Structure/Function Studies on the Activation Motif of Two Non-Mammalian Mrap1 Orthologs, and Observations on the Phylogeny of Mrap1, Including a Novel Characterization of an Mrap1 from the Chondrostean Fish, Polyodon spathula
Abstract
1. Introduction
2. Materials and Methods
2.1. Mc2r and Mrap1 Sequences
2.2. Melanocortin Peptides
2.3. cAMP Reporter Gene Assay
2.4. Statistical Analysis
3. Results
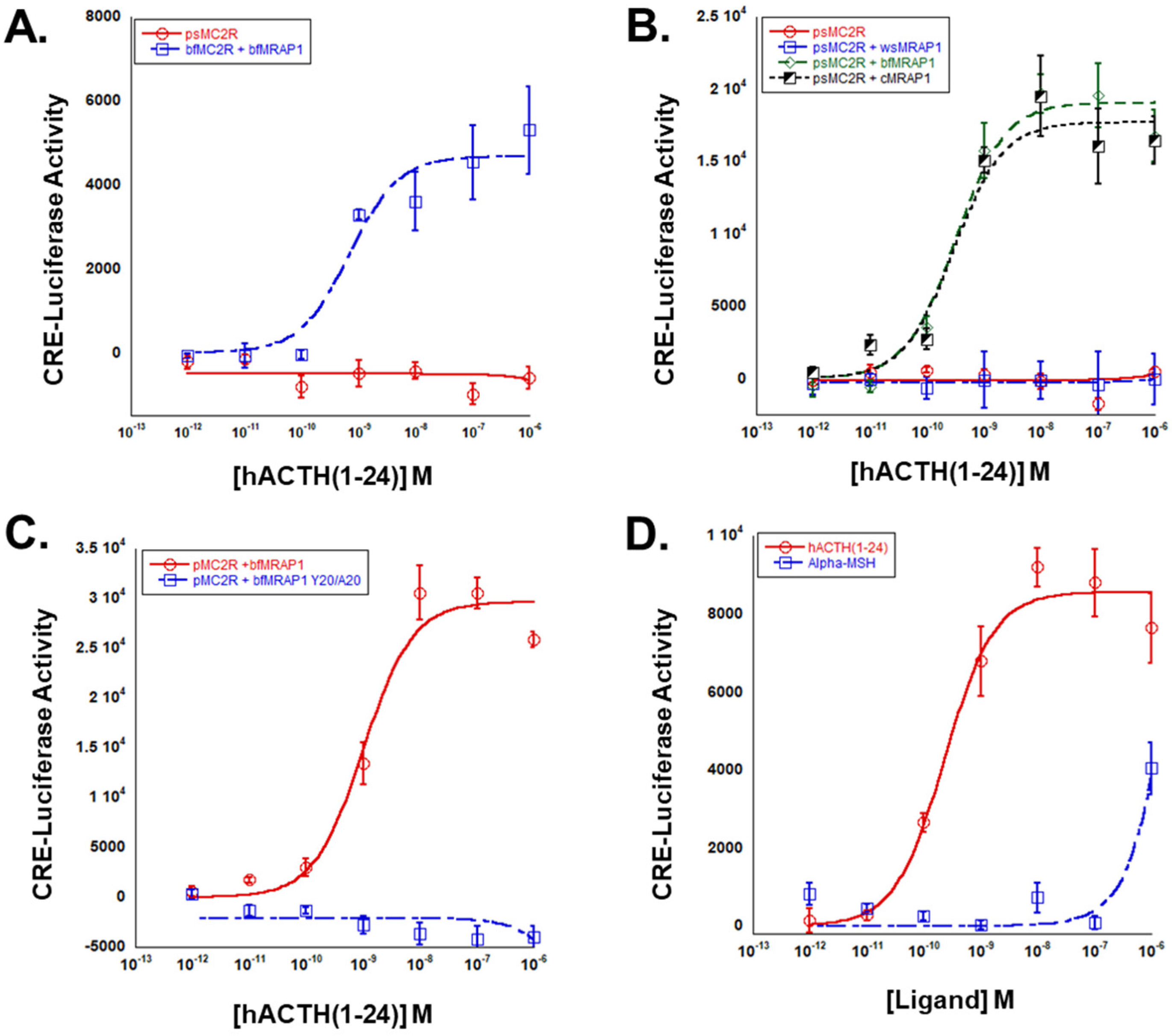
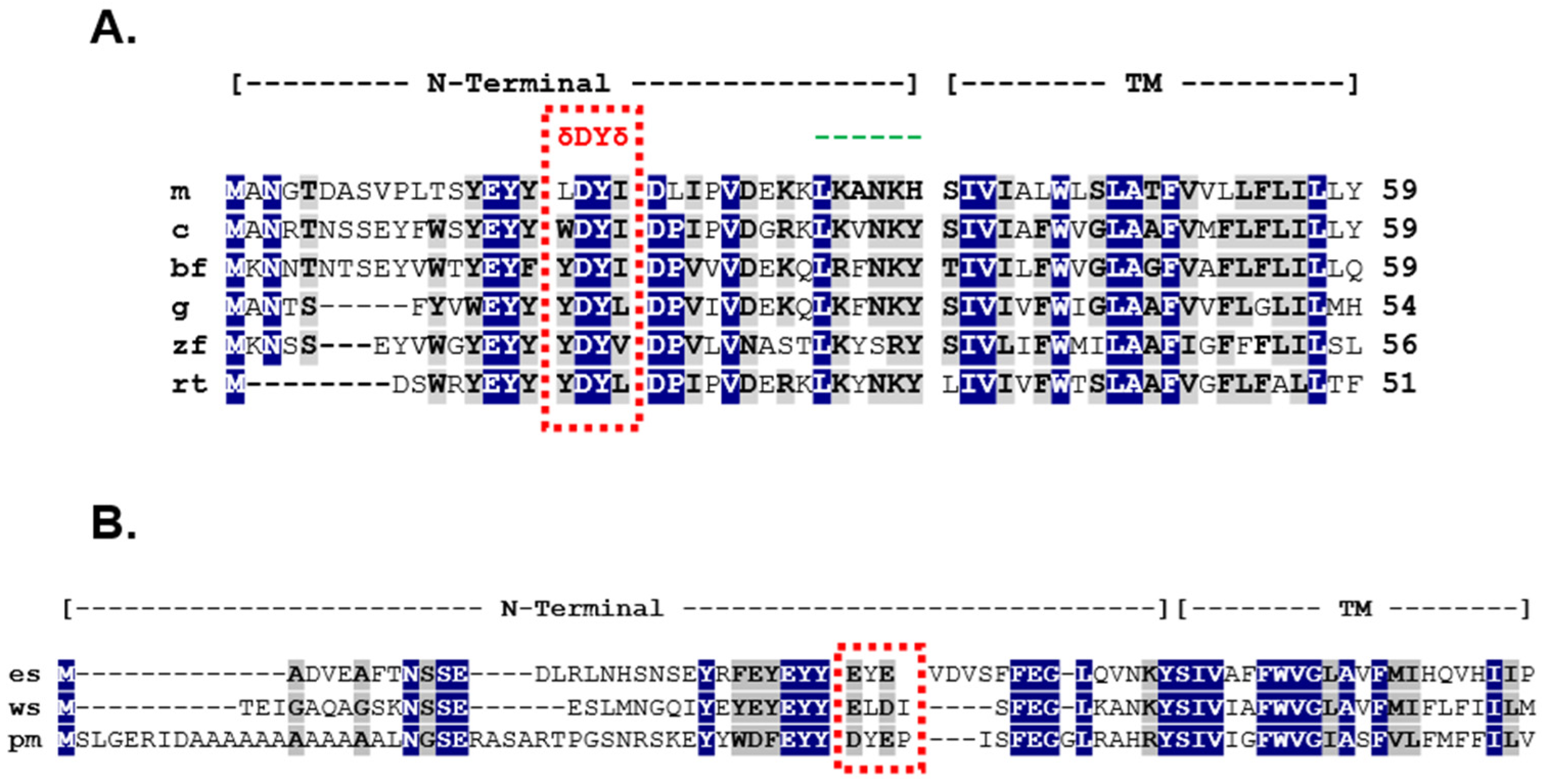
3.1. Analysis of the W18D19Y20I21 Motif in cMRAP1
3.2. Analysis of the Y18D19Y20I21 Motif in bfMrap1
3.3. Pharmacological Properties of Paddlefish Mc2r
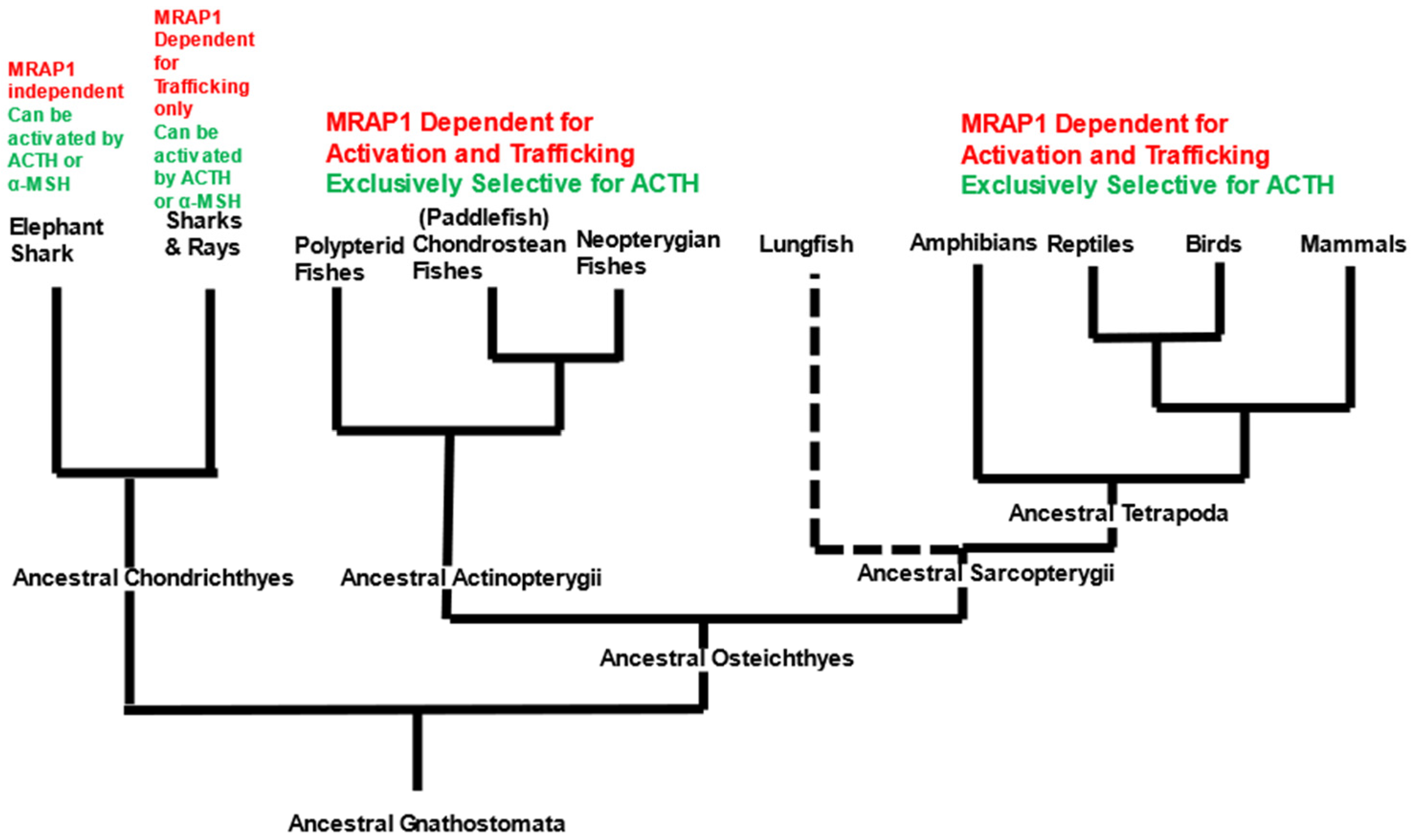
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cone, R.D. Studies on the physiological studies of the melanocortin system. Endocr. Rev. 2006, 27, 736–749. [Google Scholar] [CrossRef] [PubMed]
- Hinkle, P.M.; Sebag, J.A. Structure and function of the melanocortin2 receptor accessory protein (MRAP). Mol. Cell. Endocrinol. 2009, 300, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Webb, R.R.; Clark, A.J.L. Minireview: The melanocortin 2 receptor accessory proteins. Mol. Endocrinol. 2010, 24, 475–484. [Google Scholar] [CrossRef]
- Noon, L.A.; Franklin, J.M.; King, P.J.; Goulding, N.J.; Hunyady, L.; Clark, A.J. Failed export of the adrenocorticotropin receptor from the endoplasmic reticulum in non-adrenal cells: Evidence in support of a requirement for a specific adrenal accessory factor. J. Endocrinol. 2002, 174, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Mountjoy, K.G.; Bird, I.M.; Rainey, W.E.; Cone, R.D. ACTH induces up-regulation of ACTH receptor mRNA in mouse and human adrenocortical cell lines. Mol. Cell. Endocrinol. 1994, 99, R17–R20. [Google Scholar] [CrossRef]
- Dores, R.M.; Garcia, Y. Views on the co-evolution of the melanocortin-2 receptor, MRAPs, and the hypothalamus/pituitary/adrenal-interrenal axis. Mol. Cell. Endocrinol. 2015, 408, 12–22. [Google Scholar] [CrossRef]
- Dores, R.M.; Chapa, E. Hypothesis and Theory: Evaluating the co-evolution of the melanocortin-2 receptor (MC2R) and the accessory protein MRAP1. Front. Endocrinol. 2021, 12, 747843. [Google Scholar] [CrossRef]
- Metherell, L.A.; Chapple, J.P.; Cooray, S.; David, A.; Becker, C.; Ruschendorf, F.; Naville, D.; Begeot, M.; Khoo, B.; Nurnberg, P.; et al. Mutations in MRAP, encoding a new interacting partner of the ACTH receptor, cause familial glucocorticoid deficiency type 2. Nat. Genet. 2005, 37, 166–170. [Google Scholar] [CrossRef]
- Sebag, J.A.; Hinkle, P.M. Melanocortin-2 receptor accessory protein MRAP forms antiparallel homodimers. Proc. Natl. Acad. Sci. USA 2007, 104, 20244–20249. [Google Scholar] [CrossRef]
- Cooray, S.N.; Almiro Do Vale, I.; Leung, K.Y.; Webb, T.R.; Chapple, J.P.; Egertova, M.; Cheetham, M.E.; Elphick, M.R.; Clark, A.J. The melanocortin 2 receptor accessory protein exists as a homodimer and is essential for the function of the melanocortin 2 receptor in the mouse y1 cell line. Endocrinology 2008, 149, 1935–1941. [Google Scholar] [CrossRef]
- Sebag, J.A.; Hinkle, P.M. Regions of melanocortin 2 (MC2) receptor accessory protein necessary for dual topology and MC2 receptor trafficking and signaling. J. Biol. Chem. 2009, 284, 610–618. [Google Scholar] [CrossRef]
- Shaughnessy, C.A.; Jensen, M.F.; Dores, R.M. A basal actinopterygian melanocortin receptor: Molecular and functional characterization of an Mc2r ortholog from the Senegal bichir (Polypterus senegalus). Gen. Comp. Endocrinol. 2022, 328, 114105. [Google Scholar] [CrossRef] [PubMed]
- Hoglin, B.E.; Miner, M.; Dores, R.M. Pharmacological Properties of Whale Shark (Rhincodon typus) Melanocortin-2 Receptor and Melancortin-5 Receptor: Interaction with MRAP1 and MRAP2. Gen. Comp. Endocrinol. 2020, 293, 113463. [Google Scholar] [CrossRef] [PubMed]
- Brazeau, M.D.; Frieddman, M. The origin and early phylogenetic history of jawed vertebrates. Nature 2015, 520, 490–497. [Google Scholar] [CrossRef] [PubMed]
- Nelson, J.S. Fishes of the World; John Wiley & Sons, Inc.: New York, NY, USA, 1994. [Google Scholar]
- Carroll, R.E. Vertebrate Paleontology and Evolution; W.H. Freeman and Company: New York, NY, USA, 1988. [Google Scholar]
- Thompson, A.W.; Brent Hawkins, M.B.; Parey, E.; Wcisel, D.J.; Ota, T.; Kawasaki, K.; Funk, E.; Losilla, M.; Fitch, O.E.; Pan, Q.; et al. The bowfin genome illuminates the developmental evolution of ray-finned fishes. Nat. Genet. 2021, 53, 1373–1384. [Google Scholar] [CrossRef] [PubMed]
- Chepurny, O.G.; Holz, G.G. A novel cyclic adenosine monophosphate responsiveluciferase reporter incorporating a nonpalindromic cyclic adenosine monophosphateresponse element provides optimal performance for use in G protein coupled receptordrug discovery efforts. J. Biomol. Screen. 2007, 12, 740–746. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Reinick, C.L.; Liang, L.; Angleson, J.K.; Dores, R.M. Identification of an Mrap-independent melanocortin-2 receptor: Functional expression of the cartilaginous fish, Callorhinchus milii, melanocortin-2 receptor in CHO cells. Endocrinology 2012, 153, 4757–4765. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liang, L.; Sebag, J.A.; Eagelston, L.; Serasinghe, M.N.; Veo, K.; Reinick, C.; Angleson, J.; Hinkle, P.M.; Dores, R.M. Functional Expression of frog and rainbow trout melanocortin 2 receptors using heterologous Mrap1s. Gen. Comp. Endocrinol. 2011, 174, 5–14. [Google Scholar] [CrossRef]
- Cheng, P.; Huang, Y.; Lv, Y.; Du, H.; Ruan, Z.; Li, C.; Ye, H.; Zhang, H.; Wu, J.; Wang, C.; et al. The American Paddlefish Genome Provides Novel Insights into Chromo, somal Evolution and Bone Mineralization in Early Vertebrates. Mol. Biol. Evol. 2020, 38, 1595–1607. [Google Scholar] [CrossRef]
- Sheng, J.A.; Bales, N.J.; Myers, S.A.; Bautisa, A.I.; Mina, R.; Taben, M.H.; Handa, R.J. The Hypothalamus-Pituitary-Adrenal Axis: Development, Programming Actions insights of Hormones, and Maternal-Fetal Interactions. Front. Behav. Neurosci. 2021, 14, 601939. [Google Scholar] [CrossRef]
- Wong, M.K.-S.; Dores, R.M. Analyzing the Hypothalamus/Pituitary/Interrenal axis of the neopterygian fish, Lepisosteus oculatus: Co-localization of MC2R, MC5R, MRAP1, and MRAP2 in interrenal cells. Gen. Comp. Endocrinol. 2022. [Google Scholar] [CrossRef]
- Dores, R.M.; Liang, L.; Hollmann, R.E.; Sandhu, N.; Vijayan, M.M. Identifying the activation motif in the N-terminal of rainbow trout and zebrafish melanocortin-2 receptor accessory protein 1 (MRAP1) orthologs. Gen. Comp. Endocrinol. 2016, 234, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Agulleiro, M.A.; Sánchez, E.; Leal, E.; Cortés, R.; Fernández-Durán, B.; Guillot, R.; Gallo-Payet, N.; Dores, R.M.; Davis, P.; Cerdá-Reverter, J.M. Molecular Characterization and Functional Regulation of Melanocortin 2 Receptor (MC2R) in the Sea Bass. A Putative Role in the Adaptation to Stress. PLoS ONE 2013, 8, e65450. [Google Scholar] [CrossRef] [PubMed]
- Agulleiro, M.A.; Roy, S.; Sanchez, E.; Puchol, S.; Gallo-Payet NCerda-Reverter, J.M. Role of melanocortin receptor accessory proteins in the function of zebrafish melanocortin receptor type 2. Mol. Cell. Endocrinol. 2010, 320, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Broughton, R.E.; Betancur-R, R.; Li, C.; Arratia, G.; Orti, G. Multi-locus phylogenetic analysis reveals the pattern and tempo of bony fish evolution. PLoS Curr. 2013, 5. [Google Scholar] [CrossRef]
- Barney, E.; Dores, M.R.; McAvoy, D.; Davis, P.; Racareanu, R.-C.; Iki, A.; Hyodo, S.; Dores, R.M. Elephant shark melanocortin receptors: Novel interactions with MRAP1 and implication for the HPI axis. Gen. Comp. Endocrinol. 2019, 272, 42–51. [Google Scholar] [CrossRef]
- Dores, R.M.; Scuba-Gray, M.; McNally, B.; Davis, P.; Takahashi, A. Evaluating the interactions between red stingray (Dasyatis akajei) melanocortin receptors and elephant shark (Callorhinchus milii) Mrap1 and Mrap2 following stimulation with either stingray ACTH(1-24) or stingray Des-Acetyl-αMSH: A pharmacological study in Chinese Hamster Ovary Cells. Gen. Comp. Endocrinol. 2018, 265, 133–140. [Google Scholar]
- Dores, R.M. Hypothesis and Theory: Revisiting views on the co-evolution of the melanocortin receptors and the accessory proteins, MRAP1 and MRAP2. Front. Endocrinol. 2016, 7, 79. [Google Scholar] [CrossRef] [PubMed]
- Crespi, E.J.; Williams, T.D.; Jessop, T.S.; Delehanty, B. Life history and the ecology of stress: How do glucocorticoid hormones influence life-history variation in animals? Funct. Ecol. 2013, 27, 93–116. [Google Scholar] [CrossRef]
- Davis, P.E.; Shaughnessy, C.A.; Dores, R.M. Human melanocortin-2 receptor: Identifying a role for residues in the TM4, EC2, and TM5 domains in activation and trafficking as a result of co-expression with the accessory protein, Mrap1 in Chinese Hamster Ovary Cells. Biomolecules 2022, 12, 1422. [Google Scholar] [CrossRef]
- Mehta, T.K.; Ravi, V.; Yamasaki, S.; Lee, A.P.; Lian, M.M.; Tay, B.-H.; Tohari, S.; Yanai, S.; Tay, A.; Brenner, S.; et al. Evidence for at least six Hox clusters in the Japanese lamprey (Lethenteron japonicum). Proc. Natl. Acad. Sci. USA 2013, 110, 16044–16049. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.J.; Keinath, M.C. The sea lamprey meiotic map improves resolution of ancient vertebrate genome duplications. Genome Res. 2015, 25, 1081–1090. [Google Scholar] [CrossRef] [PubMed]
- Holland, L.Z.; Ocampo Daza, D. A new look at an old question: When did the second whole genome duplication occur in vertebrate evolution. Genome Biol. 2018, 19, 209. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.J.; Timoshevskaya, N.; Ye, C.; Holt, C.; Keinath, M.C.; Parker, H.J.; Cook, M.E.; Hess, J.E.; Narum, S.R.; Lamanna, F.; et al. The sea lamprey germline genome provides insights into programmed genome rearrangement and vertebrate evolution. Nat. Genet. 2018, 50, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Rouault, A.A.J.; Srinivasan, D.K.; Yin, T.C.; Lee, A.A.; Sebag, J.A. Melanocortin receptor accessory proteins (MRAPs): Functions in the melanocortin system and beyond. Biochim. Biophys. Acta 2017, 1864, 2322–2329. [Google Scholar] [CrossRef] [PubMed]




| Panel | Transfection with cmc2r | EC50 (M) | p |
|---|---|---|---|
| 2A | cMRAP1 (positive control) | 7.6 × 10−10 ± 2.0 × 10−10 | |
| 2A | cMRAP1−A18A19A20A21 | n.d. | |
| 2A | cMRAP1−W18/A | 5.1 × 10−10 ± 12.1 × 10−9 | 0.06 |
| 2A | cMRAP1−D19/A | 1.6 × 10−9 ± 8.1 × 10−10 | 0.07 |
| 2B | cMRAP1 (positive control) | 1.6 × 10−10 ± 7.7 × 10−10 | |
| 2B | cMRAP1−Y21/A | n.d. | |
| 2B | cMRAP1−I22/A | 1.5 × 10−9 ± 5.2 × 10−10 | 0.11 |
| 3A | bfmrap1 (positive control) | 3.6 × 10−11 ± 1.1 × 10−10 | |
| 3A | bfmrap1−a18a19a20a21 | n.d. | |
| 3A | bfmrap1−y18/a | 3.3 × 10−10 ± 1.3 × 10−10 | 0.07 |
| 3A | bfmrap1−d19/a | 6.8 × 10−11 ± 4.1 × 10−11 | 0.23 |
| 3B | bfmrap1 (positive control) | 9.5 × 10−11 ± 3.1 × 10−11 | |
| 3B | bfmrap1−y20/a | 1.9 × 10−8 ± 4.2 × 10−9 | 0.04 |
| 3B | bfmrap1−i21/a | 5.8 × 10−10 ± 1.6 × 10−10 | 0.02 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dores, R.M.; McKinley, G.; Meyers, A.; Martin, M.; Shaughnessy, C.A. Structure/Function Studies on the Activation Motif of Two Non-Mammalian Mrap1 Orthologs, and Observations on the Phylogeny of Mrap1, Including a Novel Characterization of an Mrap1 from the Chondrostean Fish, Polyodon spathula. Biomolecules 2022, 12, 1681. https://doi.org/10.3390/biom12111681
Dores RM, McKinley G, Meyers A, Martin M, Shaughnessy CA. Structure/Function Studies on the Activation Motif of Two Non-Mammalian Mrap1 Orthologs, and Observations on the Phylogeny of Mrap1, Including a Novel Characterization of an Mrap1 from the Chondrostean Fish, Polyodon spathula. Biomolecules. 2022; 12(11):1681. https://doi.org/10.3390/biom12111681
Chicago/Turabian StyleDores, Robert M., Greer McKinley, Audrey Meyers, Morgan Martin, and Ciaran A. Shaughnessy. 2022. "Structure/Function Studies on the Activation Motif of Two Non-Mammalian Mrap1 Orthologs, and Observations on the Phylogeny of Mrap1, Including a Novel Characterization of an Mrap1 from the Chondrostean Fish, Polyodon spathula" Biomolecules 12, no. 11: 1681. https://doi.org/10.3390/biom12111681
APA StyleDores, R. M., McKinley, G., Meyers, A., Martin, M., & Shaughnessy, C. A. (2022). Structure/Function Studies on the Activation Motif of Two Non-Mammalian Mrap1 Orthologs, and Observations on the Phylogeny of Mrap1, Including a Novel Characterization of an Mrap1 from the Chondrostean Fish, Polyodon spathula. Biomolecules, 12(11), 1681. https://doi.org/10.3390/biom12111681







