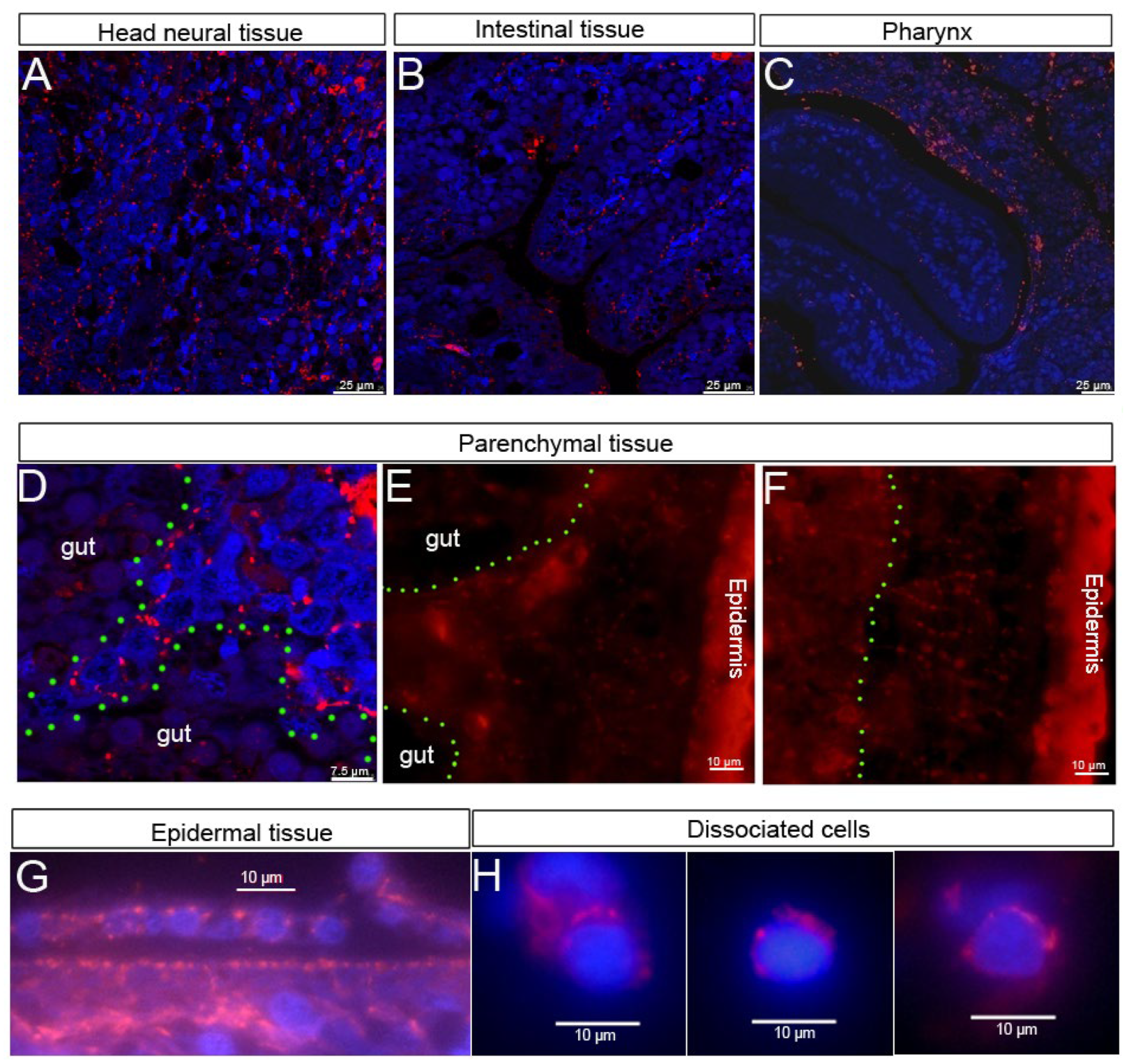
4.1. HSP60 Is Ubiquitously Expressed in All Types of Cells in Planarians
4.2. RNAi-Djhsp60 May Impair the Energy Metabolism of Planarian Stem Cells
4.3. Intestinal Cells Play an Essential Role in Planarian Regeneration and Tissue Homeostasis
4.4. Disruption of the Mitochondrial—Lysosome Axis and Activating Cathepsin L-Mediated Cell Death after RNAi May Be the Reason for Cell Death
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Reddien, P.W.; Oviedo, N.J.; Jennings, J.R.; Jenkin, J.C.; Sánchez Alvarado, A. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science 2005, 310, 1327–1330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molina, M.D.; Cebrià, F. Decoding stem cells: An overview on planarian stem cell heterogeneity and lineage progression. Biomolecules 2021, 11, 1532. [Google Scholar] [CrossRef] [PubMed]
- Orii, H.; Sakurai, T.; Watanabe, K. Distribution of the stem cells (neoblasts) in the planarian Dugesia japonica. Dev. Genes Evol. 2005, 215, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Forsthoefel, D.J.; Parka, A.E.; Newmark, P.A. Stem cell-based growth, regeneration, and remodeling of the planarian intestine. Dev. Biol. 2011, 356, 445–459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guedelhoefer, O.C., 4th; Sánchez Alvarado, A. Amputation induces stem cell mobilization to sites of injury during planarian regeneration. Development 2012, 139, 3510–3520. [Google Scholar] [CrossRef] [Green Version]
- González-Estévez, C.; Saló, E. Autophagy and apoptosis in planarians. Apoptosis 2010, 15, 279–292. [Google Scholar] [CrossRef]
- Sahu, S.; Sridhar, D.; Abnave, P.; Kosaka, N.; Thompson, J.M.; Hill, M.A.; Aboobaker, A. Ongoing repair of migration-coupled DNA damage allows planarian adult stem cells to reach wound sites. eLife 2021, 10, e63779. [Google Scholar] [CrossRef]
- Smith, G.M.; Gallo, G. The role of mitochondria in axon development and regeneration. Dev. Neurobiol. 2018, 78, 221–237. [Google Scholar] [CrossRef]
- Mercer, T.R.; Neph, S.; Dinger, M.E.; Crawford, J.; Smith, M.A.; Shearwood, A.M.; Haugen, E.; Bracken, C.P.; Rackham, O.; Stamatoyannopoulos, J.A.; et al. The human mitochondrial transcriptome. Cell 2011, 146, 645–658. [Google Scholar] [CrossRef] [Green Version]
- Chacinska, A.; Koehler, C.M.; Milenkovic, D.; Lithgow, T.; Pfanner, N. Importing mitochondrial proteins: Machineries and mechanisms. Cell 2009, 138, 628–644. [Google Scholar] [CrossRef] [Green Version]
- Llanos-González, E.; Henares-Chavarino, Á.A.; Pedrero-Prieto, C.M.; García-Carpintero, S.; Frontiñán-Rubio, J.; Sancho-Bielsa, F.J.; Alcain, F.J.; Peinado, J.R.; Rabanal-Ruíz, Y.; Durán-Prado, M. Interplay between mitochondrial oxidative disorders and proteostasis in Alzheimer’s disease. Front. Neurosci. 2020, 13, 1444. [Google Scholar] [CrossRef] [PubMed]
- De, I.; Dogra, N.; Singh, S. The mitochondrial unfoldedproteinresponse: Role in cellular homeostasis and disease. Curr. Mol. Med. 2017, 17, 587–597. [Google Scholar] [CrossRef] [PubMed]
- Böttinger, L.; Oeljeklaus, S.; Guiard, B.; Rospert, S.; Warscheid, B.; Becker, T. Mitochondrial heat shock protein (Hsp) 70 and Hsp10 cooperate in the formation of Hsp60 complexes. J. Biol. Chem. 2015, 290, 11611–11622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez, A.; Von Salzen, D.; Holguin, B.A.; Bernal, R.A. Complex destabilization in the mitochondrial chaperonin Hsp60 leads to disease. Front. Mol. Biosci. 2020, 7, 159. [Google Scholar] [CrossRef]
- Cheng, M.Y.; Hartl, F.U.; Martin, J.; Pollock, R.A.; Kalousek, F.; Neupert, W.; Hallberg, E.M.; Hallberg, R.L.; Horwich, A.L. Mitochondrial heat-shock protein hsp60 is essential for assembly of proteins imported into yeast mitochondria. Nature 1989, 337, 620–625. [Google Scholar] [CrossRef] [Green Version]
- Perezgasga, L.; Segovia, L.; Zurita, M. Molecular characterizationof the 5’ control region and of two lethal alleles affecting the hsp60 gene in Drosophila melanogaster. FEBS Lett. 1999, 456, 269–273. [Google Scholar] [CrossRef] [Green Version]
- Hansen, J.J.; Durr, A.; Cournu-Rebeix, I.; Georgopoulos, C.; Ang, D.; Nielsen, M.N.; Davoine, C.-S.; Brice, A.; Fontaine, B.; Gregersen, N.; et al. Hereditary spastic paraplegia SPG13 is associated with a mutation in the gene encoding the mitochondrial chaperonin Hsp60. Am. J. Hum. Genet. 2002, 70, 1328–1332. [Google Scholar] [CrossRef] [Green Version]
- Magen, D.; Georgopoulos, C.; Bross, P.; Ang, D.; Segev, Y.; Goldsher, D.; Nemirovski, A.; Shahar, E.; Ravid, S.; Luder, A.; et al. Mitochondrial hsp60 chaperonopathy causes an autosomal-recessive neurodegenerative disorder linked to brain hypomyelination and leukodystrophy. Am. J. Hum. Genet. 2008, 83, 30–42. [Google Scholar] [CrossRef] [Green Version]
- Bross, P.; Magnoni, R.; Bie, A.S. Molecular chaperone disorders: Defective Hsp60 in neurodegeneration. Curr. Top. Med. Chem. 2012, 12, 2491–2503. [Google Scholar] [CrossRef]
- Seo, N.-H.; Lee, E.-H.; Seo, J.-H.; Song, H.-R.; Han, M.-K. HSP60 is required for stemness and properdifferentiation of mouse embryonic stem cells. Exp. Mol. Med. 2018, 50, e459. [Google Scholar] [CrossRef] [Green Version]
- Berger, E.; Rath, E.; Yuan, D.; Waldschmitt, N.; Khaloian, S.; Allgäuer, M.; Staszewski, O.; Lobner, E.M.; Schöttl, T.; Giesbertz, P.; et al. Mitochondrial function controls intestinal epithelial stemness and proliferation. Nat. Commu. 2016, 7, 13171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, W.; Cheon, M.G.; Kim, J.E. Mitochondrial CCAR2/DBC1 is required for cell survival against rotenone-induced mitochondrial stress. Biochem. Biophys. Res. Commun. 2017, 485, 782–789. [Google Scholar] [CrossRef] [PubMed]
- Fan, F.; Duan, Y.; Yang, F.; Trexler, C.; Wang, H.; Huang, L.; Li, Y.; Tang, H.; Wang, G.; Fang, X. Deletion of heat shock protein 60 in adult mouse cardiomyocytes perturbs mitochondrial protein homeostasis and causes heart failure. Cell Death. Differ. 2020, 27, 587–600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makino, S.; Whitehead, G.G.; Lien, C.-L.; Kim, S.; Jhawar, P.; Kono, A.; Kawata, Y.; Keating, M.T. Heat-shock protein 60 is required for blastema formation and maintenance during regeneration. Proc. Natl. Acad. Sci. USA 2005, 102, 14599–14604. [Google Scholar] [CrossRef] [Green Version]
- Pei, W.; Tanaka, K.; Huang, S.C.; Xu, L.; Liu, B.; Sinclair, J.; Idol, J.; Varshney, G.K.; Huang, H.; Lin, S.; et al. Extracellular HSP60 triggers tissue regeneration and wound healing by regulating inflammation and cell proliferation. NPJ Regen. Med. 2016, 1, 16013. [Google Scholar] [CrossRef]
- Pirotte, N.; Stevens, A.-S.; Fraguas, S.; Plusquin, M.; Roten, A.V.; Belleghem, F.V.; Paesen, R.; Ameloot, M.; Cebria, F.; Artois, T.; et al. Reactive oxygen species in planarian regeneration: An upstream necessity for correct patterning and brain formation. Oxid. Med. Cell Longev. 2015, 2015, 392476. [Google Scholar] [CrossRef]
- Lévesque, M.; Guimond, J.C.; Pilote, M.; Leclerc, S.; Moldovan, F.; Roy, S. Expression of heat-shock protein 70 during limb development and regeneration in the axolotl. Dev. Dyn. 2005, 233, 1525–1534. [Google Scholar] [CrossRef]
- Wolf, J.H.; Bhatti, T.R.; Fouraschen, S.; Chakravorty, S.; Wang, L.; Kurian, S.; Salomon, D.; Olthoff, K.M.; Hancock, W.W.; Levine, M.H. Heatshockprotein 70 is required for optimal liver regeneration after partial hepatectomy in mice. Liver Transpl. 2014, 20, 376–385. [Google Scholar] [CrossRef] [Green Version]
- Conte, M.; Deri, P.; Isolani, M.E.; Mannini, L.; Batistoni, R. Characterization of hsp genes in planarian stem cells. Belg. J. Zool. 2010, 140, 137–143. [Google Scholar]
- Conte, M.; Deri, P.; Isolani, M.E.; Mannini, L.; Batistoni, R. A mortalin-like gene is crucial for planarian stem cell viability. Dev. Biol. 2009, 334, 109–118. [Google Scholar] [CrossRef] [Green Version]
- Rouhana, L.; Weiss, J.A.; Forsthoefel, D.J.; Lee, H.; King, R.S.; Inoue, T.; Shibata, N.; Agata, K.; Newmark, P.A. RNA interference by feeding in vitro-synthesized double-stranded RNA to planarians: Methodology and dynamics. Dev. Dyn. 2013, 242, 718–730. [Google Scholar] [CrossRef] [Green Version]
- Ma, K.; Zhang, Y.; Song, G.; Wu, M.; Chen, G. Identification of autophagy-related gene 7 and autophagic cell death in the planarian Dugesia japonica. Front. Physiol. 2018, 9, 1223. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Asami, M.; Higuchi, S.; Shibata, N.; Agata, K. Isolation of planarian X-ray-sensitive stem cells by fluorescence-activated cell sorting. Dev. Growth. Differ. 2006, 48, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Cebria, F.; Newmark, P.A. Planarian homologs of netrin and netrin receptor are required for proper regeneration of the central nervous system and the maintenance of nervous system architecture. Development 2005, 132, 3691–3703. [Google Scholar] [CrossRef] [Green Version]
- Stubenhaus, B.; Pellettieri, J. Detection of apoptoticcells in planarians by whole-mountTUNEL. Methods Mol. Biol. 2018, 1774, 435–444. [Google Scholar]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Roberts, A.; Pachter, L. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat. Methods 2013, 10, 71–73. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [Green Version]
- Fincher, C.T.; Wurtzel, O.; de Hoog, T.; Kravarik, K.M.; Reddien, P.W. Celltypetranscriptome atlas for the planarian Schmidtea mediterranea. Science 2018, 360, eaaq1736. [Google Scholar]
- Salvetti, A.; Rossi, L.; Deri, P.; Batistoni, R. An MCM2-related gene is expressed in proliferating cells of intact and regenerating planarians. Dev. Dyn. 2000, 218, 603–614. [Google Scholar] [CrossRef]
- Solana, J.; Kao, D.; Mihaylova, Y.; Jaber-Hijazi, F.; Malla, S.; Wilson, R.; Aboobaker, A. Defining the molecular profile of planarian pluripotent stem cells using a combinatorial RNA-seq, RNA interference and irradiation approach. Genome Biol. 2012, 13, R19. [Google Scholar] [CrossRef] [Green Version]
- Nishizaki, K.; Yoshino, T.; Orita, Y.; Nomiya, S.; Masuda, Y. TUNEL staining of inner ear structures may reflect autolysis, not apoptosis. Hear Res. 1999, 130, 131–136. [Google Scholar] [CrossRef]
- Tu, H.-C.; Ren, D.; Wang, G.X.; Chen, D.Y.; Westergard, T.D.; Kim, H.; Sasagawa, S.; Hsieh, J.J.-D.; Cheng, E.H.-Y. The p53-cathepsin axis cooperates with ROS to activate programmed necrotic death upon DNA damage. Proc. Natl. Acad. Sci. USA 2009, 106, 1093–1098. [Google Scholar] [CrossRef] [Green Version]
- Rossi, L.; Salvetti, A.; Lena, A.; Batistoni, R.; Pugliesi, C.; Loreti, E.; Gremigni, V. DjPiwi-1, a member of the PAZ-Piwi gene family, defines a subpopulation of planarian stem cells. Dev. Genes Evol. 2006, 216, 335–346. [Google Scholar] [CrossRef]
- Moreno-Ulloa, A.; Cid, A.; Rubio-Gayosso, I.; Ceballos, G.; Villarreal, F.; Ramirez-Sanchez, I. Effects of(-)-epicatechin and derivatives on nitric oxide mediated induction of mitochondrial proteins. Bioorg. Med. Chem. Lett. 2013, 23, 4441–4446. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.H.; Yoo, Y.H.; Lee, J.H.; Park, J.W. Mitochondrial NADP(+)-dependent isocitrate dehydrogenase knockdown inhitits tumorigenicity of melanoma cells. Biochem. Biophys. Res. Commun. 2014, 451, 246–251. [Google Scholar] [CrossRef]
- Ensenauer, R.; He, M.; Willard, J.M.; Goetzman, E.S.; Corydon, T.J.; Vandahl, B.B.; Mohsen, A.W.; Isaya, G.; Vockley, J. Human acyl-CoAdehydrogenase-9 plays a novel role in the mitochondrial beta-oxidation of unsaturated fatty acids. J. Biol. Chem. 2005, 280, 32309–32316. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Zhou, X.; Liu, X.; Yang, L.; Chen, Q.; Zhao, D.; Zuo, J.; Liu, W. Mitochondrial dysfunction induced by knockdown of mortalin is rescued by Parkin. Biochem. Biophys. Res. Commun. 2011, 410, 114–120. [Google Scholar] [CrossRef]
- Tsujimoto, Y. Role of Bcl-2 family proteins in apoptosis: Apoptosomes or mitochondria? Genes Cells 1998, 3, 697–707. [Google Scholar] [CrossRef]
- Kale, J.; Osterlund, E.J.; Andrews, D.W. BCL-2 family proteins: Changing partners in the dance towards death. Cell Death Differ. 2018, 25, 65–80. [Google Scholar] [CrossRef] [Green Version]
- Thommen, A.; Werner, S.; Frank, O.; Philipp, J.; Knittelfelder, O.; Quek, Y.; Fahmy, K.; Shevchenko, A.; Friedrich, B.M.; Julicher, F.; et al. Body size-dependent energy storage causes Kleiber’s law scaling of the metabolic rate in planarians. eLife 2019, 8, e38187. [Google Scholar] [CrossRef]
- Forsthoefel, D.J.; Cejda, N.I.; Khan, U.W.; Newmark, P.A. Cell-type diversity and regionalized gene expression in the planarian intestine. Elife 2020, 9, e52613. [Google Scholar] [CrossRef]
- Bijnens, K.; Jaenen, V.; Wouters, A.; Leynen, N.; Pirotte, N.; Artois, T.; Smeets, K. A spatiotemporal characterisation of redox molecules in planarians, with a focus on the role of glutathione during regeneration. Biomolecules 2021, 11, 714. [Google Scholar] [CrossRef]
- Yang, H.; Zhang, R.; Zhang, Y.; Liu, Q.; Li, Y.; Gong, J.; Hou, Y. Cathepsin-L is involved in degradation of fat body and programmed cell death in Bombyx mori. Gene 2020, 760, 144998. [Google Scholar] [CrossRef]
- Peyer, S.M.; Kremer, N.; McFall-Ngai, M.J. Involvement of a host cathepsin L in symbiont-induced cell death. Microbiologyopen 2018, 7, e00632. [Google Scholar] [CrossRef]
- Cappello, F.; Conway, de.; Macario, E.; Marasà, L.; Zummo, G.; Macario, A.J. Hsp60 expression, new locations, functions and perspectives for cancer diagnosis and therapy. Cancer Biol. Ther. 2008, 7, 801–809. [Google Scholar] [CrossRef]
- Tong, W.W.; Tong, G.H.; Kong, H.; Liu, Y. The tumor promoting roles of HSP60 and HIF2 alpha in gastric cancer cells. Tumour Biol. 2016, 37, 9849–9854. [Google Scholar] [CrossRef]
- Yun, C.W.; Kim, H.J.; Lim, J.H.; Lee, S.H. Heat shock proteins: Agents of cancer development and therapeutic targets in anti-cancer therapy. Cells 2019, 9, 60. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Taboada, E.; Rodríguez-Esteban, G.; Saló, E.; Abril, J.F. A proteomics approach to decipher the molecular nature of planarian stem cells. BMC Genom. 2011, 12, 133. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.; Li, X.; Zhang, W.; Chen, Y.; Zhu, S.; Chen, L.; Xu, R.; Lv, Y.; Wu, D.; Guo, M.; et al. HSP60-regulated mitochondrial proteostasis and protein translation promote tumor growth of ovarian cancer. Sci. Rep. 2019, 9, 12628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, X.; Guo, J.; Chen, Y.; Liu, X.; Yang, G.; Wu, Y.; Tian, Y.; Liu, N.; Yang, L.; Wei, S.; et al. The 60-kDa heat shock protein regulates energy rearrangement and protein synthesis to promote proliferation of multiple myeloma cells. Br. J. Haematol. 2020, 190, 741–752. [Google Scholar] [CrossRef] [PubMed]
- Haroon, M.M.; Lakshmanan, V.; Sarkar, S.R.; Lei, K.; Vemula, P.K.; Palakodeti, D. Mitochondrial state determines functionally divergent stem cell population in planaria. Stem Cell Rep. 2021, 16, 1302–1316. [Google Scholar] [CrossRef] [PubMed]
- Flores, N.M.; Oviedo, N.J.; Sage, J. Essential role for the planarian intestinal GATA transcription factor in stem cells and regeneration. Dev. Biol. 2016, 418, 179–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forsthoefel, D.J.; James, N.P.; Escobar, D.J.; Stary, J.M.; Vieira, A.P.; Waters, F.A.; Newmark, P.A. An RNAi screen reveals intestinal regulators of branching morphogenesis, differentiation, and stem cell proliferation in planarians. Dev. Cell. 2012, 23, 691–704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowen, I.D.; Ryder, T.A.; Thompson, J.A. The fine structure of the planarian Polycelis tenuis Iijima. II. The intestine and gastrodermal phagocytosis. Protoplasma 1974, 79, 1–17. [Google Scholar] [CrossRef]
- Shibata, N.; Umesono, Y.; Orii, H.; Sakurai, T.; Watanabe, K.; Agata, K. Expression of vasa(vas)-related genes in germline cells and totipotent somatic stem cells of planarians. Dev. Biol. 1999, 206, 73–87. [Google Scholar] [CrossRef] [Green Version]
- Zeng, A.; Li, H.; Guo, L.; Gao, X.; McKinney, S.; Wang, Y.; Yu, Z.; Park, J.; Semerad, C.; Ross, E.; et al. Prospectively isolated tetraspanin(+) neoblasts are adult pluripotent stem cells underlying planaria regeneration. Cell 2018, 173, 1593–1608. [Google Scholar] [CrossRef] [Green Version]
- Kim, R.; Emi, M.; Tanabe, K. Role of mitochondria as the gardens of cell death. Cancer Chemother. Pharmacol. 2006, 57, 545–553. [Google Scholar] [CrossRef]
- Terman, A.; Gustafsson, B.; Brunk, U.T. The lysosomal-mitochondrial axis theory of postmitotic aging and cell death. Chem. Biol. Interact. 2006, 163, 29–37. [Google Scholar] [CrossRef]
- Lin, Y.C.; Horng, L.Y.; Sung, H.C.; Wu, R.T. Sodium iodate disrupted the mitochondrial-lysosomal axis in cultured retinal pigment epithelial cells. J. Ocul. Pharmacol. Ther. 2018, 34, 500–511. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, F.J.; Servin, J.; Dang, D.; Kim, J.; Molinaro, C.; Daniels, T.; Brown-Bryan, T.A.; Imoto-Egami, M.; Casiano, C.A. Involvement of lysosomal cathepsins in the cleavage of DNA topoisomerase I during necrotic cell death. Arthritis Rheum. 2005, 52, 2133–2145. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, K.; Li, R.; Song, G.; Guo, F.; Wu, M.; Lu, Q.; Li, X.; Chen, G. Djhsp60 Is Required for Planarian Regeneration and Homeostasis. Biomolecules 2022, 12, 808. https://doi.org/10.3390/biom12060808
Ma K, Li R, Song G, Guo F, Wu M, Lu Q, Li X, Chen G. Djhsp60 Is Required for Planarian Regeneration and Homeostasis. Biomolecules. 2022; 12(6):808. https://doi.org/10.3390/biom12060808
Chicago/Turabian StyleMa, Kexue, Rui Li, Gege Song, Fangying Guo, Meng Wu, Qiong Lu, Xinwei Li, and Guangwen Chen. 2022. "Djhsp60 Is Required for Planarian Regeneration and Homeostasis" Biomolecules 12, no. 6: 808. https://doi.org/10.3390/biom12060808
APA StyleMa, K., Li, R., Song, G., Guo, F., Wu, M., Lu, Q., Li, X., & Chen, G. (2022). Djhsp60 Is Required for Planarian Regeneration and Homeostasis. Biomolecules, 12(6), 808. https://doi.org/10.3390/biom12060808




