miRNA Pattern in Hypoxic Microenvironment of Kidney Cancer—Role of PTEN
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. CRPISPR/Cas9 Mediated PTEN Knockout in Renca Cells
2.3. Healthy and Tumor Kidney Cancer Tissue
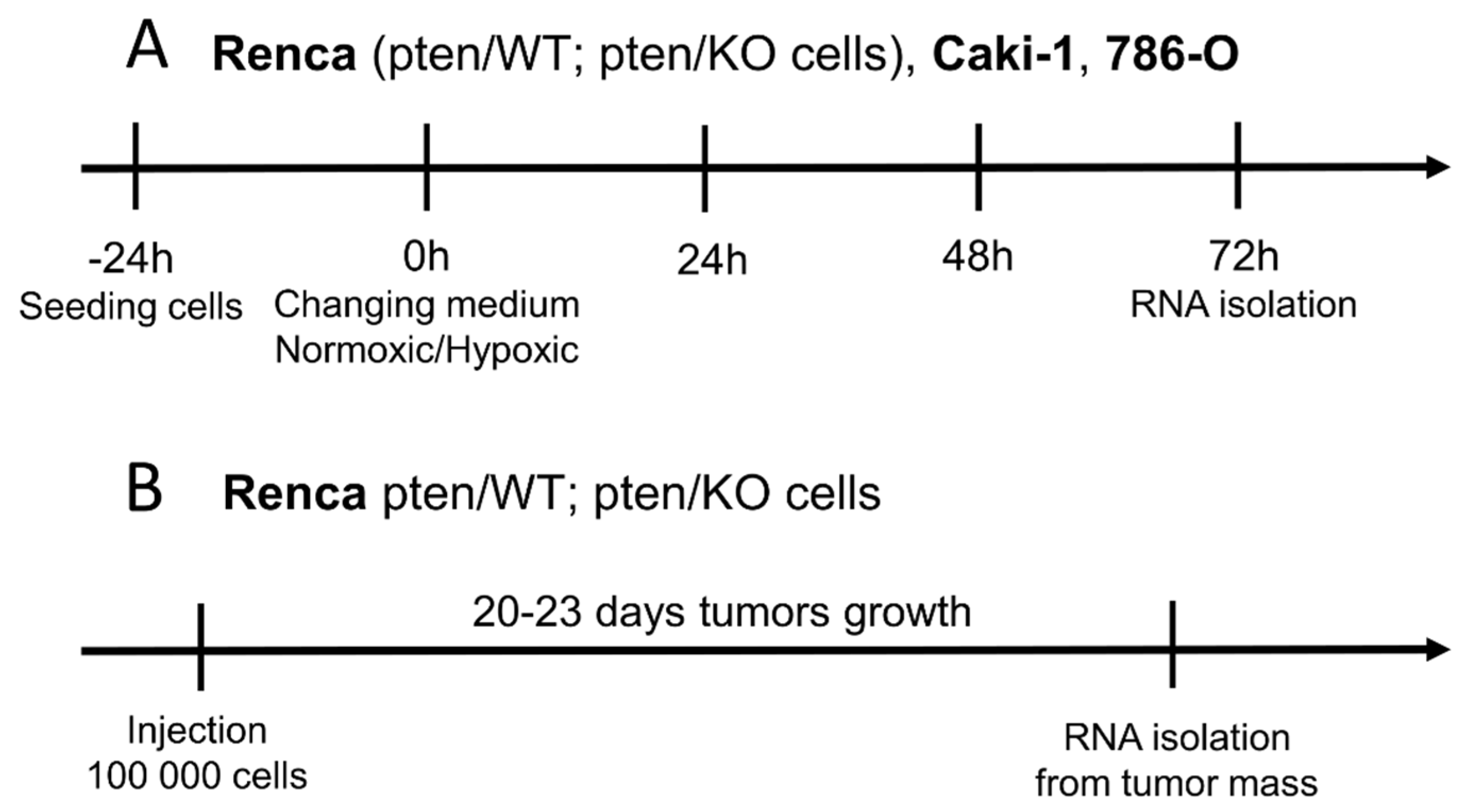
2.4. Cell Culture Methods in Normoxic and Hypoxic Conditions
2.5. In Vivo Renca pten/WT and pten/KO Tumor Implantation and Development
2.6. Total RNA Isolation
2.7. Next Generation Sequencing
2.8. Validation of DEmiRNAs Expression by qRT-PCR
2.9. Prediction of Potential Targets for DEmiRNAs
2.10. Evaluation of the Expression of Target Genes for DEmiRNAs by qRT-PCR
2.11. Western Blot
2.12. Exosomes Isolation
2.13. Culture MBr MEC FVB with Conditioned Medium from Renca Cells
2.14. Statistical Analysis
3. Results
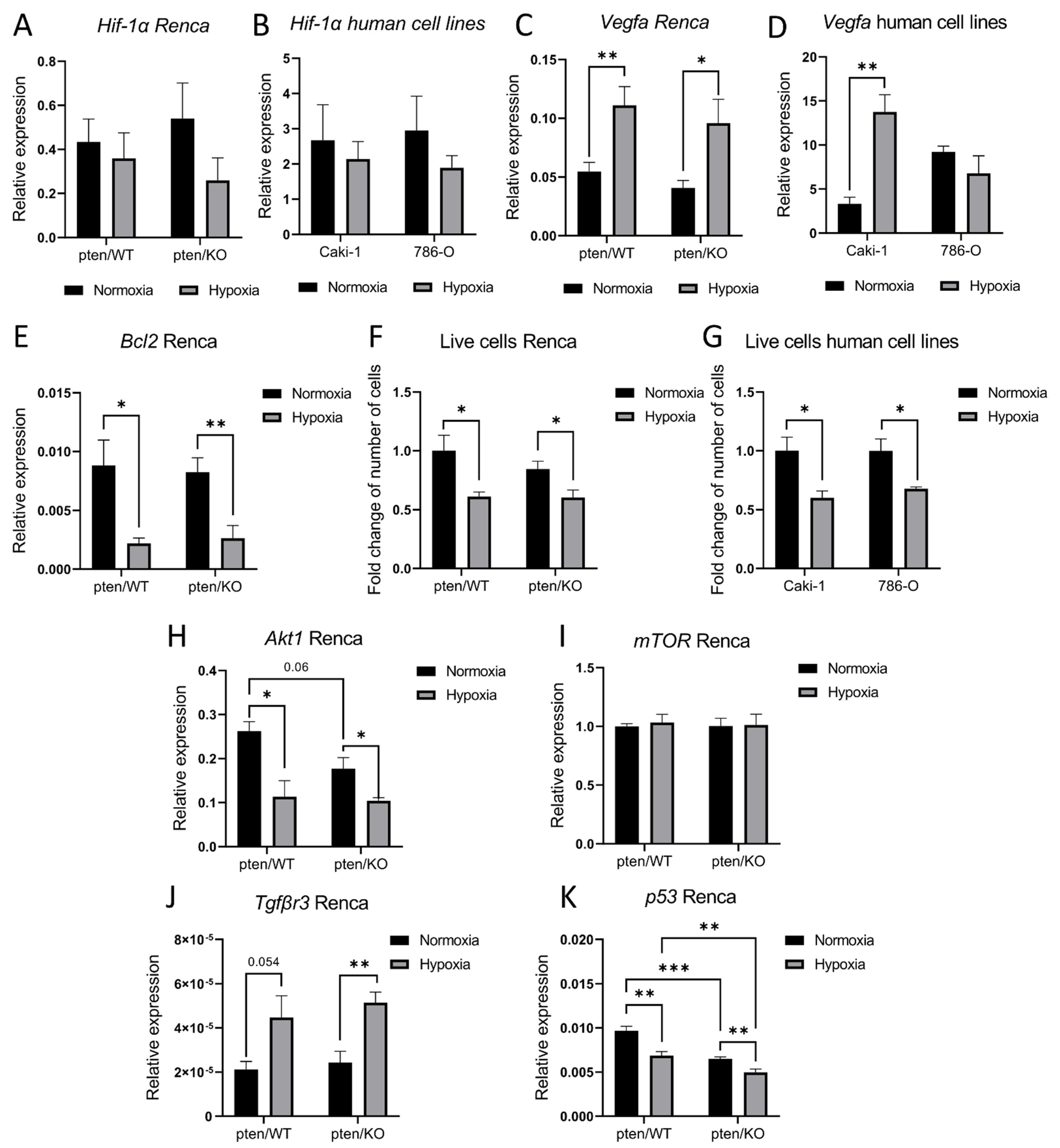
3.1. PTEN Status in Tested Models
3.2. Hypoxia-Induced miRNA Signature in Murine Kidney Cancer Models with Different PTEN Status
3.3. Identification of PTEN-Related miRNA in Hypoxic Conditions
3.4. PTEN-Dependent miRNA Pattern
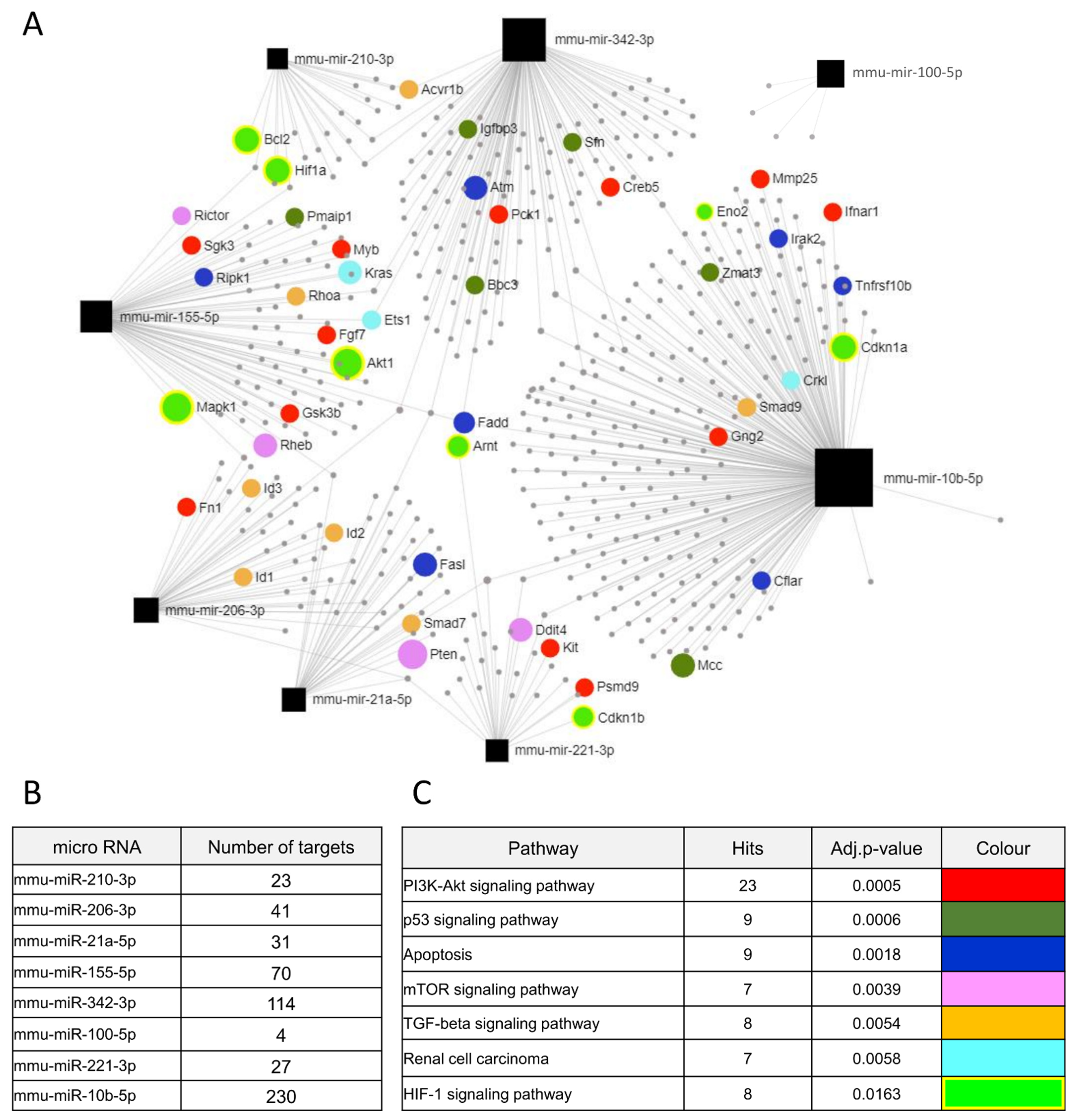
3.5. Identification of Potential Hypoxia- and PTEN-Dependent miRNAs Targets
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Garzon, R.; Fabbri, M.; Cimmino, A.; Calin, G.A.; Croce, C.M. MicroRNA expression and function in cancer. Trends Mol. Med. 2006, 12, 580–587. [Google Scholar] [CrossRef] [PubMed]
- Qu, H.; Zheng, L.; Pu, J.; Mei, H.; Xiang, X.; Zhao, X.; Li, D.; Li, S.; Mao, L.; Huang, K.; et al. miRNA-558 promotes tumorigenesis and aggressiveness of neuroblastoma cells through activating the transcription of heparanase. Hum. Mol. Genet. 2015, 24, 2539–2551. [Google Scholar] [CrossRef] [PubMed]
- Lou, W.; Liu, J.; Gao, Y.; Zhong, G.; Chen, D.; Shen, J.; Bao, C.; Xu, L.; Pan, J.; Cheng, J.; et al. MicroRNAs in cancer metastasis and angiogenesis. Oncotarget 2017, 8, 115787–115802. [Google Scholar] [CrossRef]
- Goradel, N.H.; Mohammadi, N.; Haghi-Aminjan, H.; Farhood, B.; Negahdari, B.; Sahebkar, A. Regulation of tumor angiogenesis by microRNAs: State of the art. J. Cell. Physiol. 2018, 234, 1099–1110. [Google Scholar] [CrossRef]
- Pratama, M.Y.; Pascut, D.; Massi, M.N.; Tiribelli, C. The role of microRNA in the resistance to treatment of hepatocellular carcinoma. Ann. Transl. Med. 2019, 7, 577. [Google Scholar] [CrossRef]
- Sohel, M.H. Circulating microRNAs as biomarkers in cancer diagnosis. Life Sci. 2020, 248, 117473. [Google Scholar] [CrossRef]
- Nejad, A.E.; Najafgholian, S.; Rostami, A.; Sistani, A.; Shojaeifar, S.; Esparvarinha, M.; Nedaeinia, R.; Javanmard, S.H.; Taherian, M.; Ahmadlou, M.; et al. The role of hypoxia in the tumor microenvironment and development of cancer stem cell: A novel approach to developing treatment. Cancer Cell Int. 2021, 21, 1–26. [Google Scholar] [CrossRef]
- Majmundar, A.J.; Wong, W.J.; Simon, M.C. Hypoxia-Inducible Factors and the Response to Hypoxic Stress. Mol. Cell 2010, 40, 294–309. [Google Scholar] [CrossRef]
- Hayashi, Y.; Yokota, A.; Harada, H.; Huang, G. Hypoxia/pseudohypoxia-mediated activation of hypoxia-inducible factor-1α in cancer. Cancer Sci. 2019, 110, 1510–1517. [Google Scholar] [CrossRef]
- Cowey, C.L.; Rathmell, W.K. VHL gene mutations in renal cell carcinoma: Role as a biomarker of disease outcome and drug efficacy. Curr. Oncol. Rep. 2009, 11, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Escudier, B.; Worden, F.; Kudo, M. Sorafenib: Key lessons from over 10 years of experience. Expert Rev. Anticancer Ther. 2018, 19, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Makhov, P.; Joshi, S.; Ghatalia, P.; Kutikov, A.; Uzzo, R.G.; Kolenko, V.M. Resistance to Systemic Therapies in Clear Cell Renal Cell Carcinoma: Mechanisms and Management Strategies. Mol. Cancer Ther. 2018, 17, 1355–1364. [Google Scholar] [CrossRef] [PubMed]
- Shen, G.; Li, X.; Jia, Y.-F.; Piazza, G.; Xi, Y. Hypoxia-regulated microRNAs in human cancer. Acta Pharmacol. Sin. 2013, 34, 336–341. [Google Scholar] [CrossRef] [PubMed]
- Nallamshetty, S.; Chan, S.Y.; Loscalzo, J. Hypoxia: A master regulator of microRNA biogenesis and activity. Free Radic. Biol. Med. 2013, 64, 20–30. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Wu, S.Y.; Pradeep, S.; Ivan, C.; Pecot, C.V.; Gharpure, K.M.; Nagaraja, A.S.; Armaiz-Pena, G.N.; McGuire, M.; Zand, B.; et al. Hypoxia-mediated downregulation of miRNA biogenesis promotes tumour progression. Nat. Commun. 2014, 5, 5202. [Google Scholar] [CrossRef]
- Peng, X.; Gao, H.; Xu, R.; Wang, H.; Mei, J.; Liu, C. The interplay between HIF-1α and noncoding RNAs in cancer. J. Exp. Clin. Cancer Res. 2020, 39, 27. [Google Scholar] [CrossRef]
- Tang, K.; Xu, H. Prognostic value of meta-signature miRNAs in renal cell carcinoma: An integrated miRNA expression profiling analysis. Sci. Rep. 2015, 5, 10272. [Google Scholar] [CrossRef]
- I McCormick, R.; Blick, C.; Ragoussis, J.; Schoedel, J.; Mole, D.R.; Young, A.C.; Selby, P.J.; Banks, R.; Harris, A.L. miR-210 is a target of hypoxia-inducible factors 1 and 2 in renal cancer, regulates ISCU and correlates with good prognosis. Br. J. Cancer 2013, 108, 1133–1142. [Google Scholar] [CrossRef]
- Neal, C.S.; Michael, M.Z.; Rawlings, L.H.; Van Der Hoek, M.B.; Gleadle, J.M. The VHL-dependent regulation of microRNAs in renal cancer. BMC Med. 2010, 8, 64. [Google Scholar] [CrossRef]
- Milella, M.; Falcone, I.; Conciatori, F.; Cesta Incani, U.; Del Curatolo, A.; Inzerilli, N.; Nuzzo, C.M.; Vaccaro, V.; Vari, S.; Cognetti, F.; et al. PTEN: Multiple Functions in Human Malignant Tumors. Front. Oncol. 2015, 5, 24. [Google Scholar] [CrossRef] [PubMed]
- Brenner, W.; Färber, G.; Herget, T.; Lehr, H.-A.; Hengstler, J.G.; Thüroff, J.W. Loss of tumor suppressor protein PTEN during renal carcinogenesis. Int. J. Cancer 2002, 99, 53–57. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Li, X.; Gao, Y.; Chen, L.; Gu, L.; Chen, J.; Lyu, X.; Zhang, Y.; Zhang, X. Phosphatase and tensin homolog (PTEN) expression on oncologic outcome in renal cell carcinoma: A systematic review and meta-analysis. PLoS ONE 2017, 12, e0179437. [Google Scholar] [CrossRef] [PubMed]
- Simpson, L.; Parsons, R. PTEN: Life as a Tumor Suppressor. Exp. Cell Res. 2001, 264, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Emerling, B.M.; Weinberg, F.; Liu, J.-L.; Mak, T.W.; Chandel, N.S. PTEN regulates p300-dependent hypoxia-inducible factor 1 transcriptional activity through Forkhead transcription factor 3a (FOXO3a). Proc. Natl. Acad. Sci. USA 2008, 105, 2622–2627. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhang, T.; Guo, L.; Huang, L. Regulation of PTEN expression by noncoding RNAs. J. Exp. Clin. Cancer Res. 2018, 37, 223. [Google Scholar] [CrossRef] [PubMed]
- Dillon, L.M.; Miller, T.W. Therapeutic targeting of cancers with loss of PTEN function. Curr. Drug Targets 2014, 15, 65–79. [Google Scholar] [CrossRef]
- Dart, D.A.; Uysal-Onganer, P.; Jiang, W.G. Prostate-specific PTen deletion in mice activates inflammatory microRNA expression pathways in the epithelium early in hyperplasia development. Oncogenesis 2017, 6, 400. [Google Scholar] [CrossRef]
- Yuan, T.; Yang, Y.; Chen, J.; Li, W.; Zhang, Q.; Mi, Y.; Goswami, R.; You, J.Q.; Lin, D.; Qian, M.D.; et al. Regulation of PI3K signaling in T-cell acute lymphoblastic leukemia: A novel PTEN/Ikaros/miR-26b mechanism reveals a critical targetable role for PIK3CD. Leukemia 2017, 31, 2355–2364. [Google Scholar] [CrossRef]
- Freeman, D.J.; Li, A.G.; Wei, G.; Li, H.-H.; Kertesz, N.; Lesche, R.; Whale, A.D.; Martinez-Diaz, H.; Rozengurt, N.; Cardiff, R.D.; et al. PTEN tumor suppressor regulates p53 protein levels and activity through phosphatase-dependent and -independent mechanisms. Cancer Cell 2003, 3, 117–130. [Google Scholar] [CrossRef]
- Tang, Y.; Weng, X.; Liu, C.; Li, X.; Chen, C. Hypoxia Enhances Activity and Malignant Behaviors of Colorectal Cancer Cells through the STAT3/MicroRNA-19a/PTEN/PI3K/AKT Axis. Anal. Cell. Pathol. 2021, 2021, 4132488. [Google Scholar] [CrossRef] [PubMed]
- Liang, T.; Gao, F.; Chen, J. Role of PTEN-less in cardiac injury, hypertrophy and regeneration. Cell Regen. 2021, 10, 25. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Yao, L.; Yang, J.; Wang, Z.; Du, G. PI3K/Akt and HIF-1 signaling pathway in hypoxia-ischemia (Review). Mol. Med. Rep. 2018, 18, 3547–3554. [Google Scholar] [CrossRef] [PubMed]
- Bizouarne, N.; Denis, V.; Legrand, A.; Monsigny, M.; Kieda, C.; Bizourne, N. A SV-40 immortalized murine endothelial cell line from peripheral lymph node high endothelium expresses a new α-L-fucose binding protein. Biol. Cell 1993, 79, 209–218. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef]
- Fusco, N.; Sajjadi, E.; Venetis, K.; Gaudioso, G.; Lopez, G.; Corti, C.; Rocco, E.G.; Criscitiello, C.; Malapelle, U.; Invernizzi, M. PTEN Alterations and Their Role in Cancer Management: Are We Making Headway on Precision Medicine? Genes 2020, 11, 719. [Google Scholar] [CrossRef]
- The AACR Project GENIE Consortium AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov. 2017, 7, 818–831. [CrossRef]
- Khouzam, R.A.; Rao, S.P.; Venkatesh, G.H.; Zeinelabdin, N.A.; Buart, S.; Meylan, M.; Nimmakayalu, M.; Terry, S.; Chouaib, S. An Eight-Gene Hypoxia Signature Predicts Survival in Pancreatic Cancer and Is Associated with an Immunosuppressed Tumor Microenvironment. Front. Immunol. 2021, 12, 435. [Google Scholar] [CrossRef]
- Sørensen, B.S.; Knudsen, A.; Wittrup, C.F.; Nielsen, S.; Aggerholm-Pedersen, N.; Busk, M.; Horsman, M.; Høyer, M.; Bouchelouche, P.N.; Overgaard, J.; et al. The usability of a 15-gene hypoxia classifier as a universal hypoxia profile in various cancer cell types. Radiother. Oncol. 2015, 116, 346–351. [Google Scholar] [CrossRef]
- Kong, F.; Ran, W.; Jiang, N.; Li, S.; Zhang, D.; Sun, D. Identification and characterization of differentially expressed miRNAs in HepG2 cells under normoxic and hypoxic conditions. RSC Adv. 2019, 9, 16884–16891. [Google Scholar] [CrossRef]
- Bertero, T.; Rezzonico, R.; Pottier, N.; Mari, B. Impact of MicroRNAs in the Cellular Response to Hypoxia. Int. Rev. Cell Mol. Biol. 2017, 333, 91–158. [Google Scholar] [CrossRef] [PubMed]
- Yoshino, H.; Yonemori, M.; Miyamoto, K.; Tatarano, S.; Kofuji, S.; Nohata, N.; Nakagawa, M.; Enokida, H. microRNA-210-3p depletion by CRISPR/Cas9 promoted tumorigenesis through revival of TWIST1 in renal cell carcinoma. Oncotarget 2017, 8, 20881–20894. [Google Scholar] [CrossRef] [PubMed]
- Song, B.-W.; Lee, C.Y.; Kim, R.; Kim, W.J.; Lee, H.W.; Lee, M.Y.; Kim, J.; Jeong, J.-Y.; Chang, W. Multiplexed targeting of miRNA-210 in stem cell-derived extracellular vesicles promotes selective regeneration in ischemic hearts. Exp. Mol. Med. 2021, 53, 695–708. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.-D.; Cai, F.; Liu, L.; Zhang, Y.; Yang, A.-L. microRNA-210 is involved in the regulation of postmenopausal osteoporosis through promotion of VEGF expression and osteoblast differentiation. Biol. Chem. 2015, 396, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Chio, C.-C.; Lin, J.-W.; Cheng, H.-A.; Chiu, W.-T.; Wang, Y.-H.; Wang, J.-J.; Hsing, C.-H.; Chen, R.-M. MicroRNA-210 targets antiapoptotic Bcl-2 expression and mediates hypoxia-induced apoptosis of neuroblastoma cells. Arch. Toxicol. 2012, 87, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Zhou, J.; Tong, R.; Wang, B.; Chen, X.; Zhuang, Y.; Xia, F.; Wei, X. Hypoxia-induced miR-210 contributes to apoptosis of mouse spermatocyte GC-2 cells by targeting Kruppel-like factor 7. Mol. Med. Rep. 2018, 19, 271–279. [Google Scholar] [CrossRef]
- Kohnoh, T.; Hashimoto, N.; Ando, A.; Sakamoto, K.; Miyazaki, S.; Aoyama, D.; Kusunose, M.; Kimura, M.; Omote, N.; Imaizumi, K.; et al. Hypoxia-induced modulation of PTEN activity and EMT phenotypes in lung cancers. Cancer Cell Int. 2016, 16, 33. [Google Scholar] [CrossRef]
- Shi, J.; Zhang, D.; Zhong, Z.; Zhang, W. lncRNA ROR promotes the progression of renal cell carcinoma through the miR-206/VEGF axis. Mol. Med. Rep. 2019, 20, 3782–3792. [Google Scholar] [CrossRef]
- Singh, A.; Pruett, N.; Pahwa, R.; Mahajan, A.P.; Schrump, D.S.; Hoang, C.D. MicroRNA-206 suppresses mesothelioma progression via the Ras signaling axis. Mol. Ther. Nucleic Acids 2021, 24, 669–681. [Google Scholar] [CrossRef]
- Liao, M.; Peng, L. MiR-206 may suppress non-small lung cancer metastasis by targeting CORO1C. Cell. Mol. Biol. Lett. 2020, 25, 22. [Google Scholar] [CrossRef]
- Huang, J.; Sun, C.; Wang, S.; He, Q.; Li, D. microRNA miR-10b inhibition reduces cell proliferation and promotes apoptosis in non-small cell lung cancer (NSCLC) cells. Mol. BioSyst. 2015, 11, 2051–2059. [Google Scholar] [CrossRef] [PubMed]
- Sheedy, P.; Medarova, Z. The fundamental role of miR-10b in metastatic cancer. Am. J. Cancer Res. 2018, 8, 1674–1688. [Google Scholar] [PubMed]
- Kim, J.; Siverly, A.N.; Chen, D.; Wang, M.; Yuan, Y.; Wang, Y.; Lee, H.; Zhang, J.; Muller, W.J.; Liang, H.; et al. Ablation of miR-10b Suppresses Oncogene-Induced Mammary Tumorigenesis and Metastasis and Reactivates Tumor-Suppressive Pathways. Cancer Res. 2016, 76, 6424–6435. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-C.; Liao, W.-T.; Yang, T.-Y.; Lu, H.-J.; Hsu, S.-L.; Wu, C.-C. MicroRNA-10b modulates cisplatin tolerance by targeting p53 directly in lung cancer cells. Oncol. Rep. 2021, 46, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Bhandari, V.; PCAWG Consortium; Li, C.H.; Bristow, R.G.; Boutros, P.C. Divergent mutational processes distinguish hypoxic and normoxic tumours. Nat. Commun. 2020, 11, 737. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Li, J.; Qi, Y.; Zhang, C.; Zhu, D.; Liu, D.; Zhao, S. SNHG14 confers gefitinib resistance in non-small cell lung cancer by up-regulating ABCB1 via sponging miR-206-3p. Biomed. Pharmacother. 2019, 116, 108995. [Google Scholar] [CrossRef]
- Chen, Z.; Gao, Y.-J.; Hou, R.-Z.; Ding, D.-Y.; Song, D.-F.; Wang, D.-Y.; Feng, Y. MicroRNA-206 facilitates gastric cancer cell apoptosis and suppresses cisplatin resistance by targeting MAPK2 signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 171–180. [Google Scholar]
- Wang, T.; Dong, X.-M.; Zhang, F.-L.; Zhang, J.-R. miR-206 enhances nasopharyngeal carcinoma radiosensitivity by targeting IGF1. Kaohsiung J. Med. Sci. 2017, 33, 427–432. [Google Scholar] [CrossRef]
- Nishida, N.; Yamashita, S.; Mimori, K.; Sudo, T.; Tanaka, F.; Shibata, K.; Yamamoto, H.; Ishii, H.; Doki, Y.; Mori, M. MicroRNA-10b is a Prognostic Indicator in Colorectal Cancer and Confers Resistance to the Chemotherapeutic Agent 5-Fluorouracil in Colorectal Cancer Cells. Ann. Surg. Oncol. 2012, 19, 3065–3071. [Google Scholar] [CrossRef]
- Zhen, L.; Li, J.; Zhang, M.; Yang, K. MiR-10b decreases sensitivity of glioblastoma cells to radiation by targeting AKT. J. Biol. Res. 2016, 23, 14. [Google Scholar] [CrossRef]
- Pranzini, E.; Leo, A.; Rapizzi, E.; Ramazzotti, M.; Magherini, F.; Giovannelli, L.; Caselli, A.; Cirri, P.; Taddei, M.L.; Paoli, P. miR-210-3p mediates metabolic adaptation and sustains DNA damage repair of resistant colon cancer cells to treatment with 5-fluorouracil. Mol. Carcinog. 2019, 58, 2181–2192. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, H.-Q.; Wang, J.; Yuan, Z.-Y.; Zhao, L.-J.; Wang, P.; Wang, C.-L. The drug-resistance to gefitinib in PTEN low expression cancer cells is reversed by irradiation in vitro. J. Exp. Clin. Cancer Res. 2009, 28, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Steelman, L.S.; Navolanic, P.M.; Sokolosky, M.L.; Taylor, J.R.; Lehmann, B.D.; Chappell, W.H.; Abrams, S.L.; Wong, E.W.T.; Stadelman, K.M.; Terrian, D.M.; et al. Suppression of PTEN function increases breast cancer chemotherapeutic drug resistance while conferring sensitivity to mTOR inhibitors. Oncogene 2008, 27, 4086–4095. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Wang, W.; Yao, C.; Zhang, S.; Liang, L.; Han, M.; Ren, J.; Qi, X.; Zhang, X.; Wang, S.; et al. Radiation-resistant cancer stem-like cell properties are regulated by PTEN through the activity of nuclear β-catenin in nasopharyngeal carcinoma. Oncotarget 2017, 8, 74661–74672. [Google Scholar] [CrossRef]
- Fu, X.; Wen, H.; Jing, L.; Yang, Y.; Wang, W.; Liang, X.; Nan, K.; Yao, Y.; Tian, T. MicroRNA-155-5p promotes hepatocellular carcinoma progression by suppressing PTEN through the PI3K/Akt pathway. Cancer Sci. 2017, 108, 620–631. [Google Scholar] [CrossRef]
- Wang, G.; Chen, L.; Meng, J.; Chen, M.; Zhuang, L.; Zhang, L. Overexpression of microRNA-100 predicts an unfavorable prognosis in renal cell carcinoma. Int. Urol. Nephrol. 2013, 45, 373–379. [Google Scholar] [CrossRef]
- Nagaraja, A.K.; Creighton, C.J.; Yu, Z.; Zhu, H.; Gunaratne, P.H.; Reid, J.G.; Olokpa, E.; Itamochi, H.; Ueno, N.T.; Hawkins, S.; et al. A Link between mir-100 and FRAP1/mTOR in Clear Cell Ovarian Cancer. Mol. Endocrinol. 2010, 24, 447–463. [Google Scholar] [CrossRef]
- Guo, Y.; Li, X.; Zheng, J.; Fang, J.; Pan, G.; Chen, Z. Identification of a novel immune-related microRNA prognostic model in clear cell renal cell carcinoma. Transl. Androl. Urol. 2021, 10, 888–899. [Google Scholar] [CrossRef]
- Shi, J.; Zhang, Y.; Jin, N.; Li, Y.; Wu, S.; Xu, L. MicroRNA-221-3p Plays an Oncogenic Role in Gastric Carcinoma by Inhibiting PTEN Expression. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2017, 25, 523–536. [Google Scholar] [CrossRef]
- Detassis, S.; Grasso, M.; Del Vescovo, V.; Denti, M.A. microRNAs Make the Call in Cancer Personalized Medicine. Front. Cell Dev. Biol. 2017, 5, 86. [Google Scholar] [CrossRef]
- Wu, X.-G.; Zhou, C.-F.; Zhang, Y.-M.; Yan, R.-M.; Wei, W.-F.; Chen, X.-J.; Yi, H.-Y.; Liang, L.-J.; Fan, L.-S.; Liang, L.; et al. Cancer-derived exosomal miR-221-3p promotes angiogenesis by targeting THBS2 in cervical squamous cell carcinoma. Angiogenesis 2019, 22, 397–410. [Google Scholar] [CrossRef] [PubMed]
- Deng, D.; Liang, H. Silencing MEG3 protects PC12 cells from hypoxic injury by targeting miR-21. Artif. Cells Nanomed. Biotechnol. 2020, 48, 610–619. [Google Scholar] [CrossRef] [PubMed]
- Kulshreshtha, R.; Ferracin, M.; Wojcik, S.E.; Garzon, R.; Alder, H.; Agosto-Perez, F.J.; Davuluri, R.; Liu, C.-G.; Croce, C.M.; Negrini, M.; et al. A MicroRNA Signature of Hypoxia. Mol. Cell. Biol. 2007, 27, 1859–1867. [Google Scholar] [CrossRef] [PubMed]
- Papagiannakopoulos, T.; Shapiro, A.; Kosik, K.S. MicroRNA-21 Targets a Network of Key Tumor-Suppressive Pathways in Glioblastoma Cells. Cancer Res. 2008, 68, 8164–8172. [Google Scholar] [CrossRef] [PubMed]
- Zheng, F.; He, K.; Li, X.; Zhao, D.; Sun, F.; Zhang, Y.; Nie, D.; Li, X.; Chu, W.; Sun, Y.; et al. Transient overexpression of TGFBR3 induces apoptosis in human nasopharyngeal carcinoma CNE-2Z cells. Biosci. Rep. 2013, 33, e00029. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed]




| miRNA | Assay Number |
|---|---|
| miR-210-3p | mmu481343_mir |
| miR-206-3p | mmu481645_mir |
| miR-100-5p | mmu478224_mir |
| miR-155-5p | mmu480953_mir |
| miR-100-5p | 478224_mir |
| miR-155-5p | 483064_mir |
| miR-342-3p | mmu481074_mir |
| miR-221-3p | mmu481005_mir |
| miR-10b-5p | mmu478494_mir |
| miR-21a-5p | mmu482709_mir |
| miR-25-3p | mmu483226_mir |
| miR-16-5p | mmu482960_mir |
| Step | Temperature | Time | Cycle |
|---|---|---|---|
| Enzyme activation | 95 °C | 20 s | 1 |
| Denature | 95 °C | 1 s | 40 |
| Anneal/Extend | 60 °C | 20 s |
| Gene | Taq Man Probes |
|---|---|
| Hif-1α | Mm00468869; Hs00153153 |
| Vegfa | Mm00437306; Hs00900055 |
| Akt1 | Mm00437306 |
| Tp53 | Mm01731287 |
| mTOR | Mm01731287 |
| Pten | Hs02621230 |
| Actinβ | Mm02619580 |
| GusB | Hs00939627 |
| Gene | Primers Sequences |
|---|---|
| Bcl2 | F: GACTGAGTACCTGAACCGGC |
| R: AGTTCCACAAAGGCATCCCAG | |
| Tgfβr3 | F: AGTGCTCTGAGTGCTCCCTA |
| R: TACTCCCACACAGGGGAGAC | |
| Actinβ | F: CCTAGGCACCAGGGTGTGA |
| R: GTTGGCCTTAGGGTTCAGGG |
| Step | Temperature | Duration | Cycle |
|---|---|---|---|
| UNG incubation | 50 °C | 2 min | Hold |
| Polymerase Activation | 95 °C | 10 min | Hold |
| Denature | 95 °C | 15 s | 40 |
| Anneal/extend | 60 °C | 15 s |
| Step | Temperature | Duration | Cycle |
|---|---|---|---|
| UDG activation | 50 °C | 2 min | Hold |
| Dual-Lock™ DNA polymerase | 95 °C | 2 min | Hold |
| Denature | 95 °C | 15 s | 40 |
| Anneal | 60 °C | 45 s | |
| Extend | 60 °C | 1 min |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Majewska, A.; Brodaczewska, K.; Filipiak-Duliban, A.; Kajdasz, A.; Kieda, C. miRNA Pattern in Hypoxic Microenvironment of Kidney Cancer—Role of PTEN. Biomolecules 2022, 12, 686. https://doi.org/10.3390/biom12050686
Majewska A, Brodaczewska K, Filipiak-Duliban A, Kajdasz A, Kieda C. miRNA Pattern in Hypoxic Microenvironment of Kidney Cancer—Role of PTEN. Biomolecules. 2022; 12(5):686. https://doi.org/10.3390/biom12050686
Chicago/Turabian StyleMajewska, Aleksandra, Klaudia Brodaczewska, Aleksandra Filipiak-Duliban, Arkadiusz Kajdasz, and Claudine Kieda. 2022. "miRNA Pattern in Hypoxic Microenvironment of Kidney Cancer—Role of PTEN" Biomolecules 12, no. 5: 686. https://doi.org/10.3390/biom12050686
APA StyleMajewska, A., Brodaczewska, K., Filipiak-Duliban, A., Kajdasz, A., & Kieda, C. (2022). miRNA Pattern in Hypoxic Microenvironment of Kidney Cancer—Role of PTEN. Biomolecules, 12(5), 686. https://doi.org/10.3390/biom12050686








