Assessing the Formation of Purine Lesions in Mitochondrial DNA of Cockayne Syndrome Cells
Abstract
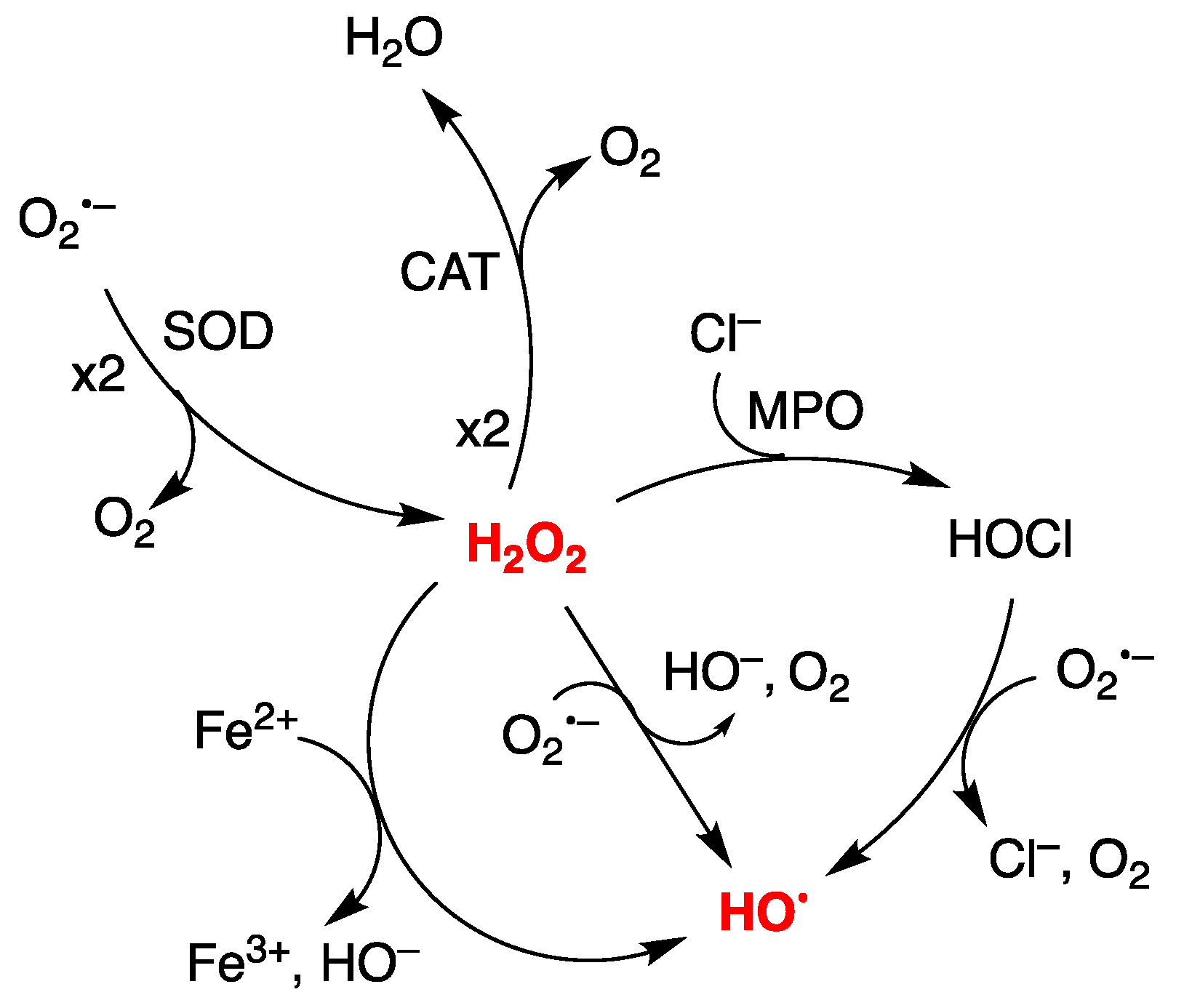
1. Introduction
2. Materials and Methods
2.1. Cell Lines and DNA Isolation
2.2. γ–Radiolysis Experiments
2.3. Enzymatic Digestion and Quantification of Modified Nucleosides by Stable Isotope LC–MS/MS
2.4. Statistical Analysis
3. Results and Discussion
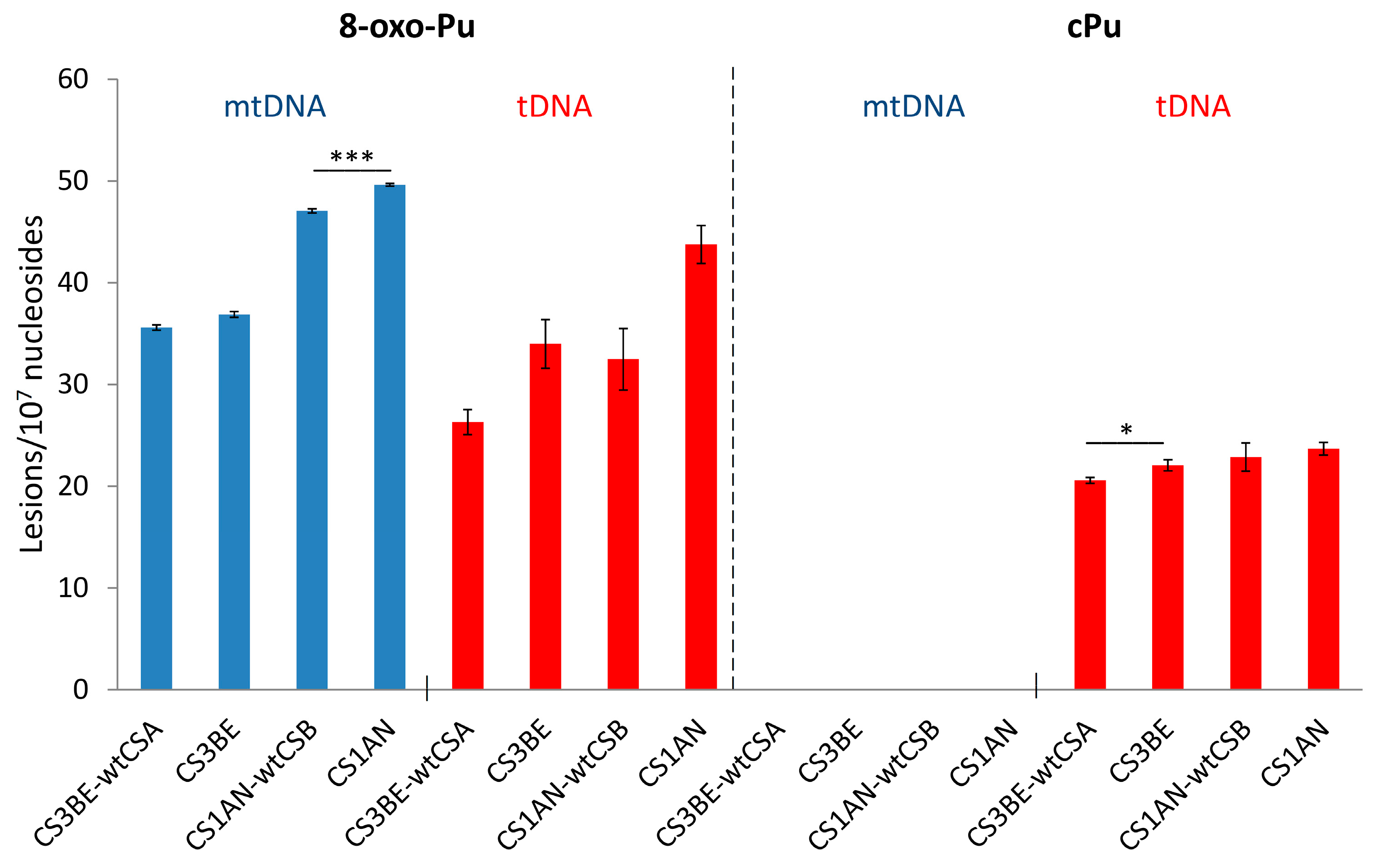
3.1. Purine mtDNA Lesions Levels in Wild Type and Defective CSA and CSB Cells
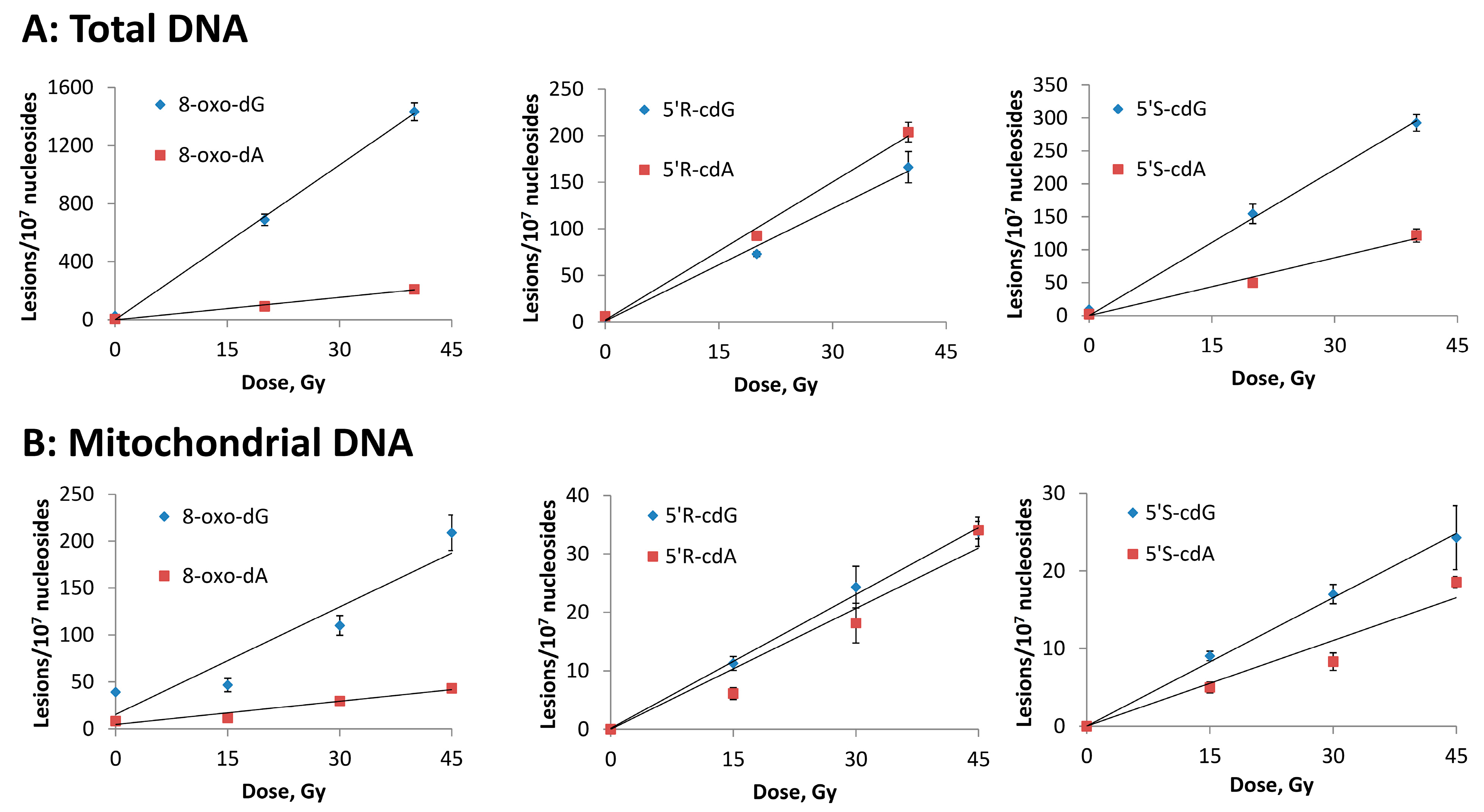
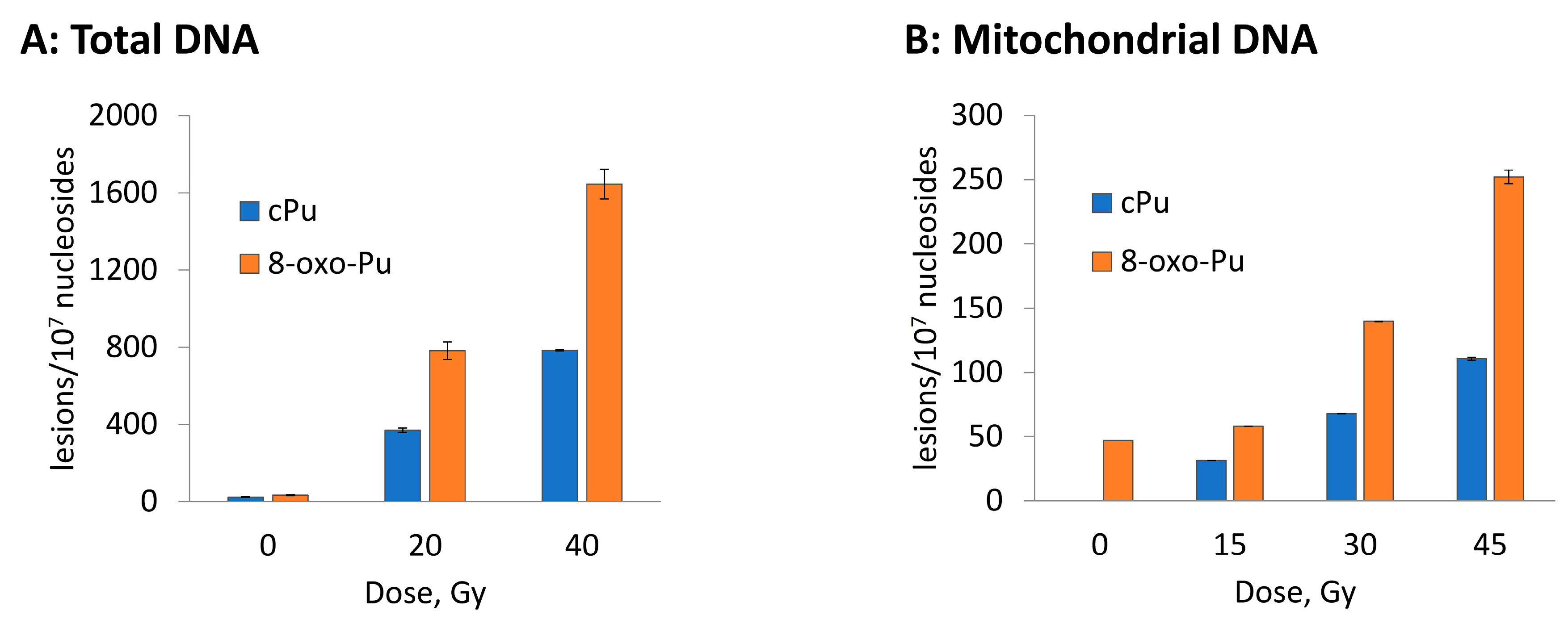
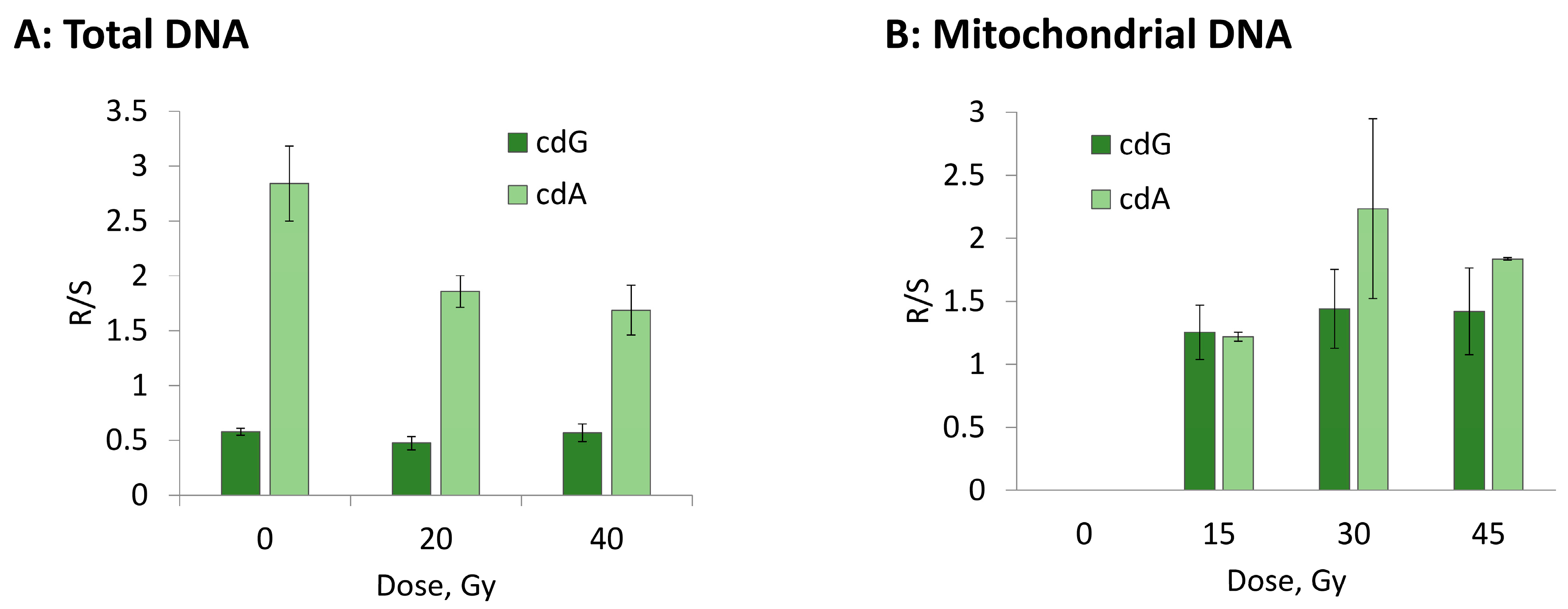
3.2. Hydroxyl Radical–Induced Formation of Purine Lesions: tDNA vs. mtDNA
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dröge, W. Free radicals in the physiological control of cell function. Physiol. Rev. 2002, 82, 47–95. [Google Scholar] [CrossRef] [PubMed]
- Shadel, G.S.; Horvath, T.L. Mitochondrial ROS signaling in organismal homeostasis. Cell 2015, 163, 560–569. [Google Scholar] [CrossRef] [PubMed]
- Sies, H.; Berndt, C.; Jones, D.P. Oxidative stress. Annu. Rev. Biochem. 2017, 86, 715–748. [Google Scholar] [CrossRef]
- Sies, H.; Jones, D.P. Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat. Rev. Mol. Cell. Biol. 2020, 21, 363–383. [Google Scholar] [CrossRef]
- Gammella, E.; Recalcati, S.; Cairo, G. Dual role of ROS as signal and stress agents: Iron tips the balance in favor of toxic effects. Oxid. Med. Cell. Longev. 2016, 2016, 1–9. [Google Scholar] [CrossRef]
- Murphy, M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009, 417, 1–13. [Google Scholar] [CrossRef]
- Solaini, G.; Baracca, A.; Lenaz, G.; Sgarbi, G. Hypoxia and mitochondrial oxidative metabolism. Biochim. Biophys. Acta 2010, 1797, 1171–1177. [Google Scholar] [CrossRef] [PubMed]
- Mailloux, R.J. Teaching the fundamentals of electron transfer reactions in mitochondria and the production and detection of reactive oxygen species. Redox Biol. 2015, 4, 381–398. [Google Scholar] [CrossRef] [PubMed]
- Mori, M.P.; Penjweini, R.; Knutson, J.R.; Wang, P.; Hwang, P.M. Mitochondria and oxygen homeostasis. FEBS J. 2021; online ahead of print. [Google Scholar] [CrossRef]
- Richter, C. Oxidative damage to mitochondrial DNA and its relationship to ageing. Int. J. Biochem. Cell Biol. 1995, 27, 647–653. [Google Scholar] [CrossRef]
- Harman, D. The biologic clock: The mitochondria? J. Am. Geriatr. Soc. 1972, 20, 145–147. [Google Scholar] [CrossRef]
- Anderson, S.; Bankier, A.T.; Barrell, B.G.; de Bruijn, M.H.; Coulson, A.R.; Drouin, J.; Eperon, I.C.; Nierlich, D.P.; Roe, B.A.; Sanger, F.; et al. Sequence and organization of the human mitochondrial genome. Nature 1981, 290, 457–465. [Google Scholar] [CrossRef] [PubMed]
- Nunnari, J.; Suomalainen, A. Mitochondria: In sickness and in health. Cell 2012, 148, 1145–1159. [Google Scholar] [CrossRef] [PubMed]
- Farge, G.; Falkenberg, M. Organization of DNA in mammalian mitochondria. Int. J. Mol. Sci. 2019, 20, 2770. [Google Scholar] [CrossRef] [PubMed]
- Mandavilli, B.S.; Santos, J.H.; Van Houten, B. Mitochondrial DNA repair and ageing. Mutat. Res. 2002, 509, 127–151. [Google Scholar] [CrossRef]
- Huang, Z.; Chen, Y.; Zhang, Y. Mitochondrial reactive oxygen species cause major oxidative mitochondrial DNA damages and repair pathways. J. Biosci. 2020, 45, 84. [Google Scholar] [CrossRef]
- Muftuoglu, M.; Mori, M.P.; de Souza–Pinto, N.C. Formation and repair of oxidative damage in the mitochondrial DNA. Mitochondrion 2014, 17, 164–181. [Google Scholar] [CrossRef]
- Kauppila, J.H.K.; Stewart, J.B. Mitochondrial DNA: Radically free of free–radical driven mutations. Biochim. Biophys. Acta 2015, 1847, 1354–1361. [Google Scholar] [CrossRef]
- Kopinski, P.K.; Singh, L.N.; Zhang, S.; Lott, M.T.; Wallace, D.C. Mitochondrial DNA variation and cancer. Nat. Rev. 2021, 21, 431–445. [Google Scholar] [CrossRef]
- D’Errico, M.; Parlanti, E.; Pascucci, B.; Filomeni, G.; Mastroberardino, P.G.; Dogliotti, E. The interplay between mitochondrial functionality and genome integrity in the prevention of human neurologic diseases. Arch. Biochem. Biophys. 2021, 710, 108977. [Google Scholar] [CrossRef]
- Chatgilialoglu, C.; Studer, A. (Eds.) Encyclopedia of Radicals in Chemistry, Biology and Materials; Wiley: Chichester, UK, 2012. [Google Scholar]
- Halliwell, B.; Gutteridge, J.M.C. Free Radicals in Biology and Medicine, 5th ed.; Oxford University Press: Oxford, UK, 2015. [Google Scholar]
- Fridovich, I. Superoxide radical and superoxide dismutases. Annu. Rev. Biochem. 1995, 64, 97–112. [Google Scholar] [CrossRef]
- Stohs, S.J.; Bagchi, D. Oxidative mechanisms in the toxicity of metal ions. Free Radic. Biol. Med. 1995, 18, 321–336. [Google Scholar] [CrossRef]
- Davies, M.J.; Hawkins, C.L. The role of myeloperoxidase in biomolecule modification, chronic inflammation, and disease. Antioxid. Redox Signal. 2020, 32, 957–981. [Google Scholar] [CrossRef] [PubMed]
- Winterbourn, C.C. Reconciling the chemistry and biology of reactive oxygen species. Nat. Chem. Biol. 2008, 4, 278–286. [Google Scholar] [CrossRef]
- Dizdaroglu, M.; Lloyd, R.S. DNA Damage, DNA Repair and Disease; Royal Society of Chemistry: Croydon, UK, 2021. [Google Scholar]
- Alexeyev, M.; Shokolenko, I.; Wilson, G.; Ledoux, S. The maintenance of mitochondrial DNA integrity—Critical analysis and update. Cold Spring Harb. Perspect. Biol. 2013, 5, a012641. [Google Scholar] [CrossRef] [PubMed]
- Shokolenko, I.; Venediktova, N.; Bochkareva, A.; Wilson, G.L.; Alexeyev, M.F. Oxidative stress induces degradation of mitochondrial DNA. Nucleic Acids Res. 2009, 37, 2539–2548. [Google Scholar] [CrossRef] [PubMed]
- Furda, A.M.; Marrangoni, A.M.; Lokshin, A.; Van Houten, B. Oxidants and not alkylating agents induce rapid mtDNA loss and mitochondrial dysfunction. DNA Repair. 2012, 11, 684–692. [Google Scholar] [CrossRef]
- Moretton, A.; Morel, F.; Macao, B.; Lachaume, P.; Ishak, L.; Lefebvre, M.; Garreau-Balandier, I.; Vernet, P.; Falkenberg, M.; Farge, G. Selective mitochondrial DNA degradation following double-strand breaks. PLoS ONE 2017, 12, e0176795. [Google Scholar] [CrossRef]
- Peeva, V.; Blei, D.; Trombly, G.; Corsi, S.; Szukszto, M.J.; Rebelo-Guiomar, P.; Gammage, P.A.; Kudin, A.P.; Becker, C.; Altmüller, J.; et al. Linear mitochondrial DNA is rapidly degraded by components of the replication machinery. Nat. Commun. 2018, 9, 1727. [Google Scholar] [CrossRef]
- Chatgilialoglu, C.; Ferreri, C.; Krokidis, M.G.; Masi, A.; Terzidis, M.A. On the relevance of hydroxyl radical to purine DNA damage. Free Radic. Res. 2021, 55, 384–404. [Google Scholar] [CrossRef]
- Chatgilialoglu, C.; Ferreri, C.; Terzidis, M.A. Purine 5′,8–cyclonucleoside lesions: Chemistry and biology. Chem. Soc. Rev. 2011, 40, 1368–1382. [Google Scholar] [CrossRef]
- Dizdaroglu, M.; Jaruga, P. Mechanisms of free radical–induced damage to DNA. Free Radic. Res. 2012, 46, 382–419. [Google Scholar] [CrossRef] [PubMed]
- Chatgilialoglu, C.; Ferreri, C.; Geacintov, N.E.; Krokidis, M.G.; Liu, Y.; Masi, A.; Shafirovich, N.; Terzidis, M.A.; Tsegay, P.S. 5′,8–Cyclopurine lesions in DNA damage: Chemical, analytical, biological and diagnostic significance. Cells 2019, 8, 513. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Ye, W.; Prestwich, E.G.; Wishnok, J.S.; Taghizadeh, K.; Dedon, P.C.; Tannenbaum, S.R. Comparative analysis of four oxidized guanine lesions from reactions of DNA with peroxynitrite, single oxygen, and γ–radiation. Chem. Res. Toxicol. 2013, 26, 195–202. [Google Scholar] [CrossRef]
- Brooks, P.J.; Wise, D.S.; Berry, D.A.; Kosmoski, J.V.; Smerdon, M.J.; Somers, R.L.; Mackie, H.; Spoonde, A.Y.; Ackerman, E.J.; Coleman, K.; et al. The oxidative DNA lesion 8,5′–(S)–cyclo–2′–deoxyadenosine is repaired by the nucleotide excision repair pathway and blocks gene expression in mammalian cells. J. Biol. Chem. 2000, 275, 22355–22362. [Google Scholar] [CrossRef]
- Kuraoka, I.; Bender, C.; Romieu, A.; Cadet, J.; Wood, R.D.; Lindahl, T. Removal of oxygen free–radical–induced 5′,8–purine cyclodeoxynucleosides from DNA by the nucleotide excision– repair pathway in human cells. Proc. Natl. Acad. Sci. USA 2000, 97, 3832–3837. [Google Scholar] [CrossRef] [PubMed]
- Pande, P.; Das, R.S.; Sheppard, C.; Kow, Y.W.; Basu, A.K. Repair efficiency of (5′S)–8,5–cyclo–2′–deoxyguanosine and (5′ S)–8,5′–cyclo–2′–deoxyadenosine depends on the complementary base. DNA Repair. 2012, 11, 926–931. [Google Scholar] [CrossRef][Green Version]
- Kropachev, K.; Ding, S.; Terzidis, M.A.; Masi, A.; Liu, Z.; Cai, Y.; Kolbanovskiy, M.; Chatgilialoglu, C.; Broyde, S.; Geacintov, N.E.; et al. Structural basis for the recognition of diastereomeric 5′,8–cyclo–2′–deoxypurine lesions by the human nucleotide excision repair system. Nucleic Acids Res. 2014, 42, 5020–5032. [Google Scholar] [CrossRef]
- Krasikova, Y.; Rechkunova, N.; Lavrik, O. Nucleotide excision repair: From molecular defects to neurological abnormalities. Int. J. Mol. Sci. 2021, 22, 6220. [Google Scholar] [CrossRef]
- Hanawalt, P.C.; Spivak, G. Transcription–coupled DNA repair: Two decades of progress and surprises. Nat. Rev. Mol. Cell. Biol. 2008, 9, 958–970. [Google Scholar] [CrossRef]
- D’Errico, M.; Pascucci, B.; Iorio, E.; Van Houten, B.; Dogliotti, E. The role of CSA and CSB protein in the oxidative stress response. Mech. Ageing Dev. 2013, 134, 261–269. [Google Scholar] [CrossRef]
- Pascucci, B.; Fragale, A.; Marabitti, V.; Leuzzi, G.; Calcagnile, A.S.; Parlanti, E.; Franchitto, A.; Dogliotti, E.; D’Errico, M. CSA and CSB play a role in the response to DNA breaks. Oncotarget 2018, 9, 11581–11591. [Google Scholar] [CrossRef] [PubMed]
- Krokidis, M.G.; D’Errico, M.; Pascucci, B.; Parlanti, E.; Masi, A.; Ferreri, C.; Chatgilialoglu, C. Oxygen–Dependent Accumulation of Purine DNA Lesions in Cockayne Syndrome Cells. Cells 2021, 9, 1671, Correction in Cells 2021, 10, 41. [Google Scholar] [CrossRef] [PubMed]
- Ferreri, C.; Sansone, A.; Krokidis, M.G.; Masi, A.; Pascucci, B.; D’Errico, M.; Chatgilialoglu, C. Effects of Oxygen Tension for Membrane Lipidome Remodeling of Cockayne Syndrome Cell Models. Cells 2022, 11, 1286. [Google Scholar] [CrossRef] [PubMed]
- Chatgilialoglu, C. Cyclopurine (cPu) lesions: What, how and why? Free Radic. Res. 2019, 53, 941–943. [Google Scholar] [CrossRef] [PubMed]
- Chatgilialoglu, C. Biomimetic Radical Chemistry and Applications. Molecules 2022, 27, 2042. [Google Scholar] [CrossRef]
- D’Errico, M.; Teson, M.; Calcagnile, A.; Nardo, T.; De Luca, N.; Lazzari, C.; Soddu, S.; Zambruno, G.; Stefanini, M.; Dogliotti, E. Differential role of transcription–coupled repair in UVB–induced response of human fibroblasts and keratinocytes. Cancer Res. 2005, 65, 232–238. [Google Scholar] [CrossRef]
- Pascucci, B.; D’Errico, M.; Romagnoli, A.; De Nuccio, C.; Savino, M.; Pietraforte, D.; Lanzafame, M.; Calcagnile, A.S.; Fortini, P.; Baccarini, S.; et al. Overexpression of parkin rescues the defective mitochondrial phenotype and the increased apoptosis of Cockayne syndrome A cells. Oncotarget 2016, 8, 102852–102867. [Google Scholar] [CrossRef][Green Version]
- Pascucci, B.; Lemma, T.; Iorio, E.; Giovannini, S.; Vaz, B.; Iavarone, I.; Calcagnile, A.; Narciso, L.; Degan, P.; Podo, F.; et al. An altered redox balance mediates the hypersensitivity of Cockayne syndrome primary fibroblasts to oxidative stress. Aging Cell 2012, 11, 520–529. [Google Scholar] [CrossRef]
- Spinks, J.W.T.; Woods, R.J. An Introduction to Radiation Chemistry, 3rd ed.; John–Wiley and Sons, Inc.: New York, NY, USA, 1990; p. 100. [Google Scholar]
- Terzidis, M.A.; Chatgilialoglu, C. An ameliorative protocol for the quantification of purine 5′,8–cyclo–2′–deoxynucleosides in oxidized DNA. Front. Chem. 2015, 3, 47. [Google Scholar] [CrossRef]
- Chatgilialoglu, C.; Krokidis, M.G.; Masi, A.; Barata–Vallejo, S.; Ferreri, C.; Terzidis, M.A.; Szreder, T.; Bobrowski, K. New insights into the reaction paths of hydroxyl radicals with purine moieties in DNA and double-stranded oligonucleotides. Molecules 2019, 24, 3860. [Google Scholar] [CrossRef]
- Chatgilialoglu, C.; Eriksson, L.A.; Krokidis, M.G.; Masi, A.; Wang, S.; Zhang, R. Oxygen dependent purine lesions in double-stranded oligodeoxynucleotides: Kinetic and computational studies highlight the mechanism for 5′,8–cyclopurine formation. J. Am. Chem. Soc. 2020, 142, 5825–5833. [Google Scholar] [CrossRef] [PubMed]
- Pascucci, B.; Spadaro, F.; Pietraforte, D.; De Nuccio, C.; Visentin, S.; Giglio, P.; Dogliotti, E.; D’Errico, M. DRP1 Inhibition Rescues Mitochondrial Integrity and Excessive Apoptosis in CS–A Disease Cell Models. Int. J. Mol. Sci. 2021, 22, 7123. [Google Scholar] [CrossRef] [PubMed]
- Chatre, L.; Biard, D.S.F.; Sarasin, A.; Ricchetti, M. Reversal of mitochondrial defects with CSB–dependent serine protease inhibitors in patient cells of the progeroid Cockayne syndrome. Proc. Natl. Acad. Sci. USA 2015, 112, E2910–E2919. [Google Scholar] [CrossRef] [PubMed]
- Wiesner, R.J.; Rüegg, C.; Morano, I. Counting target molecules by exponential polymerase chain reaction: Copy number of mitochondrial DNA in rat tissues. Biochem. Biophys. Res. Commun. 1992, 183, 553–559. [Google Scholar] [CrossRef]
- D’Erchia, A.M.; Atlante, A.; Gadaleta, G.; Pavesi, G.; Chiara, M.; De Virgilio, C.; Manzari, C.; Mastropasqua, F.; Prazzoli, G.M.; Picardi, E.; et al. Tissue–specific mtDNA abundance from exome data and its correlation with mitochondrial transcription, mass and respiratory activity. Mitochondrion 2015, 20, 13–21. [Google Scholar] [CrossRef]
- Shafirovich, V.; Kolbanovskiy, M.; Kropachev, K.; Liu, Z.; Cai, Y.; Terzidis, M.A.; Masi, A.; Chatgilialoglu, C.; Amin, S.; Dadali, A.; et al. Nucleotide excision repair and impact of site–specific 5′,8–cyclopurine and bulky DNA lesions on the physical properties of nucleosomes. Biochemistry 2019, 58, 561–574. [Google Scholar] [CrossRef] [PubMed]
- von Sonntag, C. Free–Radical–Induced DNA Damage and Its Repair. A Chemical Perspective; Springer Science: Berlin/Heidelberg, Germany, 2006. [Google Scholar]
- Buxton, G.V.; Greenstock, C.L.; Helman, W.P.; Ross, A.B. Critical review of rate constants for hydrated electrons, hydrogen atoms and hydroxyl radicals (OH/O−) in aqueous solution. J. Phys. Chem. Ref. Data 1988, 17, 513–886. [Google Scholar] [CrossRef]
- Ross, A.B.; Mallard, W.G.; Helman, W.P.; Buxton, G.V.; Huie, R.E.; Neta, P. NDRLNIST Solution Kinetic Database–Ver. 3; Notre Dame Radiation Laboratory, Notre Dame, IN and NIST Standard Reference Data: Gaithersburg, MD, USA, 1998.
- Bergeron, F.; Auvré, F.; Radicella, J.P.; Ravanat, J.-L. HO• radicals induce an unexpected high proportion of tandem base lesions refractory to repair by DNA glycosylases. Proc. Natl. Acad. Sci. USA 2010, 107, 5528–5533. [Google Scholar] [CrossRef]
- Ravanat, J.-L. Endogenous natural and radiation–induced DNA lesions: Differences and similarities and possible implications for human health and radiological protection. Radioprotection 2018, 53, 241–248. [Google Scholar] [CrossRef]
- Chatgilialoglu, C. The Two Faces of the Guanyl Radical: Molecular Context and Behavior. Molecules 2021, 26, 3511. [Google Scholar] [CrossRef]
- Terzidis, M.A.; Prisecaru, A.; Molphy, Z.; Barron, N.; Randazzo, A.; Dumont, E.; Krokidis, M.G.; Kellett, A.; Chatgilialoglu, C. Radical–induced purine lesion formation is dependent on DNA helical topology. Free Radic. Res. 2016, 50, S91–S101. [Google Scholar] [CrossRef] [PubMed]
- Krokidis, M.G.; Louka, M.; Efthimiadou, E.K.; Zervou, S.K.; Papadopoulos, K.; Hiskia, A.; Ferreri, C.; Chatgilialoglu, C. Membrane lipidome reorganization and accumulation of tissue DNA lesions in tumor–bearing mice: An exploratory study. Cancers 2019, 11, 480. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Guerrero, C.R.; Liu, S.; Amato, N.J.; Sharma, Y.; Gupta, S.; Wang, Y. Comprehensive assessment of oxidatively induced modifications of DNA in a rat model of human Wilson’s disease. Mol. Cell. Proteom. 2016, 15, 810–817. [Google Scholar] [CrossRef]
- Krokidis, M.G.; Prasinou, P.; Efthimiadou, E.K.; Boari, A.; Ferreri, C.; Chatgilialoglu, C. Effects of Aging and Disease Conditions in Brain of Tumor–Bearing Mice: Evaluation of Purine DNA Damages and Fatty Acid Pool Changes. Biomolecules 2022, 12, 1075. [Google Scholar] [CrossRef] [PubMed]
- Egler, R.A.; Fernandes, E.; Rothermund, K.; Sereika, S.; de Souza–Pinto, N.; Jaruga, P.; Dizdaroglu, M.; Prochownik, E.V. Regulation of reactive oxygen species, DNA damage, and c–Myc function by peroxiredoxin 1. Oncogene 2005, 24, 8038–8050. [Google Scholar] [CrossRef] [PubMed]
- Chatgilialoglu, C.; Bazzanini, R.; Jimenez, L.B.; Miranda, M.A. (5′S)– and (5′R)–5′,8–cyclo–2′–deoxyguanosine: Mechanistic insights on the 2′–deoxyguanosin–5′–yl radical cyclization. Chem. Res. Toxicol. 2007, 20, 1820–1824. [Google Scholar] [CrossRef]
- Boussicault, F.; Kaloudis, P.; Caminal, C.; Mulazzani, Q.G.; Chatgilialoglu, C. The fate of C5′ radicals of purine nucleosides under oxidative conditions. J. Am. Chem. Soc. 2008, 130, 8377–8385. [Google Scholar] [CrossRef]



| mtDNA | 8–oxo–dG 1 | 8–oxo–dA 1 |
|---|---|---|
| CS3BE–wtCSA | 28.30 ± 0.13 | 7.32 ± 0.13 |
| CS3BE | 29.31 ± 0.22 | 7.58 ± 0.07 |
| CS1AN–wtCSB | 39.00 ± 0.18 * | 8.08 ± 0.15 |
| CS1AN | 40.90 ± 0.22 * | 8.71 ± 0.17 |
| 8–oxo–Pu 1 | cPu 1 | |||
|---|---|---|---|---|
| mtDNA 2 | tDNA 3 | mtDNA 2 | tDNA 3 | |
| CS3BE–wtCSA | 35.62 ± 0.26 | 26.32 ± 1.22 | N/D | 20.56 ± 0.30 * |
| CS3BE | 36.89 ± 0.28 | 33.98 ± 2.40 | N/D | 22.04 ± 0.55 * |
| CS1AN–wtCSB | 47.08 ± 0.21 *** | 32.49 ± 3.04 | N/D | 22.84 ± 1.39 |
| CS1AN | 49.61 ± 0.13 *** | 43.73 ± 1.86 | N/D | 23.67 ± 0.62 |
| Lesion | tDNA | mtDNA |
|---|---|---|
| 8–oxo–dG | 35.2 | 3.8 |
| 8–oxo–dA | 5.1 | 0.8 |
| 5′R–cdG | 4.0 | 0.8 |
| 5′R–cdA | 4.9 | 0.8 |
| 5′S–cdG | 7.1 | 0.5 |
| 5′S–cdA | 3.0 | 0.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chatgilialoglu, C.; Krokidis, M.G.; Masi, A.; Barata-Vallejo, S.; Ferreri, C.; Pascucci, B.; D’Errico, M. Assessing the Formation of Purine Lesions in Mitochondrial DNA of Cockayne Syndrome Cells. Biomolecules 2022, 12, 1630. https://doi.org/10.3390/biom12111630
Chatgilialoglu C, Krokidis MG, Masi A, Barata-Vallejo S, Ferreri C, Pascucci B, D’Errico M. Assessing the Formation of Purine Lesions in Mitochondrial DNA of Cockayne Syndrome Cells. Biomolecules. 2022; 12(11):1630. https://doi.org/10.3390/biom12111630
Chicago/Turabian StyleChatgilialoglu, Chryssostomos, Marios G. Krokidis, Annalisa Masi, Sebastian Barata-Vallejo, Carla Ferreri, Barbara Pascucci, and Mariarosaria D’Errico. 2022. "Assessing the Formation of Purine Lesions in Mitochondrial DNA of Cockayne Syndrome Cells" Biomolecules 12, no. 11: 1630. https://doi.org/10.3390/biom12111630
APA StyleChatgilialoglu, C., Krokidis, M. G., Masi, A., Barata-Vallejo, S., Ferreri, C., Pascucci, B., & D’Errico, M. (2022). Assessing the Formation of Purine Lesions in Mitochondrial DNA of Cockayne Syndrome Cells. Biomolecules, 12(11), 1630. https://doi.org/10.3390/biom12111630












