Allostery Modulates Interactions between Proteasome Core Particles and Regulatory Particles
Abstract
1. Introduction
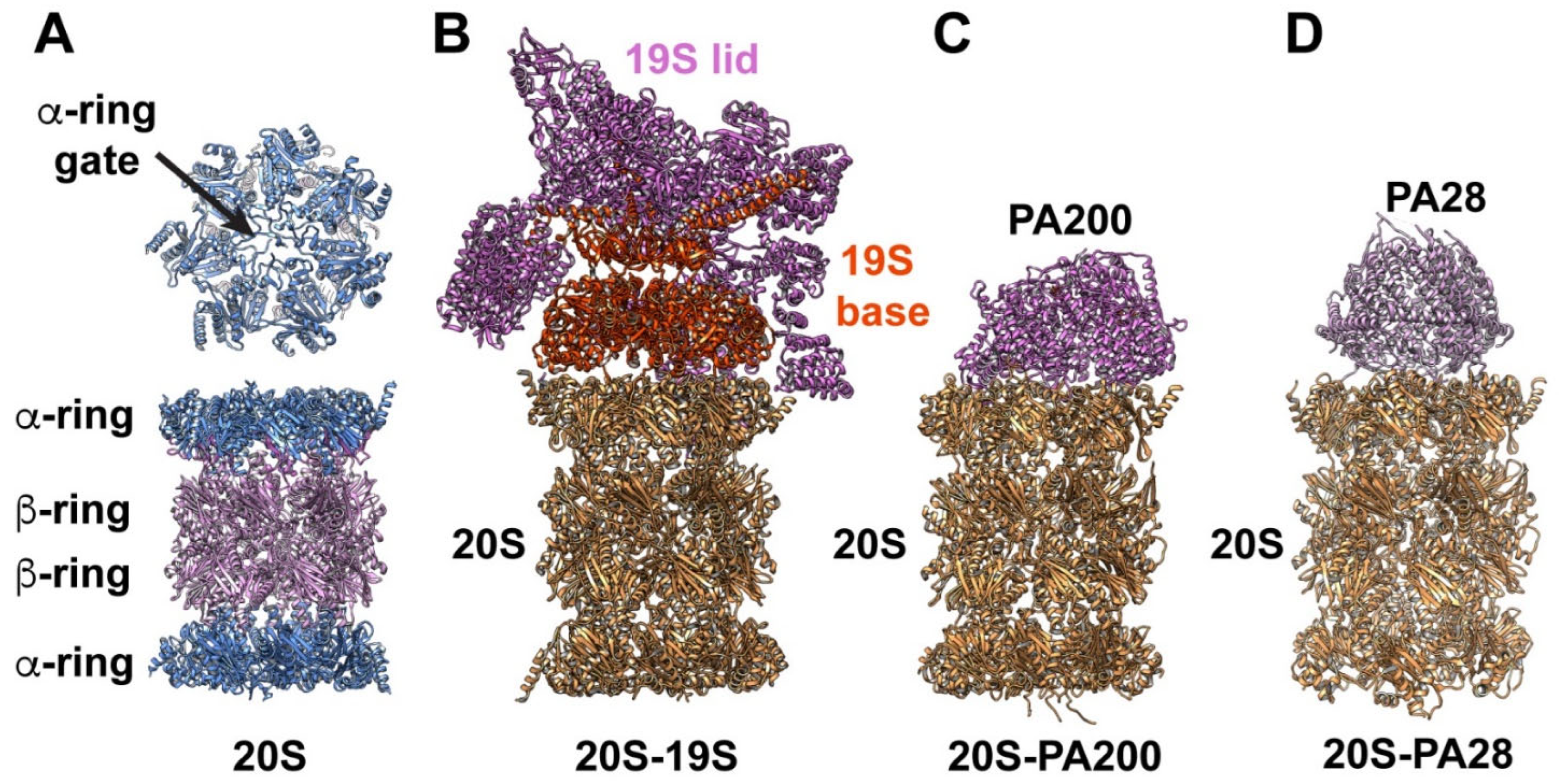
2. Proteasome Structure
3. Beyond the Classic Proteasome
4. Proteasome Allostery Supports Functional Optimization
5. Allostery by Substrates and Their Components
5.1. Protein Substrates
5.2. Proteolysis Active Site Inhibitors
5.3. Control by Ubiquitin Chains and Ubiquitin Binding Proteins-USP14/Ubp6
5.4. Processivity of Degradation
5.5. Activation by Phosphorylation
6. Allostery in RP/CP Assembly
7. Plausible Inferences and Further Speculations
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, J.; Nussinov, R. Allostery: An Overview of Its History, Concepts, Methods, and Applications. PLoS Comput. Biol. 2016, 12, e1004966. [Google Scholar] [CrossRef] [PubMed]
- Voet, D.; Voet, J.G. Biochemistry; Wiley: Hoboken, NJ, USA, 2004. [Google Scholar]
- Gerhart, J. From feedback inhibition to allostery: The enduring example of aspartate transcarbamoylase. FEBS J. 2014, 281, 612–620. [Google Scholar] [CrossRef] [PubMed]
- Gerhart, J.C.; Schachman, H.K. Distinct subunits for the regulation and catalytic activity of aspartate transcarbamylase. Biochemistry 1965, 4, 1054–1062. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Jain, A.; McDonald, L.R.; Gambogi, C.; Lee, A.L.; Dokholyan, N.V. Mapping allosteric communications within individual proteins. Nat. Commun. 2020, 11, 3862. [Google Scholar] [CrossRef] [PubMed]
- Faure, A.J.; Domingo, J.; Schmiedel, J.M.; Hidalgo-Carcedo, C.; Diss, G.; Lehner, B. Mapping the energetic and allosteric landscapes of protein binding domains. Nature 2022, 604, 175–183. [Google Scholar] [CrossRef]
- Greene, E.R.; Dong, K.C.; Martin, A. Understanding the 26S proteasome molecular machine from a structural and conformational dynamics perspective. Curr. Opin. Struct. Biol. 2020, 61, 33–41. [Google Scholar] [CrossRef]
- Dong, Y.; Zhang, S.; Wu, Z.; Li, X.; Wang, W.L.; Zhu, Y.; Stoilova-McPhie, S.; Lu, Y.; Finley, D.; Mao, Y. Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome. Nature 2019, 565, 49–55. [Google Scholar] [CrossRef]
- Groll, M.; Ditzel, L.; Lowe, J.; Stock, D.; Bochtler, M.; Bartunik, H.D.; Huber, R. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature 1997, 386, 463–471. [Google Scholar] [CrossRef]
- Groll, M.; Bajorek, M.; Kohler, A.; Moroder, L.; Rubin, D.M.; Huber, R.; Glickman, M.H.; Finley, D. A gated channel into the proteasome core particle. Nat. Struct. Biol. 2000, 7, 1062–1067. [Google Scholar] [CrossRef]
- Stadtmueller, B.M.; Hill, C.P. Proteasome activators. Mol. Cell. 2011, 41, 8–19. [Google Scholar] [CrossRef]
- Eisele, M.R.; Reed, R.G.; Rudack, T.; Schweitzer, A.; Beck, F.; Nagy, I.; Pfeifer, G.; Plitzko, J.M.; Baumeister, W.; Tomko, R.J., Jr.; et al. Expanded Coverage of the 26S Proteasome Conformational Landscape Reveals Mechanisms of Peptidase Gating. Cell. Rep. 2018, 24, 1301–1315.e5. [Google Scholar] [CrossRef] [PubMed]
- Morozov, A.V.; Karpov, V.L. Proteasomes and Several Aspects of Their Heterogeneity Relevant to Cancer. Front. Oncol. 2019, 9, 761. [Google Scholar] [CrossRef] [PubMed]
- Cascio, P.; Call, M.; Petre, B.M.; Walz, T.; Goldberg, A.L. Properties of the hybrid form of the 26S proteasome containing both 19S and PA28 complexes. EMBO J. 2002, 21, 2636–2645. [Google Scholar] [CrossRef] [PubMed]
- Kopp, F.; Dahlmann, B.; Kuehn, L. Reconstitution of hybrid proteasomes from purified PA700-20 S complexes and PA28alphabeta activator: Ultrastructure and peptidase activities. J. Mol. Biol. 2001, 313, 465–471. [Google Scholar] [CrossRef]
- Tanahashi, N.; Murakami, Y.; Minami, Y.; Shimbara, N.; Hendil, K.B.; Tanaka, K. Hybrid proteasomes. Induction by interferon-gamma and contribution to ATP-dependent proteolysis. J. Biol. Chem. 2000, 275, 14336–14345. [Google Scholar] [CrossRef] [PubMed]
- Cascio, P. PA28alphabeta: The enigmatic magic ring of the proteasome? Biomolecules 2014, 4, 566–584. [Google Scholar] [CrossRef] [PubMed]
- Makarova, T.M.; Bogdanov, A.A. The Ribosome as an Allosterically Regulated Molecular Machine. Biochemistry 2017, 82, 1557–1571. [Google Scholar] [CrossRef]
- Guzel, P.; Kurkcuoglu, O. Identification of potential allosteric communication pathways between functional sites of the bacterial ribosome by graph and elastic network models. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 3131–3141. [Google Scholar] [CrossRef]
- Wang, L.; Pulk, A.; Wasserman, M.R.; Feldman, M.B.; Altman, R.B.; Cate, J.H.; Blanchard, S.C. Allosteric control of the ribosome by small-molecule antibiotics. Nat. Struct. Mol. Biol. 2012, 19, 957–963. [Google Scholar] [CrossRef]
- Kleijnen, M.F.; Roelofs, J.; Park, S.; Hathaway, N.A.; Glickman, M.; King, R.W.; Finley, D. Stability of the proteasome can be regulated allosterically through engagement of its proteolytic active sites. Nat. Struct. Mol. Biol. 2007, 14, 1180–1188. [Google Scholar] [CrossRef]
- Osmulski, P.A.; Hochstrasser, M.; Gaczynska, M. A tetrahedral transition state at the active sites of the 20S proteasome is coupled to opening of the alpha-ring channel. Structure 2009, 17, 1137–1147. [Google Scholar] [CrossRef] [PubMed]
- Haselbach, D.; Schrader, J.; Lambrecht, F.; Henneberg, F.; Chari, A.; Stark, H. Long-range allosteric regulation of the human 26S proteasome by 20S proteasome-targeting cancer drugs. Nat. Commun. 2017, 8, 15578. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Demartino, G.N. Variably modulated gating of the 26S proteasome by ATP and polyubiquitin. Biochem. J. 2009, 421, 397–404. [Google Scholar] [CrossRef] [PubMed]
- Bech-Otschir, D.; Helfrich, A.; Enenkel, C.; Consiglieri, G.; Seeger, M.; Holzhutter, H.G.; Dahlmann, B.; Kloetzel, P.M. Polyubiquitin substrates allosterically activate their own degradation by the 26S proteasome. Nat. Struct. Mol. Biol. 2009, 16, 219–225. [Google Scholar] [CrossRef]
- Peth, A.; Besche, H.C.; Goldberg, A.L. Ubiquitinated proteins activate the proteasome by binding to Usp14/Ubp6, which causes 20S gate opening. Mol. Cell 2009, 36, 794–804. [Google Scholar] [CrossRef] [PubMed]
- Hanna, J.; Hathaway, N.A.; Tone, Y.; Crosas, B.; Elsasser, S.; Kirkpatrick, D.S.; Leggett, D.S.; Gygi, S.P.; King, R.W.; Finley, D. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell 2006, 127, 99–111. [Google Scholar] [CrossRef] [PubMed]
- Hung, K.Y.S.; Klumpe, S.; Eisele, M.R.; Elsasser, S.; Tian, G.; Sun, S.; Moroco, J.A.; Cheng, T.C.; Joshi, T.; Seibel, T.; et al. Allosteric control of Ubp6 and the proteasome via a bidirectional switch. Nat. Commun. 2022, 13, 838. [Google Scholar] [CrossRef]
- Zhang, S.; Zou, S.; Yin, D.; Zhao, L.; Finley, D.; Wu, Z.; Mao, Y. USP14-regulated allostery of the human proteasome by time-resolved cryo-EM. Nature 2022, 605, 567–574. [Google Scholar] [CrossRef]
- Sears, C.; Olesen, J.; Rubin, D.; Finley, D.; Maniatis, T. NF-kappa B p105 processing via the ubiquitin-proteasome pathway. J Biol. Chem. 1998, 273, 1409–1419. [Google Scholar] [CrossRef]
- Zhang, M.; Coffino, P. Repeat sequence of Epstein-Barr virus-encoded nuclear antigen 1 protein interrupts proteasome substrate processing. J. Biol. Chem. 2004, 279, 8635–8641. [Google Scholar] [CrossRef]
- Hoyt, M.A.; Zich, J.; Takeuchi, J.; Zhang, M.; Govaerts, C.; Coffino, P. Glycine-alanine repeats impair proper substrate unfolding by the proteasome. EMBO J. 2006, 25, 1720–1729. [Google Scholar] [CrossRef] [PubMed]
- Reichard, E.L.; Chirico, G.G.; Dewey, W.J.; Nassif, N.D.; Bard, K.E.; Millas, N.E.; Kraut, D.A. Substrate Ubiquitination Controls the Unfolding Ability of the Proteasome. J. Biol. Chem. 2016, 291, 18547–18561. [Google Scholar] [CrossRef] [PubMed]
- Cundiff, M.D.; Hurley, C.M.; Wong, J.D.; Boscia, J.A.; Bashyal, A.; Rosenberg, J.; Reichard, E.L.; Nassif, N.D.; Brodbelt, J.S.; Kraut, D.A. Ubiquitin receptors are required for substrate-mediated activation of the proteasome’s unfolding ability. Sci. Rep. 2019, 9, 14506. [Google Scholar] [CrossRef] [PubMed]
- VerPlank, J.J.S.; Goldberg, A.L. Regulating protein breakdown through proteasome phosphorylation. Biochem. J. 2017, 474, 3355–3371. [Google Scholar] [CrossRef]
- Bellelli, A.; Brunori, M. Hemoglobin allostery: Variations on the theme. Biochim. Biophys. Acta 2011, 1807, 1262–1272. [Google Scholar] [CrossRef][Green Version]
- Monod, J.; Wyman, J.; Changeux, J.P. On the Nature of Allosteric Transitions: A Plausible Model. J. Mol. Biol. 1965, 12, 88–118. [Google Scholar] [CrossRef]
- Maupin-Furlow, J.A.; Humbard, M.A.; Kirkland, P.A.; Li, W.; Reuter, C.J.; Wright, A.J.; Zhou, G. Proteasomes from structure to function: Perspectives from Archaea. Curr. Top. Dev. Biol. 2006, 75, 125–169. [Google Scholar] [CrossRef]
- Benaroudj, N.; Goldberg, A.L. PAN, the proteasome-activating nucleotidase from archaebacteria, is a protein-unfolding molecular chaperone. Nat. Cell Biol. 2000, 2, 833–839. [Google Scholar] [CrossRef]
- Yu, Z.; Yu, Y.; Wang, F.; Myasnikov, A.G.; Coffino, P.; Cheng, Y. Allosteric coupling between alpha-rings of the 20S proteasome. Nat. Commun. 2020, 11, 4580. [Google Scholar] [CrossRef]
- Smith, D.M.; Chang, S.C.; Park, S.; Finley, D.; Cheng, Y.; Goldberg, A.L. Docking of the proteasomal ATPases’ carboxyl termini in the 20S proteasome’s alpha ring opens the gate for substrate entry. Mol. Cell 2007, 27, 731–744. [Google Scholar] [CrossRef]
- Smith, D.M.; Kafri, G.; Cheng, Y.; Ng, D.; Walz, T.; Goldberg, A.L. ATP binding to PAN or the 26S ATPases causes association with the 20S proteasome, gate opening, and translocation of unfolded proteins. Mol. Cell 2005, 20, 687–698. [Google Scholar] [CrossRef] [PubMed]
- Rabl, J.; Smith, D.M.; Yu, Y.; Chang, S.C.; Goldberg, A.L.; Cheng, Y. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol. Cell 2008, 30, 360–368. [Google Scholar] [CrossRef] [PubMed]
- Rennella, E.; Huang, R.; Yu, Z.; Kay, L.E. Exploring long-range cooperativity in the 20S proteasome core particle from Thermoplasma acidophilum using methyl-TROSY-based NMR. Proc. Natl. Acad. Sci. USA 2020, 117, 5298–5309. [Google Scholar] [CrossRef] [PubMed]
- Adams, G.M.; Falke, S.; Goldberg, A.L.; Slaughter, C.A.; DeMartino, G.N.; Gogol, E.P. Structural and functional effects of PA700 and modulator protein on proteasomes. J. Mol. Biol. 1997, 273, 646–657. [Google Scholar] [CrossRef] [PubMed]
- Ruschak, A.M.; Kay, L.E. Proteasome allostery as a population shift between interchanging conformers. Proc. Natl. Acad. Sci. USA 2012, 109, E3454–E3462. [Google Scholar] [CrossRef]
- Asano, S.; Fukuda, Y.; Beck, F.; Aufderheide, A.; Forster, F.; Danev, R.; Baumeister, W. Proteasomes. A molecular census of 26S proteasomes in intact neurons. Science 2015, 347, 439–442. [Google Scholar] [CrossRef]
- Guo, X. Localized Proteasomal Degradation: From the Nucleus to Cell Periphery. Biomolecules 2022, 12, 229. [Google Scholar] [CrossRef]
- Schmidt, M.; Finley, D. Regulation of proteasome activity in health and disease. Biochim. Biophys. Acta 2014, 1843, 13–25. [Google Scholar] [CrossRef]
- Gaczynska, M.; Osmulski, P.A. Harnessing proteasome dynamics and allostery in drug design. Antioxid. Redox Signal. 2014, 21, 2286–2301. [Google Scholar] [CrossRef]
- Cromm, P.M.; Crews, C.M. The Proteasome in Modern Drug Discovery: Second Life of a Highly Valuable Drug Target. ACS Cent. Sci. 2017, 3, 830–838. [Google Scholar] [CrossRef]
- Abdel-Magid, A.F. Allosteric modulators: An emerging concept in drug discovery. ACS Med. Chem. Lett. 2015, 6, 104–107. [Google Scholar] [CrossRef] [PubMed]
- Guarnera, E.; Berezovsky, I.N. Allosteric drugs and mutations: Chances, challenges, and necessity. Curr. Opin. Struct. Biol. 2020, 62, 149–157. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coffino, P.; Cheng, Y. Allostery Modulates Interactions between Proteasome Core Particles and Regulatory Particles. Biomolecules 2022, 12, 764. https://doi.org/10.3390/biom12060764
Coffino P, Cheng Y. Allostery Modulates Interactions between Proteasome Core Particles and Regulatory Particles. Biomolecules. 2022; 12(6):764. https://doi.org/10.3390/biom12060764
Chicago/Turabian StyleCoffino, Philip, and Yifan Cheng. 2022. "Allostery Modulates Interactions between Proteasome Core Particles and Regulatory Particles" Biomolecules 12, no. 6: 764. https://doi.org/10.3390/biom12060764
APA StyleCoffino, P., & Cheng, Y. (2022). Allostery Modulates Interactions between Proteasome Core Particles and Regulatory Particles. Biomolecules, 12(6), 764. https://doi.org/10.3390/biom12060764





