Inhibition of SARS-CoV-2 Viral Channel Activity Using FDA-Approved Channel Modulators Independent of Variants
Abstract
1. Introduction
2. Materials and Methods
2.1. COVID-19 Patient Blood and Nasal Swab Samples
2.2. UV-Inactivated SARS-CoV-2 Variants
2.3. Fluorescence K+ Assay and Orf3a/E Channel Activity Detection
2.4. Ion Channel Modulators
2.5. Antigen Testing of Blood Samples
2.6. Data Analysis
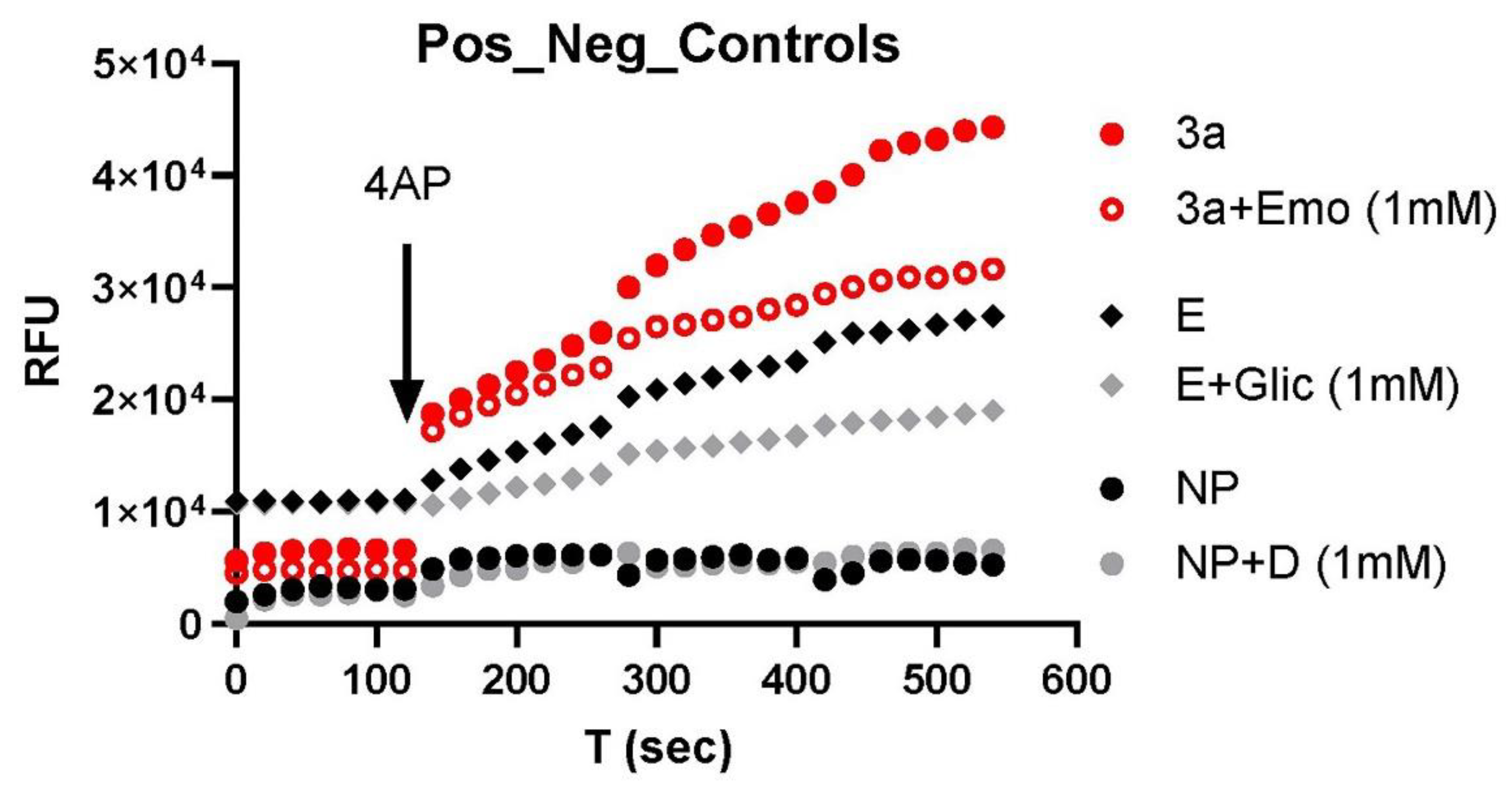
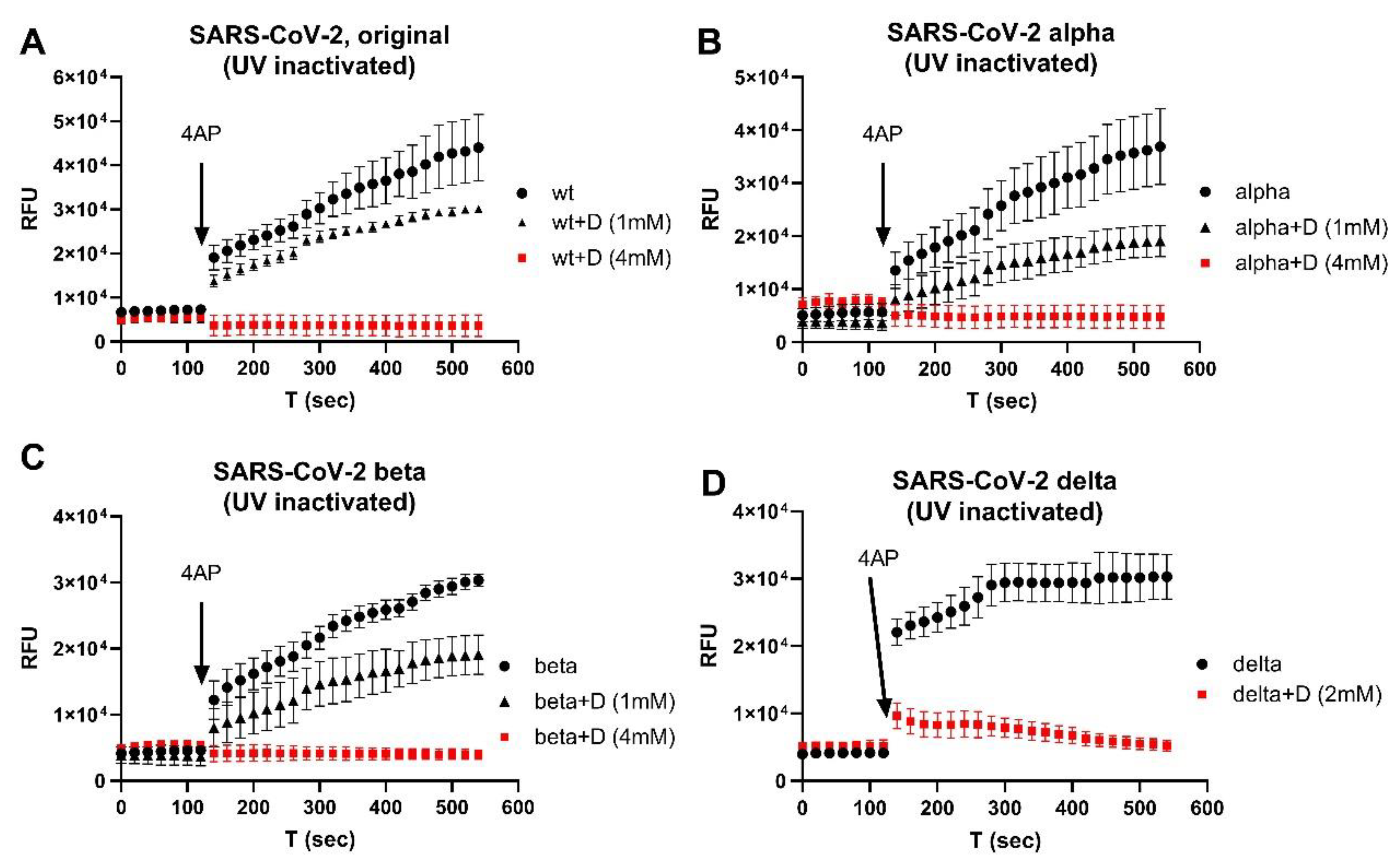
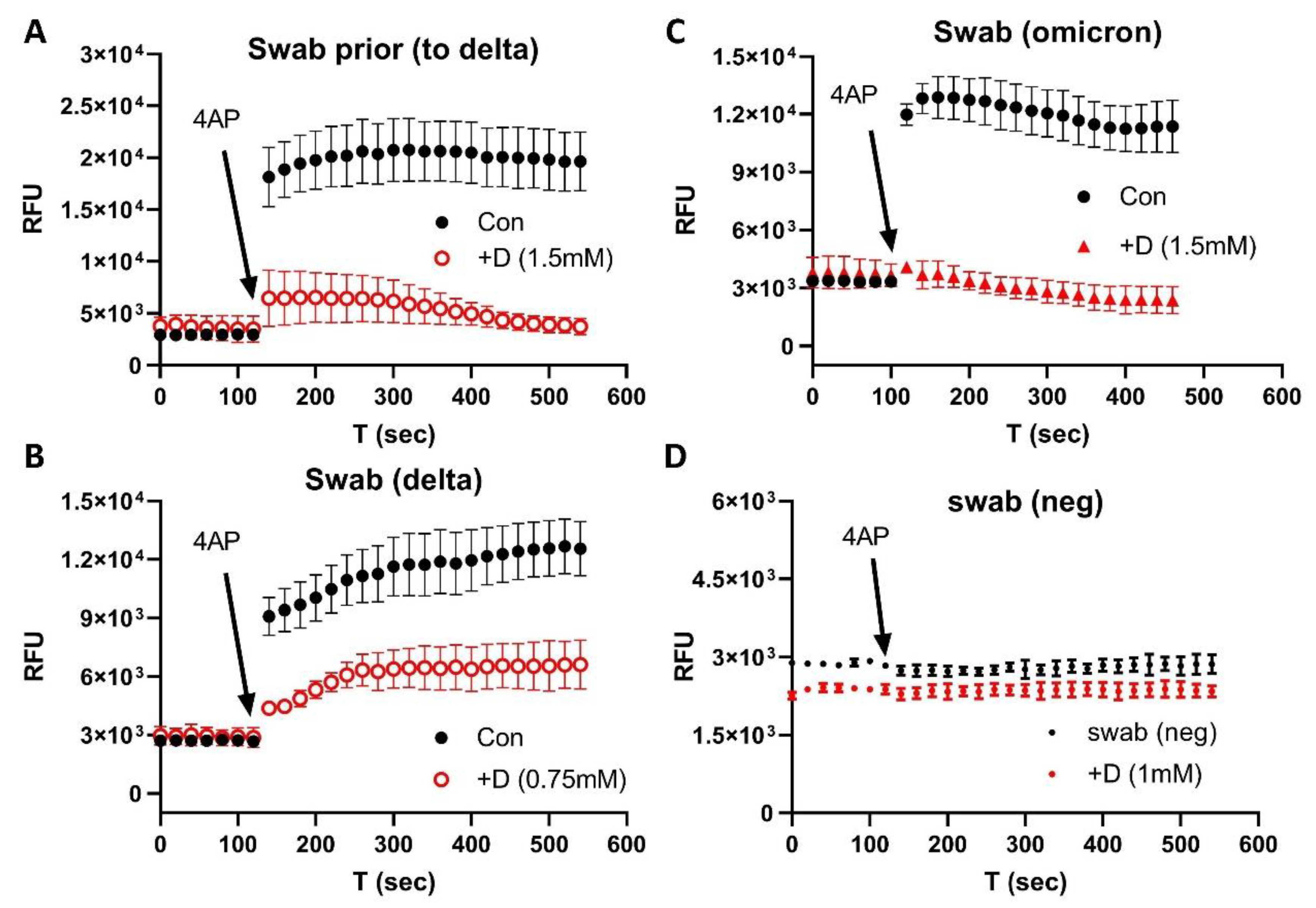
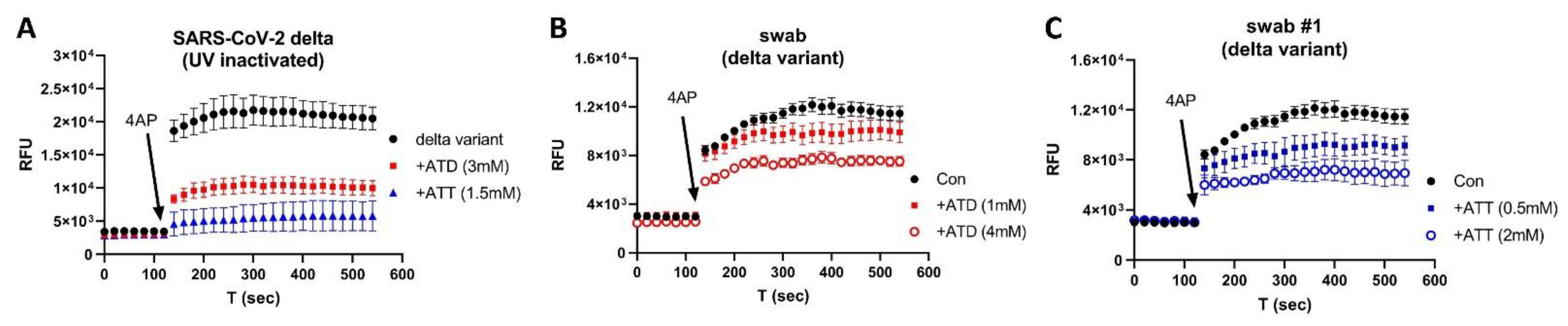
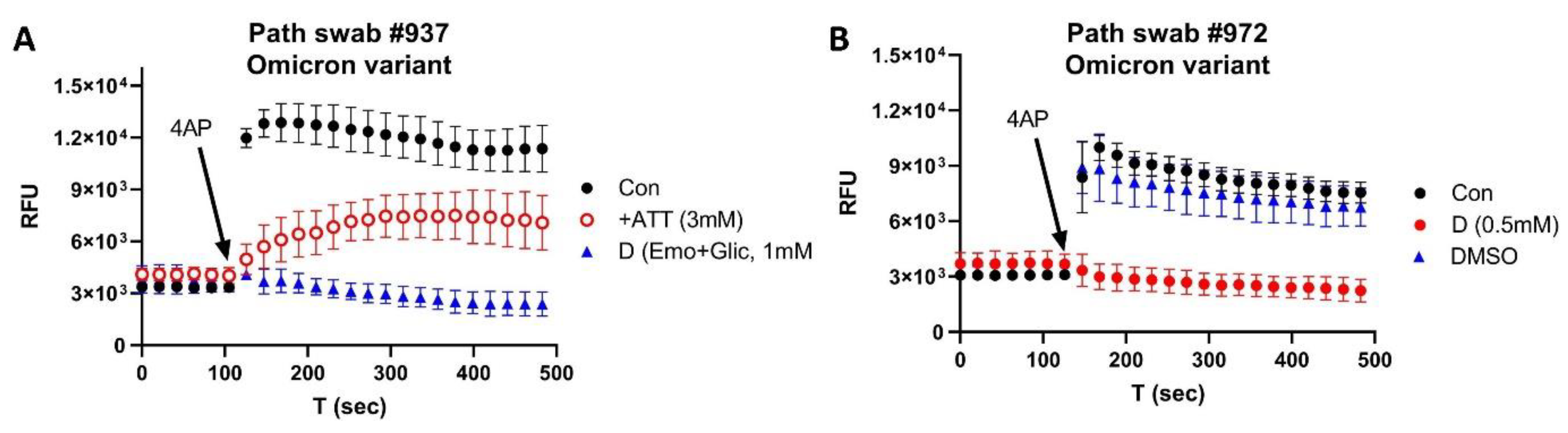
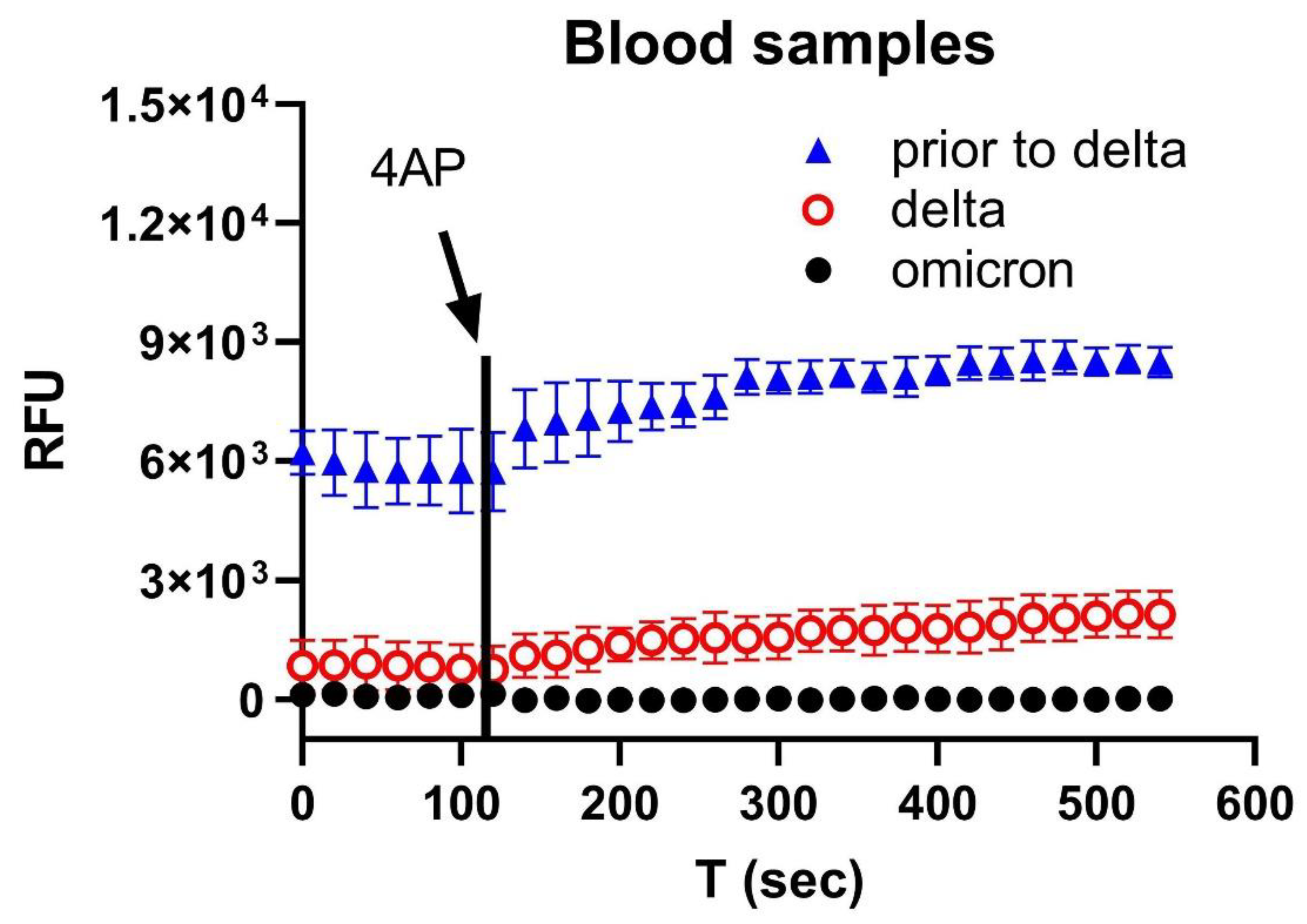
3. Results
4. Discussion
Limitations of the Work
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- CDC-Variants. US COVID-19 Cases Caused by Variants. Available online: https://wwwcdcgov/coronavirus/2019-ncov/transmission/variant-caseshtml (accessed on 5 September 2022).
- Cao, Y.; Yisimayi, A.; Jian, F.; Song, W.; Xiao, T.; Wang, L.; Du, S.; Wang, J.; Li, Q.; Chen, X.; et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature 2022, 608, 593–602. [Google Scholar] [CrossRef] [PubMed]
- Tegally, H.; Moir, M.; Everatt, J.; Giovanetti, M.; Scheepers, C.; Wilkinson, E.; Subramoney, K.; Moyo, S.; Amoako, D.G.; Baxter, C.; et al. Continued Emergence and Evolution of Omicron in South Africa: New BA.4 and BA.5 lineages. medRxiv 2022, arXiv:2022.05.01.22274406. [Google Scholar]
- Tuekprakhon, A.; Nutalai, R.; Dijokaite-Guraliuc, A.; Zhou, D.; Ginn, H.M.; Selvaraj, M.; Liu, C.; Mentzer, A.J.; Supasa, P.; Duyvesteyn, H.M.E.; et al. Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum. Cell 2022, 185, 2422–2433.e13. [Google Scholar] [CrossRef] [PubMed]
- Kimura, I.; Yamasoba, D.; Tamura, T.; Nao, N.; Oda, Y.; Mitoma, S.; Ito, J.; Nasser, H.; Zahradnik, J.; Uriu, K.; et al. Virological characteristics of the novel SARS-CoV-2 Omicron variants including BA.2.12.1, BA.4 and BA.5. bioRxiv 2022, arXiv:2022.05.26.493539. [Google Scholar]
- Hammond, J.; Leister-Tebbe, H.; Gardner, A.; Abreu, P.; Bao, W.; Wisemandle, W.; Baniecki, M.; Hendrick, V.M.; Damle, B.; Simón-Campos, A.; et al. Oral Nirmatrelvir for High-Risk, Nonhospitalized Adults with Covid-19. N. Engl. J. Med. 2022, 386, 1397–1408. [Google Scholar] [CrossRef]
- Hentzien, M.; Autran, B.; Piroth, L.; Yazdanpanah, Y.; Calmy, A. A monoclonal antibody stands out against omicron subvariants: A call to action for a wider access to bebtelovimab. Lancet Infect. Dis. 2022, 22, 1278. [Google Scholar] [CrossRef]
- Takashita, E.; Yamayoshi, S.; Simon, V.; van Bakel, H.; Sordillo, E.M.; Pekosz, A.; Fukushi, S.; Suzuki, T.; Maeda, K.; Halfmann, P.; et al. Efficacy of Antibodies and Antiviral Drugs against Omicron BA.2.12.1, BA.4, and BA.5 Subvariants. N. Engl. J. Med. 2022, 387, 468–470. [Google Scholar] [CrossRef]
- CDC-Forcasting. Observed and Forcasted Weekly COVID-19 Deaths in the United States. Available online: https://covidcdcgov/covid-data-tracker/#forecasting (accessed on 5 September 2022).
- Singh Tomar, P.P.; Arkin, I.T. SARS-CoV-2 E protein is a potential ion channel that can be inhibited by Gliclazide and Memantine. Biochem. Biophys. Res. Commun. 2020, 530, 10–14. [Google Scholar] [CrossRef]
- Schwarz, S.; Wang, K.; Yu, W.; Sun, B.; Schwarz, W. Emodin inhibits current through SARS-associated coronavirus 3a protein. Antiviral Res. 2011, 90, 64–69. [Google Scholar] [CrossRef]
- Pervushin, K.; Tan, E.; Parthasarathy, K.; Lin, X.; Jiang, F.L.; Yu, D.; Vararattanavech, A.; Soong, T.W.; Liu, D.X.; Torres, J. Structure and inhibition of the SARS coronavirus envelope protein ion channel. PLoS Pathog. 2009, 5, e1000511. [Google Scholar] [CrossRef]
- Wilson, L.; McKinlay, C.; Gage, P.; Ewart, G. SARS coronavirus E protein forms cation-selective ion channels. Virology 2004, 330, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Wilson, L.; Gage, P.; Ewart, G. Hexamethylene amiloride blocks E protein ion channels and inhibits coronavirus replication. Virology 2006, 353, 294–306. [Google Scholar] [CrossRef] [PubMed]
- Nieto-Torres, J.L.; Verdiá-Báguena, C.; Jimenez-Guardeño, J.M.; Regla-Nava, J.A.; Castaño-Rodriguez, C.; Fernandez-Delgado, R.; Torres, J.; Aguilella, V.M.; Enjuanes, L. Severe acute respiratory syndrome coronavirus E protein transports calcium ions and activates the NLRP3 inflammasome. Virology 2015, 485, 330–339. [Google Scholar] [CrossRef]
- Mandala, V.S.; McKay, M.J.; Shcherbakov, A.A.; Dregni, A.J.; Kolocouris, A.; Hong, M. Structure and drug binding of the SARS-CoV-2 envelope protein transmembrane domain in lipid bilayers. Nat. Struct. Mol. Biol. 2020, 27, 1202–1208. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.-G.; Sizemore, G.; Smoot, K.; Perrotta, P. Detecting SARS-CoV-2 Orf3a and E Ion Channel Activity in COVID-19 Blood Samples. J. Clin. Transl. Sci. 2021, 5, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Weaver, C.D.; Harden, D.; Dworetzky, S.I.; Robertson, B.; Knox, R.J. A thallium-sensitive, fluorescence-based assay for detecting and characterizing potassium channel modulators in mammalian cells. J. Biomol. Screen. 2004, 9, 671–677. [Google Scholar] [CrossRef] [PubMed]
- Raphemot, R.; Kadakia, R.J.; Olsen, M.L.; Banerjee, S.; Days, E.; Smith, S.S.; Weaver, C.D.; Denton, J.S. Development and validation of fluorescence-based and automated patch clamp-based functional assays for the inward rectifier potassium channel Kir4.1. Assay Drug Dev. Technol. 2013, 11, 532–543. [Google Scholar] [CrossRef]
- Phutthasophit, K.; Buddhari, D.; Chinnawirotpisan, P.; Joonlasak, K.; Manasatienkij, W.; Huang, A.; Kaewkao, T.; Mahayos, N.; Khontong, R.; Iamsirithaworn, S.; et al. Coding-Complete Genome Sequences of Alpha and Delta SARS-CoV-2 Variants from Kamphaeng Phet Province, Thailand, from May to July 2021. Microbiol. Resour. Announc. 2021, 10, e0087721. [Google Scholar] [CrossRef]
- Melloul, M.; Chouati, T.; Hemlali, M.; Alaoui Amine, S.; Touil, N.; Elannaz, H.; Ennibi, K.; Youbi, M.; Merabet, M.; Bellefquih, A.M.; et al. Genome Sequences of the Delta Variant (B.1.617.2) and the Kappa Variant (B.1.617.1) Detected in Morocco. Microbiol. Resour. Announc. 2021, 10, e0072721. [Google Scholar] [CrossRef]
- Kannan, S.R.; Spratt, A.N.; Sharma, K.; Chand, H.S.; Byrareddy, S.N.; Singh, K. Omicron SARS-CoV-2 variant: Unique features and their impact on pre-existing antibodies. J. Autoimmun. 2022, 126, 102779. [Google Scholar] [CrossRef]
- Gangavarapu, K.; Latif, A.A.; Mullen, J.L.; Alkuzweny, M.; Hufbauer, E.; Tsueng, G.; Haag, E.; Zeller, M.; Aceves, C.M.; Zaiets, K.; et al. Outbreak.info genomic reports: Scalable and dynamic surveillance of SARS-CoV-2 variants and mutations. medRxiv 2022, arXiv:2022.01.27.22269965. [Google Scholar]
- Verdiá-Báguena, C.; Nieto-Torres, J.L.; Alcaraz, A.; Dediego, M.L.; Enjuanes, L.; Aguilella, V.M. Analysis of SARS-CoV E protein ion channel activity by tuning the protein and lipid charge. Biochim. Biophys. Acta 2013, 1828, 2026–2031. [Google Scholar] [CrossRef] [PubMed]
- Nieto-Torres, J.L.; DeDiego, M.L.; Verdiá-Báguena, C.; Jimenez-Guardeño, J.M.; Regla-Nava, J.A.; Fernandez-Delgado, R.; Castaño-Rodriguez, C.; Alcaraz, A.; Torres, J.; Aguilella, V.M.; et al. Severe Acute Respiratory Syndrome Coronavirus Envelope Protein Ion Channel Activity Promotes Virus Fitness and Pathogenesis. PLoS Pathog. 2014, 10, e1004077. [Google Scholar] [CrossRef] [PubMed]
- Castaño-Rodriguez, C.; Honrubia, J.M.; Gutiérrez-Álvarez, J.; DeDiego, M.L.; Nieto-Torres, J.L.; Jimenez-Guardeño, J.M.; Regla-Nava, J.A.; Fernandez-Delgado, R.; Verdia-Báguena, C.; Queralt-Martín, M.; et al. Role of Severe Acute Respiratory Syndrome Coronavirus Viroporins E, 3a, and 8a in Replication and Pathogenesis. mBio 2018, 9, e02325-17. [Google Scholar] [CrossRef]
- Abdool Karim, S.S.; de Oliveira, T. New SARS-CoV-2 Variants—Clinical, Public Health, and Vaccine Implications. N. Engl. J. Med. 2021, 384, 1866–1868. [Google Scholar] [CrossRef]
- Peacock, T.P.; Penrice-Randal, R.; Hiscox, J.A.; Barclay, W.S. SARS-CoV-2 one year on: Evidence for ongoing viral adaptation. J. Gen. Virol. 2021, 102, 1584. [Google Scholar] [CrossRef]
- Chen, R.E.; Zhang, X.; Case, J.B.; Winkler, E.S.; Liu, Y.; VanBlargan, L.A.; Liu, J.; Errico, J.M.; Xie, X.; Suryadevara, N.; et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021, 27, 717–726. [Google Scholar] [CrossRef]
- Fillatre, P.; Dufour, M.-J.; Behillil, S.; Vatan, R.; Reusse, P.; Gabellec, A.; Velmans, N.; Montagne, C.; Geffroy, S.; Droumaguet, E.; et al. A new SARS-CoV-2 variant poorly detected by RT-PCR on nasopharyngeal samples, with high lethality. medRxiv 2021, arXiv:2021.05.05.21256690. [Google Scholar] [CrossRef]
- FDA-Omicron. Omicron Variant: Impact on Antigen Diagnostic Tests (As of 12/28/2021). Available online: https://wwwfdagov/medical-devices/coronavirus-covid-19-and-medical-devices/sars-cov-2-viral-mutations-impact-covid-19-tests#omicronvariantimpact (accessed on 5 September 2021).
- FDA-Omicron-Testing. SARS-CoV-2 Viral Mutations: Impact on COVID-19 Tests. Available online: https://wwwfdagov/medical-devices/coronavirus-covid-19-and-medical-devices/sars-cov-2-viral-mutations-impact-covid-19-tests#omicron-reduced (accessed on 5 September 2022).
- Guidelines, C.-T. Therapeutic Management of Nonhospitalized Adults With COVID-19. Available online: https://wwwcovid19treatmentguidelinesnihgov/management/clinical-management-of-adults/nonhospitalized-adults--therapeutic-management/ (accessed on 5 September 2022).
- Robinson, J. PANORAMIC trial to enlist 17,500 more patients as researchers add second COVID-19 antiviral. Pharm. J. 2022. Available online: https://pharmaceutical-journal.com/article/news/panoramic-trial-enlists-17500-more-patients-as-researchers-add-second-covid-19-antiviral (accessed on 5 September 2020).
- Zhou, Y.; Gammeltoft, K.A.; Ryberg, L.A.; Pham, L.V.; Fahnøe, U.; Binderup, A.; Hernandez, C.R.D.; Offersgaard, A.; Fernandez-Antunez, C.; Peters, G.H.J.; et al. Nirmatrelvir Resistant SARS-CoV-2 Variants with High Fitness in Vitro. bioRxiv 2022, arXiv:2022.06.06.494921. [Google Scholar]
- Jochmans, D.; Liu, C.; Donckers, K.; Stoycheva, A.; Boland, S.; Stevens, S.K.; De Vita, C.; Vanmechelen, B.; Maes, P.; Trüeb, B.; et al. The substitutions L50F, E166A and L167F in SARS-CoV-2 3CLpro are selected by a protease inhibitor—In vitro—And confer resistance to nirmatrelvir. bioRxiv 2022, arXiv:2022.06.07.495116. [Google Scholar]
- Westendorf, K.; Žentelis, S.; Wang, L.; Foster, D.; Vaillancourt, P.; Wiggin, M.; Lovett, E.; van der Lee, R.; Hendle, J.; Pustilnik, A.; et al. LY-CoV1404 (bebtelovimab) potently neutralizes SARS-CoV-2 variants. Cell Rep. 2022, 39, 110812. [Google Scholar] [CrossRef] [PubMed]
- Gordon, C.J.; Tchesnokov, E.P.; Woolner, E.; Perry, J.K.; Feng, J.Y.; Porter, D.P.; Götte, M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020, 295, 6785–6797. [Google Scholar] [CrossRef] [PubMed]
- Toots, M.; Yoon, J.J.; Cox, R.M.; Hart, M.; Sticher, Z.M.; Makhsous, N.; Plesker, R.; Barrena, A.H.; Reddy, P.G.; Mitchell, D.G.; et al. Characterization of orally efficacious influenza drug with high resistance barrier in ferrets and human airway epithelia. Sci. Transl. Med. 2019, 11, eaax5866. [Google Scholar] [CrossRef] [PubMed]
- FDA-Evusheld. Tixagevimab and Cilgavimab (Evusheld) for Pre-Exposure Prophylaxis of COVID-19. JAMA 2022, 327, 384–385. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Li, Z.; Ho, J.; Guo, Y.; Yeh, A.Y.; Liu, M.; Wang, M.; Yu, J.; Sheng, Z.; Huang, Y.; et al. Resistance of SARS-CoV-2 Omicron Subvariant BA.4.6 to Antibody Neutralization. bioRxiv 2022, arXiv:2022.09.05.506628. [Google Scholar]
- Nieva, J.L.; Madan, V.; Carrasco, L. Viroporins: Structure and biological functions. Nat. Rev. Microbiol. 2012, 10, 563–574. [Google Scholar] [CrossRef]
- Farag, N.S.; Breitinger, U.; Breitinger, H.G.; El Azizi, M.A. Viroporins and inflammasomes: A key to understand virus-induced inflammation. Int. J. Biochem. Cell Biol. 2020, 122, 105738. [Google Scholar] [CrossRef]
- Nieto-Torres, J.L.; Verdiá-Báguena, C.; Castaño-Rodriguez, C.; Aguilella, V.M.; Enjuanes, L. Relevance of Viroporin Ion Channel Activity on Viral Replication and Pathogenesis. Viruses 2015, 7, 3552–3573. [Google Scholar] [CrossRef]
- Redondo, N.; Zaldívar-López, S.; Garrido, J.J.; Montoya, M. SARS-CoV-2 Accessory Proteins in Viral Pathogenesis: Knowns and Unknowns. Front. Immunol. 2021, 12, 708264. [Google Scholar] [CrossRef]
- Ren, Y.; Shu, T.; Wu, D.; Mu, J.; Wang, C.; Huang, M.; Han, Y.; Zhang, X.-Y.; Zhou, W.; Qiu, Y.; et al. The ORF3a protein of SARS-CoV-2 induces apoptosis in cells. Cell. Mol. Immunol. 2020, 17, 881–883. [Google Scholar] [CrossRef]
- Lu, W.; Xu, K.; Sun, B. SARS Accessory Proteins ORF3a and 9b and Their Functional Analysis. In Molecular Biology of the SARS-Coronavirus; Lal, S.K., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 167–175. [Google Scholar]
- Siu, K.-L.; Yuen, K.-S.; Castano-Rodriguez, C.; Ye, Z.-W.; Yeung, M.-L.; Fung, S.-Y.; Yuan, S.; Chan, C.-P.; Yuen, K.-Y.; Enjuanes, L.; et al. Severe acute respiratory syndrome Coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC. FASEB J. 2019, 33, 8865–8877. [Google Scholar] [CrossRef] [PubMed]
- Xia, B.; Shen, X.; He, Y.; Pan, X.; Liu, F.L.; Wang, Y.; Yang, F.; Fang, S.; Wu, Y.; Duan, Z.; et al. SARS-CoV-2 envelope protein causes acute respiratory distress syndrome (ARDS)-like pathological damages and constitutes an antiviral target. Cell Res. 2021, 31, 847–860. [Google Scholar] [CrossRef] [PubMed]
- Polina, I.; Sung, J.H.; Jhun, B.S.; O-uchi, J. Abstract 15857: SARS-Cov-2 Genes Encode Ito-like Potassium Channels: Linkage Between Viroporins and High Mortality Rate in COVID-19 Patients with Pre-existing Cardiovascular Diseases. Circulation 2020, 142, A15857. [Google Scholar] [CrossRef]
- Kong, M.; Ba, M.; Ren, C.; Yu, L.; Dong, S.; Yu, G.; Liang, H. An updated meta-analysis of amantadine for treating dyskinesia in Parkinson’s disease. Oncotarget 2017, 8, 57316–57326. [Google Scholar] [CrossRef] [PubMed]
- Pahwa, R.; Tanner, C.M.; Hauser, R.A.; Sethi, K.; Isaacson, S.; Truong, D.; Struck, L.; Ruby, A.E.; McClure, N.L.; Went, G.T.; et al. Amantadine extended release for levodopa-induced dyskinesia in Parkinson’s disease (EASED Study). Mov. Disord. 2015, 30, 788–795. [Google Scholar] [CrossRef] [PubMed]
- Tokimatsu, I.; Nasu, M. Anti-influenza A viral drug-amantadine. Nihon Rinsho 2000, 58, 2288–2292. [Google Scholar]
- Hubsher, G.; Haider, M.; Okun, M.S. Amantadine. The journey from fighting flu to treating Parkinson disease. Neurology 2012, 78, 1096–1099. [Google Scholar] [CrossRef]
- Chizhmakov, I.V.; Geraghty, F.M.; Ogden, D.C.; Hayhurst, A.; Antoniou, M.; Hay, A.J. Selective proton permeability and pH regulation of the influenza virus M2 channel expressed in mouse erythroleukaemia cells. J. Physiol. 1996, 494, 329–336. [Google Scholar] [CrossRef]
- Thour, A.; Marwaha, R. Amitriptyline. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- Alam, U.; Sloan, G.; Tesfaye, S. Treating Pain in Diabetic Neuropathy: Current and Developmental Drugs. Drugs 2020, 80, 363–384. [Google Scholar] [CrossRef]
- Bendtsen, L.; Jensen, R.; Olesen, J. Amitriptyline, a combined serotonin and noradrenaline re-uptake inhibitor, reduces exteroceptive suppression of temporal muscle activity in patients with chronic tension-type headache. Electroencephalogr. Clin. Neurophysiol. Electromyogr. Mot. Control 1996, 101, 418–422. [Google Scholar] [CrossRef]
- McKinnon, N.K.; Reeves, D.C.; Akabas, M.H. 5-HT3 receptor ion size selectivity is a property of the transmembrane channel, not the cytoplasmic vestibule portals. J. Gen. Physiol. 2011, 138, 453–466. [Google Scholar] [CrossRef] [PubMed]
- Birch, P.J.; Dekker, L.V.; James, I.F.; Southan, A.; Cronk, D. Strategies to identify ion channel modulators: Current and novel approaches to target neuropathic pain. Drug Discov. Today 2004, 9, 410–418. [Google Scholar] [CrossRef]
- Colombo, E.; Francisconi, S.; Faravelli, L.; Izzo, E.; Pevarello, P. Ion channel blockers for the treatment of neuropathic pain. Future Med. Chem. 2010, 2, 803–842. [Google Scholar] [CrossRef] [PubMed]
- Zuliani, V.; Rivara, M.; Fantini, M.; Costantino, G. Sodium channel blockers for neuropathic pain. Expert Opin. Ther. Pat. 2010, 20, 755–779. [Google Scholar] [CrossRef]
- Kale, V.P.; Amin, S.G.; Pandey, M.K. Targeting ion channels for cancer therapy by repurposing the approved drugs. Biochim. Biophys. Acta BBA 2015, 1848, 2747–2755. [Google Scholar] [CrossRef]
- Thireau, J.; Pasquie, J.L.; Martel, E.; Le Guennec, J.Y.; Richard, S. New drugs vs. old concepts: A fresh look at antiarrhythmics. Pharmacol. Ther. 2011, 132, 125–145. [Google Scholar] [CrossRef]
- Darbar, D.; Roden, D.M. Future of antiarrhythmic drugs. Curr. Opin. Cardiol. 2006, 21, 361–367. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, H.-G.; Sizemore, G.; Martinez, I.; Perrotta, P. Inhibition of SARS-CoV-2 Viral Channel Activity Using FDA-Approved Channel Modulators Independent of Variants. Biomolecules 2022, 12, 1673. https://doi.org/10.3390/biom12111673
Yu H-G, Sizemore G, Martinez I, Perrotta P. Inhibition of SARS-CoV-2 Viral Channel Activity Using FDA-Approved Channel Modulators Independent of Variants. Biomolecules. 2022; 12(11):1673. https://doi.org/10.3390/biom12111673
Chicago/Turabian StyleYu, Han-Gang, Gina Sizemore, Ivan Martinez, and Peter Perrotta. 2022. "Inhibition of SARS-CoV-2 Viral Channel Activity Using FDA-Approved Channel Modulators Independent of Variants" Biomolecules 12, no. 11: 1673. https://doi.org/10.3390/biom12111673
APA StyleYu, H.-G., Sizemore, G., Martinez, I., & Perrotta, P. (2022). Inhibition of SARS-CoV-2 Viral Channel Activity Using FDA-Approved Channel Modulators Independent of Variants. Biomolecules, 12(11), 1673. https://doi.org/10.3390/biom12111673







