Regulatory Crosstalk between Physiological Low O2 Concentration and Notch Pathway in Early Erythropoiesis
Abstract
1. Introduction
2. Materials and Methods
2.1. CD34+ Cell Isolation
2.2. Generation of Lentiviral Vectors and Viral Particles
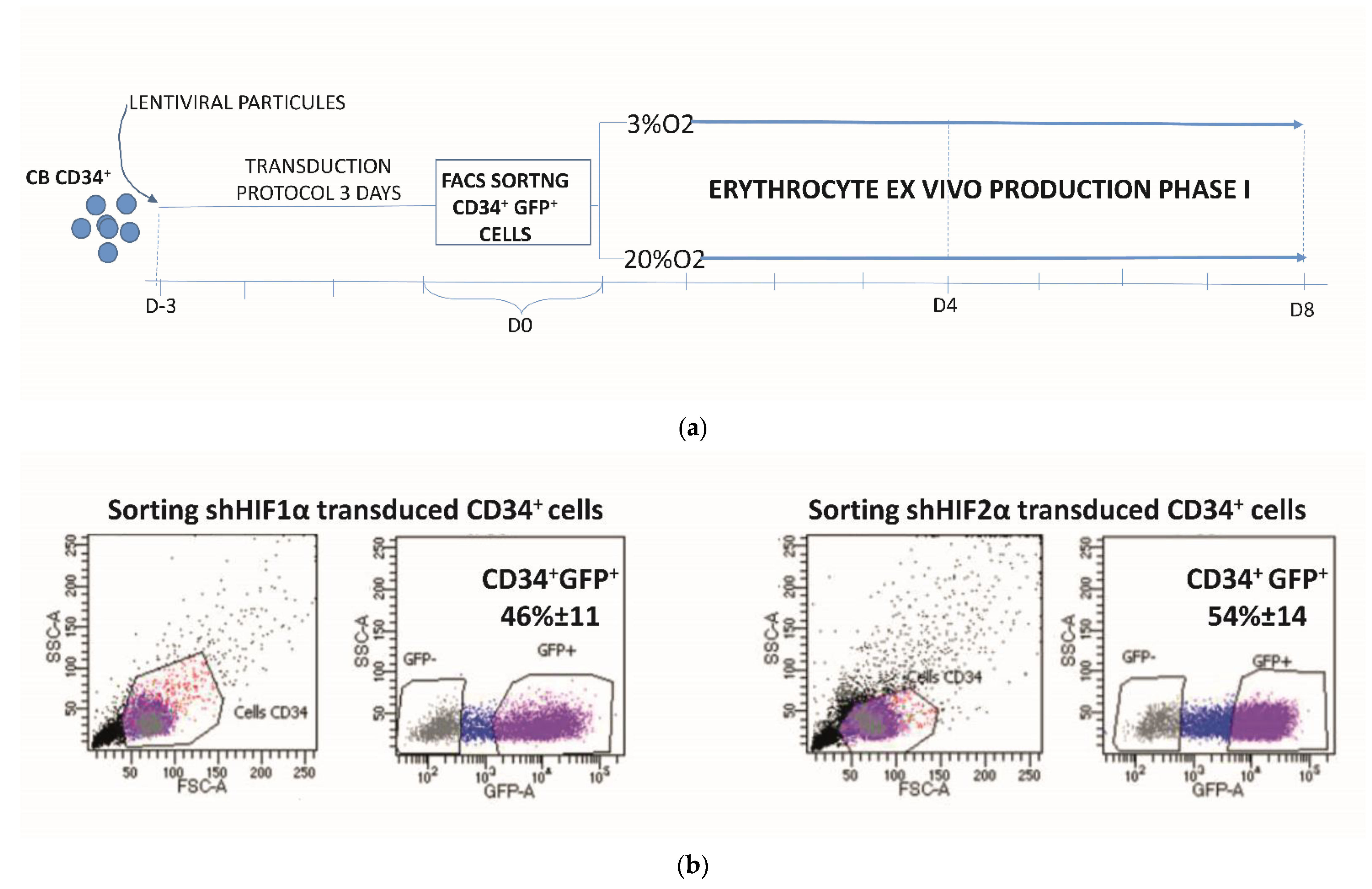
2.3. Transduction Protocol for CD34+ Cells
2.4. Erythroid Cell Cultures
2.5. CFC Assay
2.6. Phenotypic Analysis
2.7. RNA Isolation and Quantitative Real-Time PCR
2.8. CFSE Staining
2.9. Propidium Iodide Cell Viability Assay
2.10. Statistical Analysis
3. Results
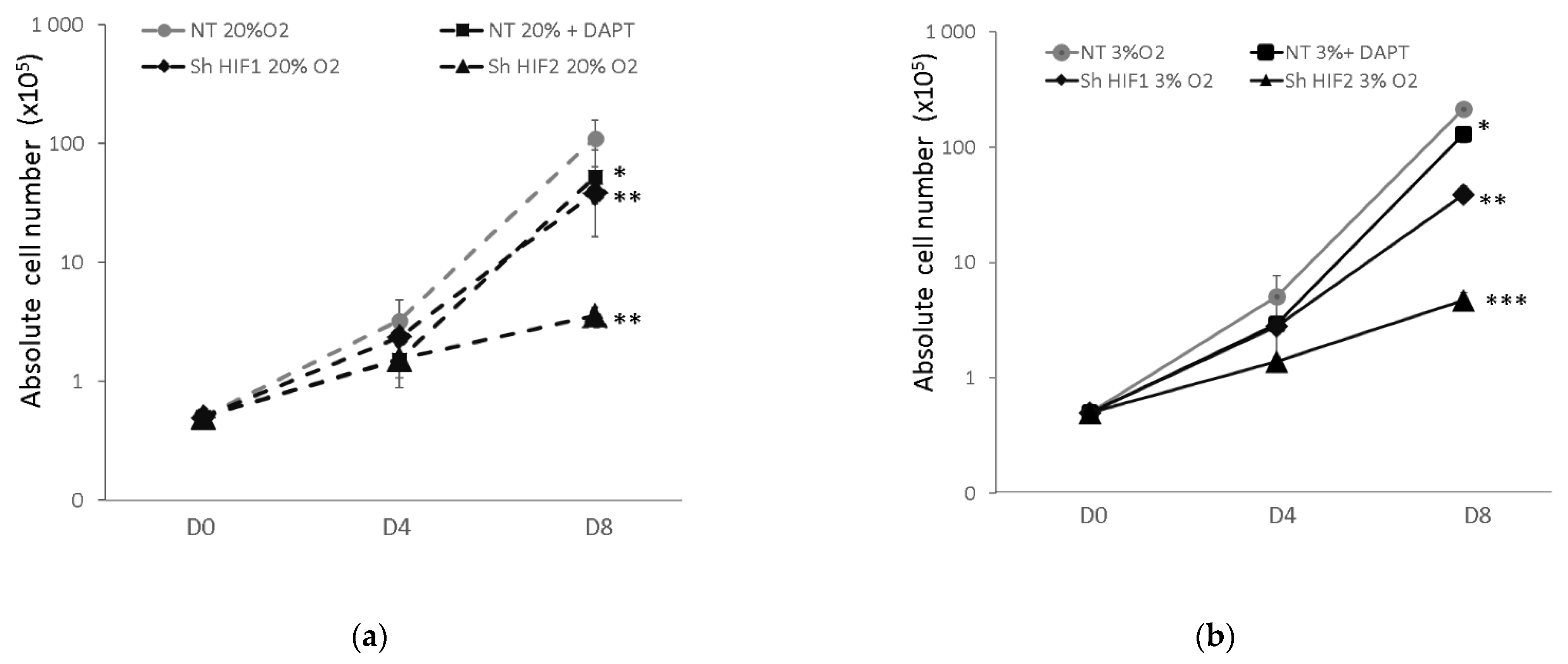
3.1. Erythroid Cell Expansion Is Abrogated When Notch and HIF-Associated Pathways Are Inhibited
3.2. Erythroid Progenitor Amplification Is Differently Modified When the HIF-Mediated or Notch Pathway Are Inhibited
3.3. Erythroid Differentiation Is Altered When HIF-Mediated but Not Notch Pathways Are Inhibited
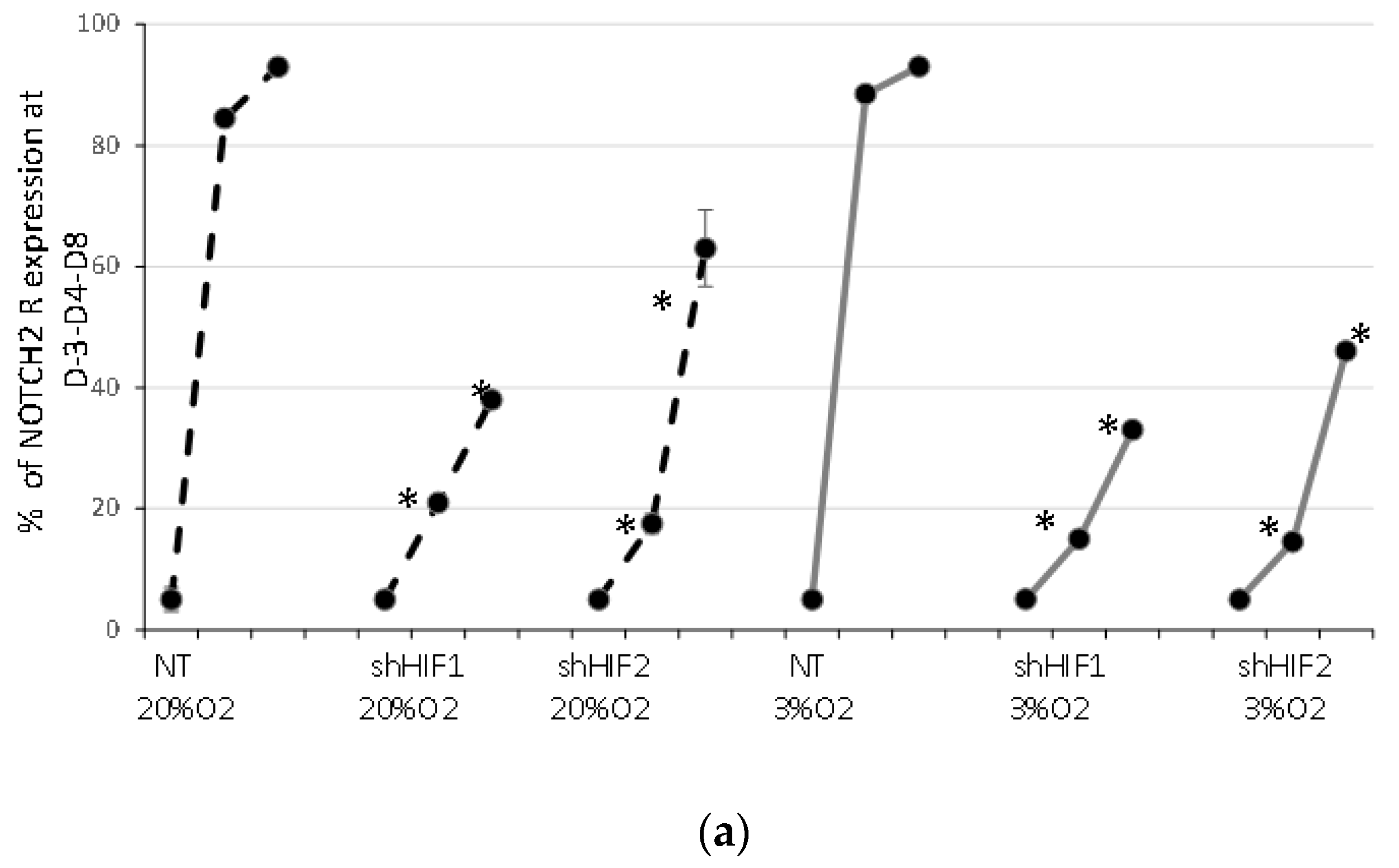
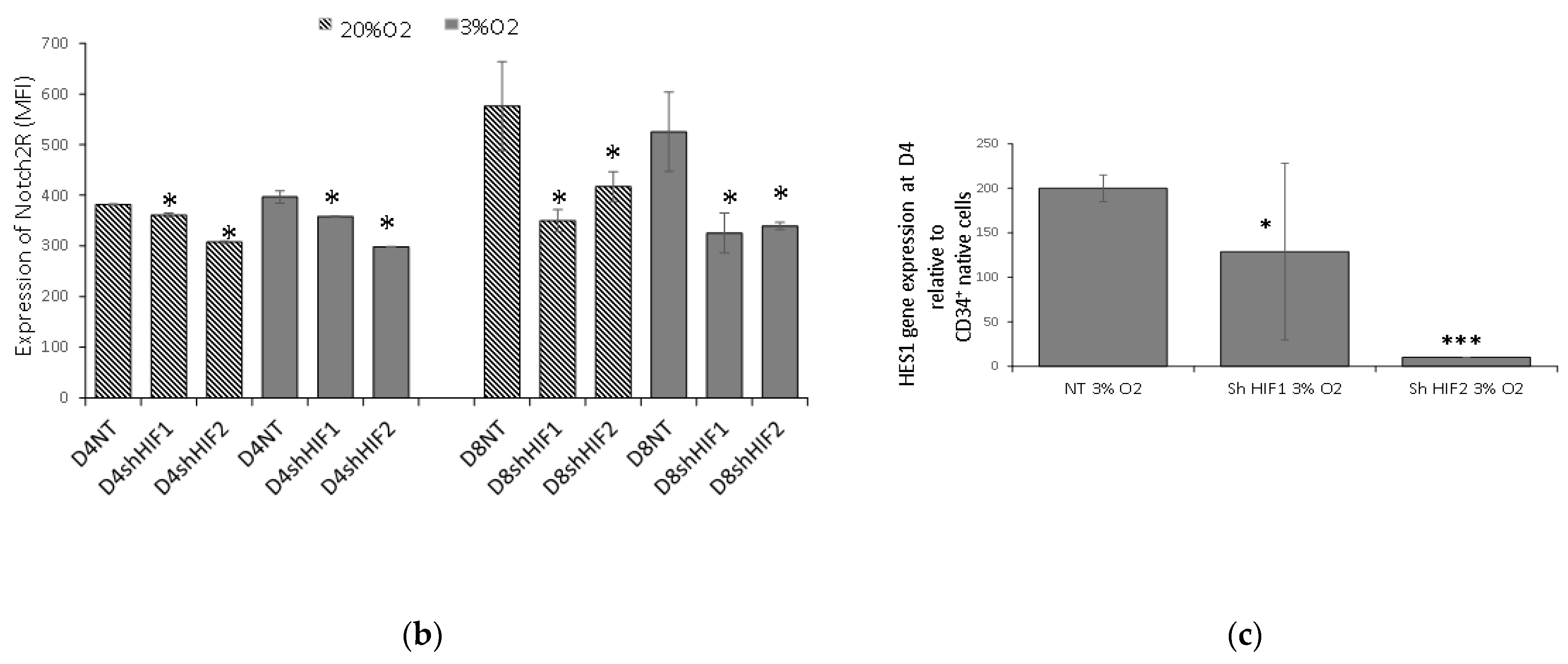
3.4. Notch Signaling Is Modified When HIF1α and HIF2α Are Silenced
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ivanovic, Z. Hypoxia or in situ normoxia: The stem cell paradigm. J. Cell. Physiol. 2009, 219, 271–275. [Google Scholar] [CrossRef] [PubMed]
- Vlaski, M.; Lafarge, X.; Chevaleyre, J.; Duchez, P.; Boiron, J.-M.; Ivanovic, Z. Low oxygen concentration as a general physiologic regulator of erythropoiesis beyond the EPO-related downstream tuning and a tool for the optimization of red blood cell production ex vivo. Exp. Hematol. 2009, 37, 573–584. [Google Scholar] [CrossRef] [PubMed]
- Bradley, T.R.; Hodgson, G.S.; Rosendaal, M. The effect of oxygen tension on haemopoietic and fibroblast cell proliferation in vitro. J. Cell. Physiol. 1978, 97, 517–522. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Broxmeyer, H.E. Comparative influences of phytohemagglutinin-stimulated leukocyte conditioned medium, hemin, prostaglandin E, and low oxygen tension on colony formation by erythroid progenitor cells in normal human bone marrow. Exp. Hematol. 1985, 13, 989–993. [Google Scholar]
- Koller, M.R.; Bender, J.G.; Papoutsakis, E.T.; Miller, W.M. Effects of synergistic cytokine combinations, low oxygen, and irradiated stroma on the expansion of human cord blood progenitors. Blood 1992, 80, 403–411. [Google Scholar] [CrossRef]
- Ivanović, Z.; Bartolozzi, B.; Bernabei, P.A.; Cipolleschi, M.G.; Rovida, E.; Milenković, P.; Praloran, V.; Sbarba, P.D. Incubation of murine bone marrow cells in hypoxia ensures the maintenance of marrow-repopulating ability together with the expansion of committed progenitors. Br. J. Haematol. 2000, 108, 424–429. [Google Scholar] [CrossRef]
- Cipolleschi, M.G.; D’Ippolito, G.; Bernabei, P.A.; Caporale, R.; Nannini, R.; Mariani, M.; Fabbiani, M.; Rossi-Ferrini, P.; Olivotto, M.; Sbarba, P.D. Severe hypoxia enhances the formation of erythroid bursts from human cord blood cells and the maintenance of BFU-E in vitro. Exp. Hematol. 1997, 25, 1187–1194. [Google Scholar]
- Rich, I.N.; Kubanek, B. The effect of reduced oxygen tension on colony formation of erythropoietic cellsin vitro. Br. J. Haematol. 1982, 52, 579–588. [Google Scholar] [CrossRef]
- Pennathur-Das, R.; Levitt, L. Augmentation of in vitro human marrow erythropoiesis under physiological oxygen tensions is mediated by monocytes and T lymphocytes. Blood 1987, 69, 899–907. [Google Scholar] [CrossRef]
- Schofield, C.; Ratcliffe, P. Oxygen sensing by HIF hydroxylases. Nat. Rev. Mol. Cell Biol. 2004, 5, 343–354. [Google Scholar] [CrossRef]
- Pedersen, M.; Löfstedt, T.; Sun, J.; Holmquist-Mengelbier, L.; Påhlman, S.; Rönnstrand, L. Stem cell factor induces HIF-1α at normoxia in hematopoietic cells. Biochem. Biophys. Res. Commun. 2008, 377, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Semenza, G.L. HIF-1 and mechanisms of hypoxia sensing. Curr. Opin. Cell Biol. 2001, 13, 167–171. [Google Scholar] [CrossRef]
- Landor, S.K.-J.; Lendahl, U. The interplay between the cellular hypoxic response and Notch signaling. Exp. Cell Res. 2017, 356, 146–151. [Google Scholar] [CrossRef] [PubMed]
- Depping, R.; Jelkmann, W.; Kosyna, F.K. Nuclear-cytoplasmatic shuttling of proteins in control of cellular oxygen sensing. Klin. Wochenschr. 2015, 93, 599–608. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.C.; Shibuya, M. A variant of nuclear localization signal of bipartite-type is required for the nuclear translocation of hypoxia inducible factors (1α, 2α and 3α). Oncogene 2001, 20, 1435–1444. [Google Scholar] [CrossRef]
- Kojika, S.; Griffin, J.D. Notch receptors and hematopoiesis. Exp. Hematol. 2001, 29, 1041–1052. [Google Scholar] [CrossRef]
- Duncan, A.W.; Rattis, F.M.; Dimascio, L.N.; Congdon, K.L.; Pazianos, G.; Zhao, C.; Yoon, K.; Cook, J.M.; Willert, K.; Gaiano, N.; et al. Integration of Notch and Wnt signaling in hematopoietic stem cell maintenance. Nat. Immunol. 2005, 6, 314–322. [Google Scholar] [CrossRef]
- Araki, D.; Fu, J.F.; Huntsman, H.; Cordes, S.; Seifuddin, F.; Alvarado, L.J.; Cheruku, P.S.; Cash, A.; Traba, J.; Li, Y.; et al. NOTCH-mediated ex vivo expansion of human hematopoietic stem and progenitor cells by culture under hypoxia. Stem Cell Rep. 2021, 16, 2336–2350. [Google Scholar] [CrossRef]
- Sugimoto, A.; Yamamoto, M.; Suzuki, M.; Inoue, T.; Nakamura, S.; Motoda, R.; Yamasaki, F.; Orita, K. Delta-4 Notch ligand promotes erythroid differentiation of human umbilical cord blood CD34+ cells. Exp. Hematol. 2006, 34, 424–432. [Google Scholar] [CrossRef]
- Dando, J.S.; Tavian, M.; Catelain, C.; Poirault, S.; Bennaceur-Griscelli, A.; Sainteny, F.; Vainchenker, W.; Péault, B.; Lauret, E. Notch/Delta4 Interaction in Human Embryonic Liver CD34+ CD38− Cells: Positive Influence on BFU-E Production and LTC-IC Potential Maintenance. Stem Cells 2005, 23, 550–560. [Google Scholar] [CrossRef]
- Walker, L.; Lynch, M.; Silverman, S.; Fraser, J.; Boulter, J.; Weinmaster, G.; Gasson, J.C. The Notch/Jagged Pathway Inhibits Proliferation of Human Hematopoietic Progenitors In Vitro. Stem Cells 1999, 17, 162–171. [Google Scholar] [CrossRef] [PubMed]
- Tachikawa, Y.; Matsushima, T.; Abe, Y.; Sakano, S.; Yamamoto, M.; Nishimura, J.; Nawata, H.; Takayanagi, R.; Muta, K. Pivotal role of Notch signaling in regulation of erythroid maturation and proliferation. Eur. J. Haematol. 2006, 77, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Diez, H.; Fischer, A.; Winkler, A.; Hu, C.-J.; Hatzopoulos, A.K.; Breier, G.; Gessler, M. Hypoxia-mediated activation of Dll4-Notch-Hey2 signaling in endothelial progenitor cells and adoption of arterial cell fate. Exp. Cell Res. 2007, 313, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Oh, P.; Lobry, C.; Gao, J.; Tikhonova, A.; Loizou, E.; Manent, J.; van Handel, B.; Ibrahim, S.; Greve, J.; Mikkola, H.; et al. In Vivo Mapping of Notch Pathway Activity in Normal and Stress Hematopoiesis. Cell Stem Cell 2013, 13, 190–204. [Google Scholar] [CrossRef]
- Walker, L.; Carlson, A.; Tan-Pertel, H.T.; Weinmaster, G.; Gasson, J. The Notch Receptor and Its Ligands Are Selectively Expressed During Hematopoietic Development in the Mouse. Stem Cells 2001, 19, 543–552. [Google Scholar] [CrossRef]
- Ivanovic, Z.; Hermitte, F.; de la Grange, P.B.; Dazey, B.; Belloc, F.; Lacombe, F.; Vezon, G.; Praloran, V. Simultaneous Maintenance of Human Cord Blood SCID-Repopulating Cells and Expansion of Committed Progenitors at Low O2 Concentration (3%). Stem Cells 2004, 22, 716–724. [Google Scholar] [CrossRef]
- Rouault-Pierre, K.; Onieva, L.L.; Foster, K.; Afonso, F.D.A.; Lamrissi-Garcia, I.; Sanchez, M.S.; Mitter, R.; Ivanovic, Z.; de Verneuil, H.; Gribben, J.; et al. HIF-2α Protects Human Hematopoietic Stem/Progenitors and Acute Myeloid Leukemic Cells from Apoptosis Induced by Endoplasmic Reticulum Stress. Cell Stem Cell 2013, 13, 549–563. [Google Scholar] [CrossRef]
- Giarratana, M.-C.; Kobari, L.; Lapillonne, H.; Chalmers, D.J.; Kiger, L.; Cynober, T.; Marden, M.C.; Wajcman, H.; Douay, L. Ex vivo generation of fully mature human red blood cells from hematopoietic stem cells. Nat. Biotechnol. 2005, 23, 69–74. [Google Scholar] [CrossRef]
- Bony, V.; Gane, P.; Bailly, P.; Cartron, J.-P. Time-course expression of polypeptides carrying blood group antigens during human erythroid differentiation. Br. J. Haematol. 1999, 107, 263–274. [Google Scholar] [CrossRef]
- Guitart, A.V.; Debeissat, C.; Hermitte, F.; Villacreces, A.; Ivanovic, Z.; Boeuf, H.; Praloran, V. Very low oxygen concentration (0.1%) reveals two FDCP-Mix cell subpopulations that differ by their cell cycling, differentiation and p27KIP1 expression. Cell Death Differ. 2010, 18, 174–182. [Google Scholar] [CrossRef]
- Crowley, L.C.; Scott, A.P.; Marfell, B.J.; Boughaba, J.A.; Chojnowski, G.; Waterhouse, N.J. Measuring Cell Death by Propidium Iodide Uptake and Flow Cytometry. Cold Spring Harb. Protoc. 2016, 7, pdb-prot087163. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Phillips, R.A.; Iscove, N.N.; Egan, S.E. Expression of notch receptors, notch ligands, and fringe genes in hematopoiesis. Exp. Hematol. 2000, 28, 527–534. [Google Scholar] [CrossRef][Green Version]
- Zeuner, A.; Francescangeli, F.; Signore, M.; Venneri, M.A.; Pedini, F.; Felli, N.; Pagliuca, A.; Conticello, C.; De Maria, R. The Notch2–Jagged1 interaction mediates stem cell factor signaling in erythropoiesis. Cell Death Differ. 2010, 18, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Hattangadi, S.M.; Wong, P.; Zhang, L.; Flygare, J.; Lodish, H.F. From stem cell to red cell: Regulation of erythropoiesis at multiple levels by multiple proteins, RNAs, and chromatin modifications. Blood 2011, 118, 6258–6268. [Google Scholar] [CrossRef] [PubMed]
- Scortegagna, M.; Morris, M.A.; Oktay, Y.; Bennett, M.; Garcia, J.A. The HIF family member EPAS1/HIF-2α is required for normal hematopoiesis in mice. Blood 2003, 102, 1634–1640. [Google Scholar] [CrossRef] [PubMed]
- Gruber, M.; Hu, C.-J.; Johnson, R.S.; Brown, E.J.; Keith, B.; Simon, M.C. Acute postnatal ablation of Hif-2 α results in anemia. Proc. Natl. Acad. Sci. USA 2007, 104, 2301–2306. [Google Scholar] [CrossRef]
- Rankin, E.; Biju, M.P.; Liu, Q.; Unger, T.L.; Rha, J.; Johnson, R.; Simon, M.C.; Keith, B.; Haase, V.H. Hypoxia-inducible factor–2 (HIF-2) regulates hepatic erythropoietin in vivo. J. Clin. Investig. 2007, 117, 1068–1077. [Google Scholar] [CrossRef]
- Warnecke, C.; Zaborowska, Z.; Kurreck, J.; Erdmann, V.A.; Frei, U.; Wiesener, M.; Eckardt, K. Differentiating the functional role of hypoxia-inducible factor (HIF)-1α and HIF-2α (EPAS-1) by the use of RNA interference: Erythropoietin is a HIF-2α target gene in Hep3B and Kelly cells. FASEB J. 2004, 18, 1462–1464. [Google Scholar] [CrossRef]
- Ratcliffe, P.J. HIF-1 and HIF-2: Working alone or together in hypoxia? J. Clin. Investig. 2007, 117, 862–865. [Google Scholar] [CrossRef]
- Gustafsson, M.V.; Zheng, X.; Pereira, T.; Gradin, K.; Jin, S.; Lundkvist, J.; Ruas, J.; Poellinger, L.; Lendahl, U.; Bondesson, M. Hypoxia Requires Notch Signaling to Maintain the Undifferentiated Cell State. Dev. Cell 2005, 9, 617–628. [Google Scholar] [CrossRef]
- Henning, K.; Heering, J.; Schwanbeck, R.; Schroeder, T.; Helmbold, H.; Schäfer, H.; Deppert, W.; Kim, E.; Just, U. Notch1 activation reduces proliferation in the multipotent hematopoietic progenitor cell line FDCP-mix through a p53-dependent pathway but Notch1 effects on myeloid and erythroid differentiation are independent of p53. Cell Death Differ. 2007, 15, 398–407. [Google Scholar] [CrossRef] [PubMed]
- Myllymäki, M.N.M.; Määttä, J.; Dimova, E.Y.; Izzi, V.; Väisänen, T.; Myllyharju, J.; Koivunen, P.; Serpi, R. Notch Downregulation and Extramedullary Erythrocytosis in Hypoxia-Inducible Factor Prolyl 4-Hydroxylase 2-Deficient Mice. Mol. Cell. Biol. 2017, 37, e00529-16. [Google Scholar] [CrossRef] [PubMed]
- Bray, S. Notch signalling in context. Nat. Rev. Mol. Cell Biol. 2016, 17, 722–735. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Linke, S.; Dias, J.M.; Zheng, X.; Gradin, K.; Wallis, T.P.; Hamilton, B.R.; Gustafsson, M.; Ruas, J.L.; Wilkins, S.; et al. Interaction with factor inhibiting HIF-1 defines an additional mode of cross-coupling between the Notch and hypoxia signaling pathways. Proc. Natl. Acad. Sci. USA 2008, 105, 3368–3373. [Google Scholar] [CrossRef]
- Main, H.; Lee, K.L.; Yang, H.; Haapa-Paananen, S.; Edgren, H.; Jin, S.; Sahlgren, C.; Kallioniemi, O.; Poellinger, L.; Lim, B.; et al. Interactions between Notch- and hypoxia-induced transcriptomes in embryonic stem cells. Exp. Cell Res. 2010, 316, 1610–1624. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Labat, V.; Bayard, E.N.v.T.d.; Refeyton, A.; Huart, M.; Avalon, M.; Debeissat, C.; Rodriguez, L.; de la Grange, P.B.; Ivanovic, Z.; Vlaski-Lafarge, M. Regulatory Crosstalk between Physiological Low O2 Concentration and Notch Pathway in Early Erythropoiesis. Biomolecules 2022, 12, 540. https://doi.org/10.3390/biom12040540
Labat V, Bayard ENvTd, Refeyton A, Huart M, Avalon M, Debeissat C, Rodriguez L, de la Grange PB, Ivanovic Z, Vlaski-Lafarge M. Regulatory Crosstalk between Physiological Low O2 Concentration and Notch Pathway in Early Erythropoiesis. Biomolecules. 2022; 12(4):540. https://doi.org/10.3390/biom12040540
Chicago/Turabian StyleLabat, Véronique, Eva Nguyen van Thanh dit Bayard, Alice Refeyton, Mathilde Huart, Maryse Avalon, Christelle Debeissat, Laura Rodriguez, Philippe Brunet de la Grange, Zoran Ivanovic, and Marija Vlaski-Lafarge. 2022. "Regulatory Crosstalk between Physiological Low O2 Concentration and Notch Pathway in Early Erythropoiesis" Biomolecules 12, no. 4: 540. https://doi.org/10.3390/biom12040540
APA StyleLabat, V., Bayard, E. N. v. T. d., Refeyton, A., Huart, M., Avalon, M., Debeissat, C., Rodriguez, L., de la Grange, P. B., Ivanovic, Z., & Vlaski-Lafarge, M. (2022). Regulatory Crosstalk between Physiological Low O2 Concentration and Notch Pathway in Early Erythropoiesis. Biomolecules, 12(4), 540. https://doi.org/10.3390/biom12040540






