Transcriptomic Biomarker Signatures for Discrimination of Oral Cancer Surgical Margins
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Group and Sample Collection
2.2. Gene Expression Profiling and Bioinformatics
2.3. Sample Classification and Feature Selection
2.4. External Validation Dataset
3. Results
3.1. Differential Gene Expression Profiling
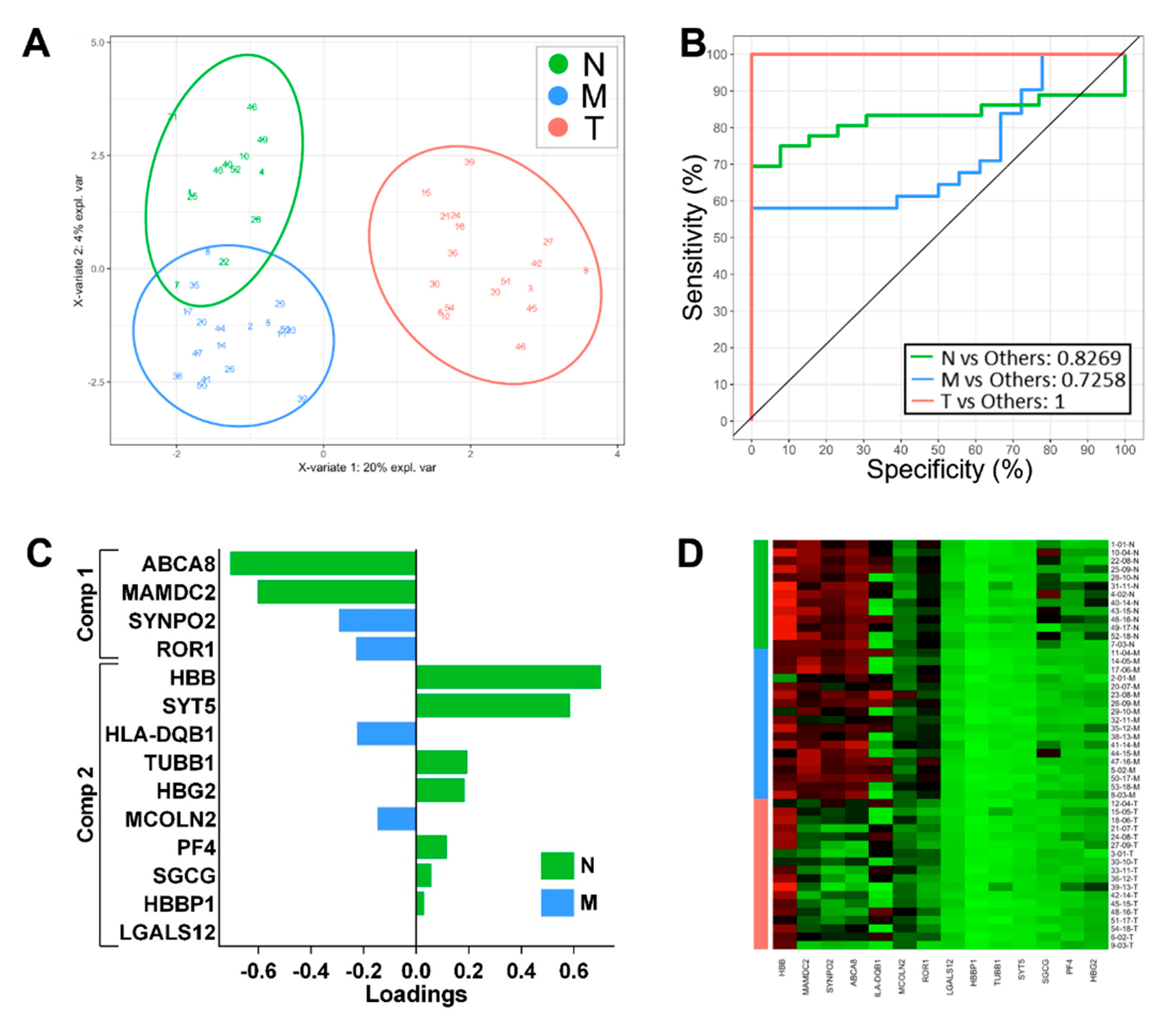
3.2. Identification of Classifying Biomarkers for Tumour, Margin and Normal Tissues
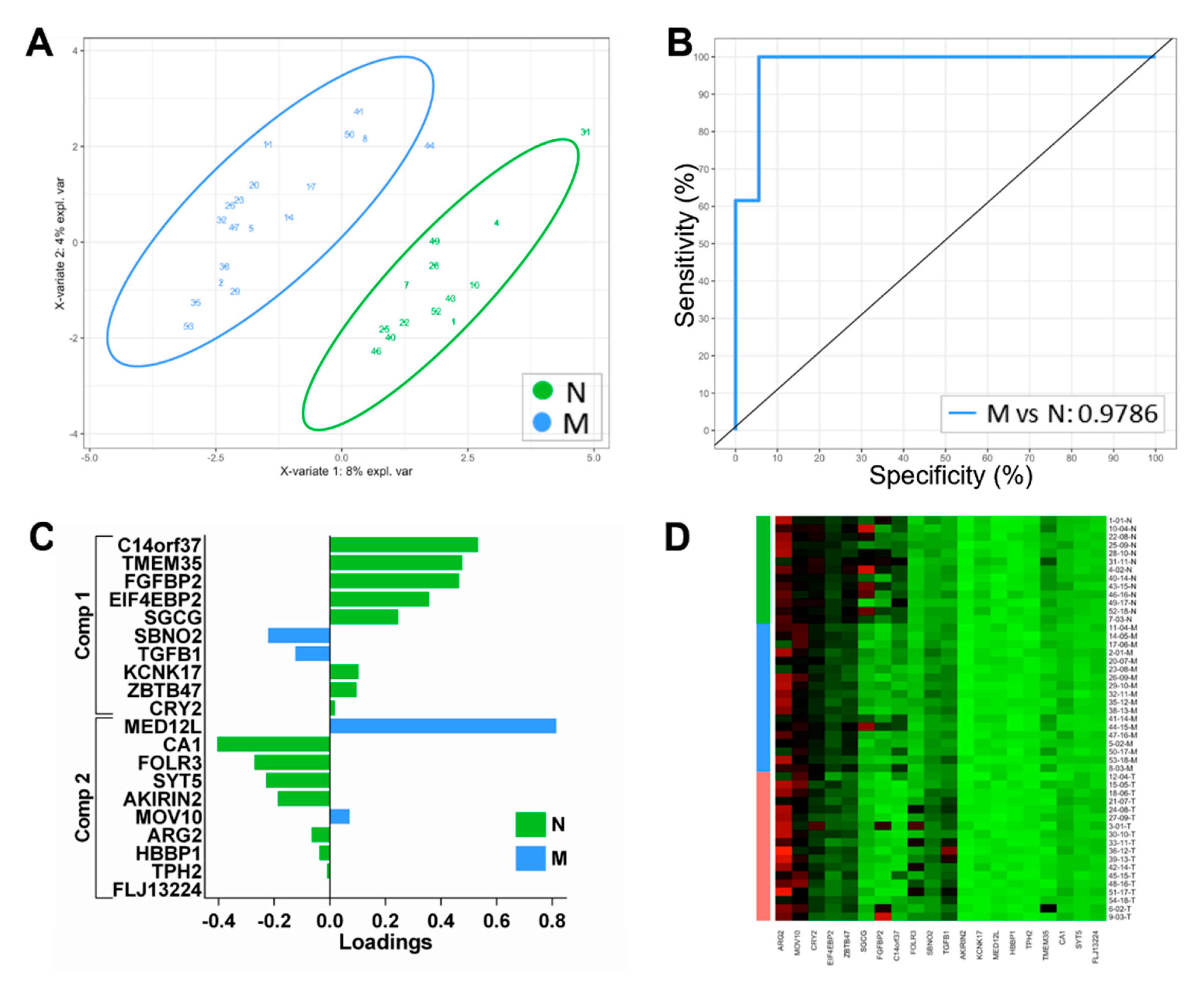
3.3. Development of a Biomarker Signature for Classification of Tumour Margins
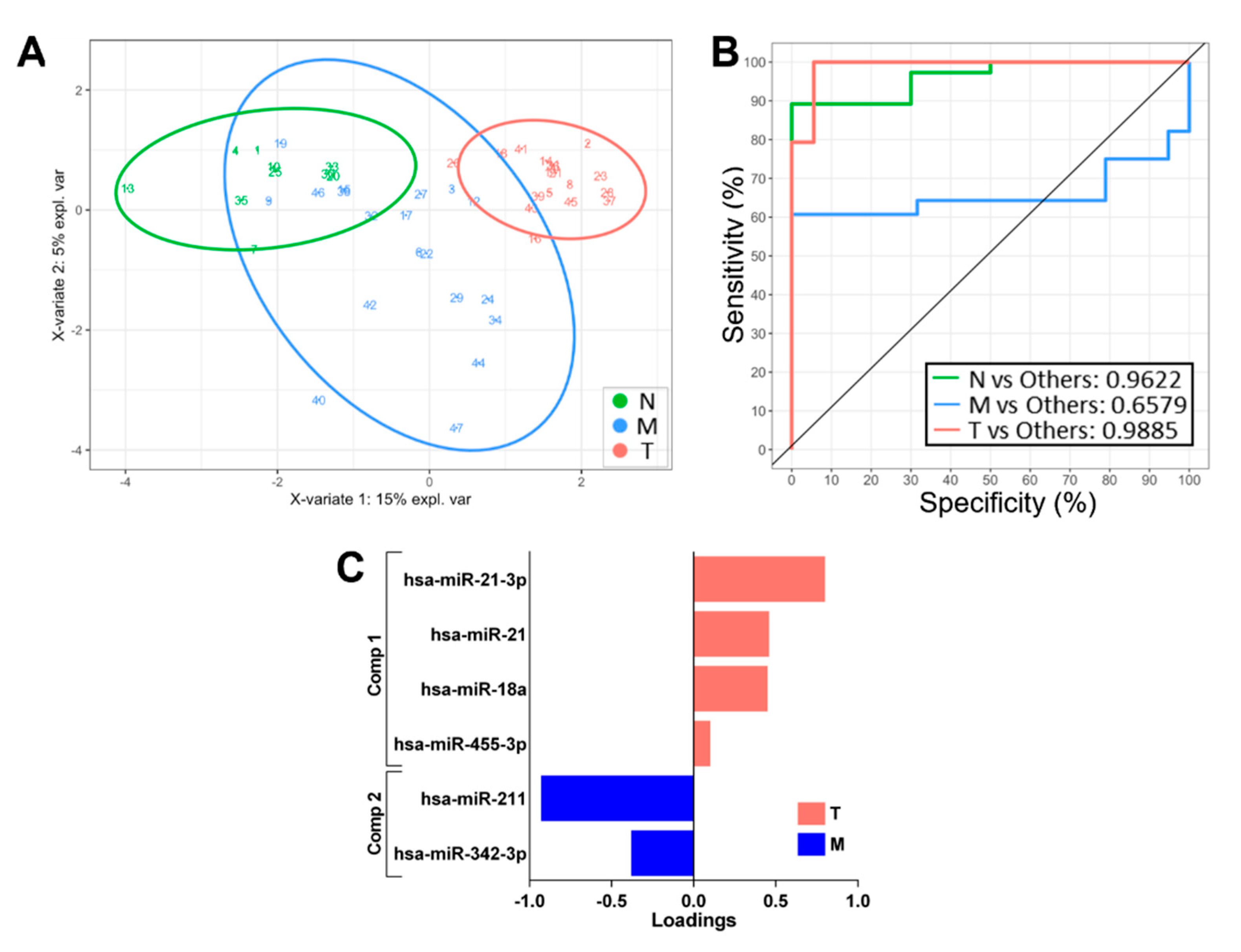
3.4. Validation in External Datasets
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gupta, B.; Johnson, N.W.; Kumar, N. Global Epidemiology of Head and Neck Cancers: A Continuing Challenge. Oncology 2016, 91, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Cronin, K.A.; Lake, A.J.; Scott, S.; Sherman, R.L.; Noone, A.M.; Howlader, N.; Henley, S.J.; Anderson, R.N.; Firth, A.U.; Ma, J.; et al. Annual Report to the Nation on the Status of Cancer, Part I: National Cancer Statistics. Cancer 2018, 124, 2785–2800. [Google Scholar] [CrossRef] [PubMed]
- Gatta, G.; Botta, L.; Sánchez, M.J.; Anderson, L.A.; Pierannunzio, D.; Licitra, L.; on behalf of the EUROCARE Working Group. Prognoses and Improvement for Head and Neck Cancers Diagnosed in Europe in Early 2000s: The EUROCARE-5 Population-Based Study. Eur. J. Cancer 2015, 51, 2130–2143. [Google Scholar] [CrossRef]
- Binahmed, A.; Nason, R.W.; Abdoh, A.A. The Clinical Significance of the Positive Surgical Margin in Oral Cancer. Oral Oncol. 2007, 43, 780–784. [Google Scholar] [CrossRef] [PubMed]
- González-García, R.; Naval-Gías, L.; Román-Romero, L.; Sastre-Pérez, J.; Rodríguez-Campo, F.J. Local Recurrences and Second Primary Tumors from Squamous Cell Carcinoma of the Oral Cavity: A Retrospective Analytic Study of 500 Patients. Head Neck 2009, 31, 1168–1180. [Google Scholar] [CrossRef]
- Braakhuis, B.J.M.; Bloemena, E.; Leemans, C.R.; Brakenhoff, R.H. Molecular Analysis of Surgical Margins in Head and Neck Cancer: More than a Marginal Issue. Oral Oncol. 2010, 46, 485–491. [Google Scholar] [CrossRef]
- Wood, H.M.; Daly, C.; Chalkley, R.; Senguven, B.; Ross, L.; Egan, P.; Chengot, P.; Graham, J.; Sethi, N.; Ong, T.K.; et al. The Genomic Road to Invasion-Examining the Similarities and Differences in the Genomes of Associated Oral Pre-Cancer and Cancer Samples. Genome Med. 2017, 9, 53. [Google Scholar] [CrossRef]
- McDermott, J.E.; Wang, J.; Mitchell, H.; Webb-Robertson, B.J.; Hafen, R.; Ramey, J.; Rodland, K.D. Challenges in Biomarker Discovery: Combining Expert Insights with Statistical Analysis of Complex Omics Data. Expert Opin. Med. Diagn. 2013, 7, 37–51. [Google Scholar] [CrossRef]
- Szymańska, E.; Saccenti, E.; Smilde, A.K.; Westerhuis, J.A. Double-Check: Validation of Diagnostic Statistics for PLS-DA Models in Metabolomics Studies. Metab. Off. J. Metab. Soc. 2012, 8, 3–16. [Google Scholar] [CrossRef]
- Christin, C.; Hoefsloot, H.C.J.; Smilde, A.K.; Hoekman, B.; Suits, F.; Bischoff, R.; Horvatovich, P. A Critical Assessment of Feature Selection Methods for Biomarker Discovery in Clinical Proteomics. Mol. Cell. Proteom. 2013, 12, 263–276. [Google Scholar] [CrossRef]
- Boulesteix, A.L.; Strimmer, K. Partial Least Squares: A Versatile Tool for the Analysis of High-Dimensional Genomic Data. Brief. Bioinform. 2007, 8, 32–44. [Google Scholar] [CrossRef] [PubMed]
- Lê Cao, K.A.; Boitard, S.; Besse, P. Sparse PLS Discriminant Analysis: Biologically Relevant Feature Selection and Graphical Displays for Multiclass Problems. BMC Bioinform. 2011, 12, 253. [Google Scholar] [CrossRef] [PubMed]
- Farah, C.S.; Dalley, A.J.; Nguyen, P.; Batstone, M.; Kordbacheh, F.; Perry-Keene, J.; Fielding, D. Improved Surgical Margin Definition by Narrow Band Imaging for Resection of Oral Squamous Cell Carcinoma: A Prospective Gene Expression Profiling Study. Head Neck 2016, 38, 832–839. [Google Scholar] [CrossRef] [PubMed]
- Farah, C.S.; Fox, S.A.; Dalley, A.J. Integrated MiRNA-MRNA Spatial Signature for Oral Squamous Cell Carcinoma: A Prospective Profiling Study of Narrow Band Imaging Guided Resection. Sci. Rep. 2018, 8, 823. [Google Scholar] [CrossRef] [PubMed]
- Farah, C.S. Narrow Band Imaging-Guided Resection of Oral Cavity Cancer Decreases Local Recurrence and Increases Survival. Oral Dis. 2018, 24, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Irizarry, R.A.; Gentleman, R.; Martinez-Murillo, F.; Spencer, F. A Model-Based Background Adjustment for Oligonucleotide Expression Arrays. J. Am. Stat. Assoc. 2004, 99, 909–917. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and Integrative Analysis of Large Gene Lists Using DAVID Bioinformatics Resources. Nat. Protoc. Lond. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Rouillard, A.D.; Gundersen, G.W.; Fernandez, N.F.; Wang, Z.; Monteiro, C.D.; McDermott, M.G.; Ma’ayan, A. The Harmonizome: A Collection of Processed Datasets Gathered to Serve and Mine Knowledge about Genes and Proteins. Database 2016, 2016. [Google Scholar] [CrossRef]
- Rohart, F.; Gautier, B.; Singh, A.; Cao, K.A.L. MixOmics: An R Package for ‘omics Feature Selection and Multiple Data Integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef]
- Reis, P.P.; Waldron, L.; Perez-Ordonez, B.; Pintilie, M.; Galloni, N.N.; Xuan, Y.; Cervigne, N.K.; Warner, G.C.; Makitie, A.A.; Simpson, C.; et al. A Gene Signature in Histologically Normal Surgical Margins Is Predictive of Oral Carcinoma Recurrence. BMC Cancer 2011, 11, 437. [Google Scholar] [CrossRef]
- Zou, H.; Hastie, T. Regularization and Variable Selection via the Elastic Net. J. R. Stat. Soc. Ser. B Stat. Methodol. 2005, 67, 301–320. [Google Scholar] [CrossRef]
- Lohavanichbutr, P.; Méndez, E.; Holsinger, F.C.; Rue, T.C.; Zhang, Y.; Houck, J.; Upton, M.P.; Futran, N.; Schwartz, S.M.; Wang, P.; et al. A 13-Gene Signature Prognostic of HPV-Negative OSCC: Discovery and External Validation. Clin. Cancer Res. 2013, 19, 1197–1203. [Google Scholar] [CrossRef] [PubMed]
- Ziober, A.F.; Patel, K.R.; Alawi, F.; Gimotty, P.; Weber, R.S.; Feldman, M.M.; Chalian, A.A.; Weinstein, G.S.; Hunt, J.; Ziober, B.L. Identification of a Gene Signature for Rapid Screening of Oral Squamous Cell Carcinoma. Clin. Cancer Res. 2006, 12, 5960–5971. [Google Scholar] [CrossRef] [PubMed]
- Anderson, C.R.; Sisson, K.; Moncrieff, M. A Meta-Analysis of Margin Size and Local Recurrence in Oral Squamous Cell Carcinoma. Oral Oncol. 2015, 51, 464–469. [Google Scholar] [CrossRef]
- Pierik, A.S.; Leemans, C.R.; Brakenhoff, R.H. Resection Margins in Head and Neck Cancer Surgery: An Update of Residual Disease and Field Cancerization. Cancers 2021, 13, 2635. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Park, B.C.; Soon Kang, J.; Cheon, Y.; Lee, S.; Jae Maeng, P. MAM Domain Containing 2 Is a Potential Breast Cancer Biomarker That Exhibits Tumour-Suppressive Activity. Cell Prolif. 2020, 53, e12883. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.S.; Quadery Tonmoy, M.I.; Islam, M.N.; Islam, M.S.; Afif, I.K.; Singha Roy, A.; Fariha, A.; Al Reza, H.; Bahadur, N.M.; Rahaman, M.M. MicroRNAs Expression Analysis Shows Key Affirmation of Synaptopodin-2 as a Novel Prognostic and Therapeutic Biomarker for Colorectal and Cervical Cancers. Heliyon 2021, 7, e07347. [Google Scholar] [CrossRef]
- Ye, H.; Yu, T.; Temam, S.; Ziober, B.L.; Wang, J.; Schwartz, J.L.; Mao, L.; Wong, D.T.; Zhou, X. Transcriptomic Dissection of Tongue Squamous Cell Carcinoma. BMC Genom. 2008, 9, 69. [Google Scholar] [CrossRef]
- Menck, K.; Heinrichs, S.; Baden, C.; Bleckmann, A. The WNT/ROR Pathway in Cancer: From Signaling to Therapeutic Intervention. Cells 2021, 10, 142. [Google Scholar] [CrossRef] [PubMed]
- Nema, S.; Kallianpur, S.; Kumar, A.; Nema, R.; Vishwakarma, S.; Nema, S.K. Do Patients with Oral Squamous Cell Carcinoma Express Receptor Tyrosine Kinase-like Orphan Receptor 1? Results of an Observational Study. J. Oral Maxillofac. Pathol. 2021, 25, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Balakrishnan, A.; Goodpaster, T.; Randolph-Habecker, J.; Hoffstrom, B.G.; Jalikis, F.G.; Koch, L.K.; Berger, C.; Kosasih, P.L.; Rajan, A.; Sommermeyer, D.; et al. Analysis of ROR1 Protein Expression in Human Cancer and Normal Tissues. Clin. Cancer Res. 2017, 23, 3061–3071. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Wang, Y.H.; Kalaitzidis, D.; Ramachandran, J.; Eda, H.; Sykes, D.B.; Raje, N.; Scadden, D.T. Endogenous Transmembrane Protein UT2 Inhibits PSTAT3 and Suppresses Hematological Malignancy. J. Clin. Investig. 2016, 126, 1300–1310. [Google Scholar] [CrossRef] [PubMed]
- Alonso, S.; Mayol, X.; Nonell, L.; Salvans, S.; Pascual, M.; Pera, M.; on behalf of the Colorectal Cancer Research Group (Hospital del Mar Medical Research Institute). Peripheral Blood Leucocytes Show Differential Expression of Tumour Progression-Related Genes in Colorectal Cancer Patients Who Have a Postoperative Intra-Abdominal Infection: A Prospective Matched Cohort Study. Colorectal Dis. 2017, 19, O115–O125. [Google Scholar] [CrossRef] [PubMed]
- Galliera, E.; Tacchini, L.; Corsi Romanelli, M.M. Matrix Metalloproteinases as Biomarkers of Disease: Updates and New Insights. Clin. Chem. Lab. Med. 2015, 53, 349–355. [Google Scholar] [CrossRef] [PubMed]
- Kuriakose, M.A.; Chen, W.T.; He, Z.M.; Sikora, A.G.; Zhang, P.; Zhang, Z.Y.; Qiu, W.L.; Hsu, D.F.; McMunn-Coffran, C.; Brown, S.M.; et al. Selection and Validation of Differentially Expressed Genes in Head and Neck Cancer. Cell. Mol. Life Sci. 2004, 61, 1372–1383. [Google Scholar] [CrossRef] [PubMed]
- Ginos, M.A.; Page, G.P.; Michalowicz, B.S.; Patel, K.J.; Volker, S.E.; Pambuccian, S.E.; Ondrey, F.G.; Adams, G.L.; Gaffney, P.M. Identification of a Gene Expression Signature Associated with Recurrent Disease in Squamous Cell Carcinoma of the Head and Neck. Cancer Res. 2004, 64, 55–63. [Google Scholar] [CrossRef]
- Ohmura, G.; Tsujikawa, T.; Yaguchi, T.; Kawamura, N.; Mikami, S.; Sugiyama, J.; Nakamura, K.; Kobayashi, A.; Iwata, T.; Nakano, H.; et al. Aberrant Myosin 1b Expression Promotes Cell Migration and Lymph Node Metastasis of HNSCC. Mol. Cancer Res. 2015, 13, 721–731. [Google Scholar] [CrossRef]
- Yao, Y.; Yan, Z.; Lian, S.; Wei, L.; Zhou, C.; Feng, D.; Zhang, Y.; Yang, J.; Li, M.; Chen, Y. Prognostic Value of Novel Immune-Related Genomic Biomarkers Identified in Head and Neck Squamous Cell Carcinoma. J. Immunother. Cancer 2020, 8, e000444. [Google Scholar] [CrossRef]
- Li, Z.; Chen, C.; Wang, J.; Wei, M.; Liu, G.; Qin, Y.; She, L.; Liu, Y.; Huang, D.; Tian, Y.; et al. Overexpressed PLAU and Its Potential Prognostic Value in Head and Neck Squamous Cell Carcinoma. PeerJ 2021, 9, e10746. [Google Scholar] [CrossRef]
- Chen, G.; Sun, J.; Xie, M.; Yu, S.; Tang, Q.; Chen, L. PLAU Promotes Cell Proliferation and Epithelial-Mesenchymal Transition in Head and Neck Squamous Cell Carcinoma. Front. Genet. 2021, 12, 651882. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; He, S.; Li, Y.; Xu, Z. Identification of Therapeutic and Prognostic Biomarkers of Lamin C (LAMC) Family Members in Head and Neck Squamous Cell Carcinoma. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2020, 26, e925735. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.M.; Yao, Y.L.; Liu, W.; Shen, X.M.; Shi, L.J.; Wu, L. MicroRNA-134 Inhibits Tumor Stem Cell Migration and Invasion in Oral Squamous Cell Carcinomas via Downregulation of PI3K-Akt Signaling Pathway by Inhibiting LAMC2 Expression. Cancer Biomark. 2020, 29, 51–67. [Google Scholar] [CrossRef]
- Meireles Da Costa, N.; Mendes, F.A.; Pontes, B.; Nasciutti, L.E.; Ribeiro Pinto, L.F.; Palumbo Júnior, A. Potential Therapeutic Significance of Laminin in Head and Neck Squamous Carcinomas. Cancers 2021, 13, 1890. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Wang, M.; Wang, M.; Yao, L.; Li, X.; Dong, H.; Li, M.; Sun, T.; Liu, X.; Liu, Y.; et al. Role of Proton-Coupled Monocarboxylate Transporters in Cancer: From Metabolic Crosstalk to Therapeutic Potential. Front. Cell Dev. Biol. 2020, 8, 651. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Qin, X. Comprehensive Analysis of Transcriptome Data for Identifying Biomarkers and Therapeutic Targets in Head and Neck Squamous Cell Carcinoma. Ann. Transl. Med. 2020, 8, 282. [Google Scholar] [CrossRef]
- Khammanivong, A.; Sorenson, B.S.; Ross, K.F.; Dickerson, E.B.; Hasina, R.; Lingen, M.W.; Herzberg, M.C. Involvement of Calprotectin (S100A8/A9) in Molecular Pathways Associated with HNSCC. Oncotarget 2016, 7, 14029–14047. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, Q.; Ma, C.; Kemmner, W. Wdr66 Is a Novel Marker for Risk Stratification and Involved in Epithelial-Mesenchymal Transition of Esophageal Squamous Cell Carcinoma. BMC Cancer 2013, 13, 137. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Liu, H.; Xia, S.L.; Zhang, X.; Bai, H.; Yang, Q.; Li, J.; Gao, L.; Jin, F.; Wei, M.J.; et al. PTEN Downregulates WD Repeat-containing Protein 66 in Salivary Adenoid Cystic Carcinoma. Oncol. Rep. 2019, 41, 1827–1836. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fox, S.A.; Vacher, M.; Farah, C.S. Transcriptomic Biomarker Signatures for Discrimination of Oral Cancer Surgical Margins. Biomolecules 2022, 12, 464. https://doi.org/10.3390/biom12030464
Fox SA, Vacher M, Farah CS. Transcriptomic Biomarker Signatures for Discrimination of Oral Cancer Surgical Margins. Biomolecules. 2022; 12(3):464. https://doi.org/10.3390/biom12030464
Chicago/Turabian StyleFox, Simon A., Michael Vacher, and Camile S. Farah. 2022. "Transcriptomic Biomarker Signatures for Discrimination of Oral Cancer Surgical Margins" Biomolecules 12, no. 3: 464. https://doi.org/10.3390/biom12030464
APA StyleFox, S. A., Vacher, M., & Farah, C. S. (2022). Transcriptomic Biomarker Signatures for Discrimination of Oral Cancer Surgical Margins. Biomolecules, 12(3), 464. https://doi.org/10.3390/biom12030464






