Computational Investigation of 1, 3, 4 Oxadiazole Derivatives as Lead Inhibitors of VEGFR 2 in Comparison with EGFR: Density Functional Theory, Molecular Docking and Molecular Dynamics Simulation Studies
Abstract
1. Introduction
2. Materials and Methods
2.1. Density Functional Theory
2.2. Molecular Docking
2.2.1. Selection of Protein Targets
2.2.2. Softwares Required
2.2.3. Preparation of Protein
2.2.4. Preparation of Ligand and Molecular Docking
Visualization
Validation
2.2.5. Molecular Dynamics Simulations
2.2.6. Cell Viability Assay
2.2.7. ADMET Properties
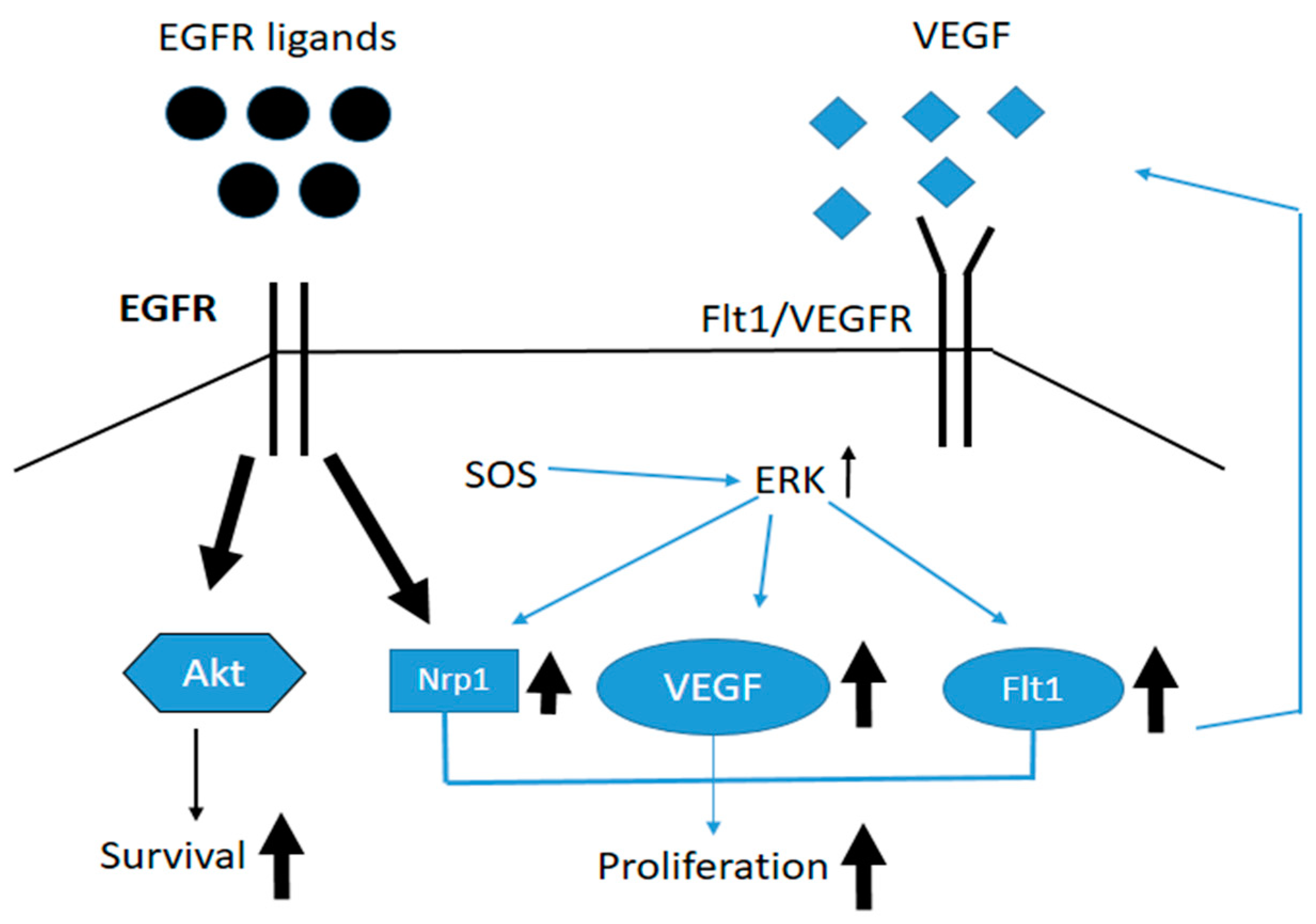
3. Results and Discussion
3.1. Synthesis of 1, 3, 4-Oxadiazole Amide Derivatives
3.2. Density Functional Theory Calculations (DFTs)
Results of DFT Studies: Global and Local Descriptors
3.3. Molecular Docking
3.3.1. Structure Activity Relationship (SAR) of 1, 3, 4-Oxadiazole Amide Derivatives
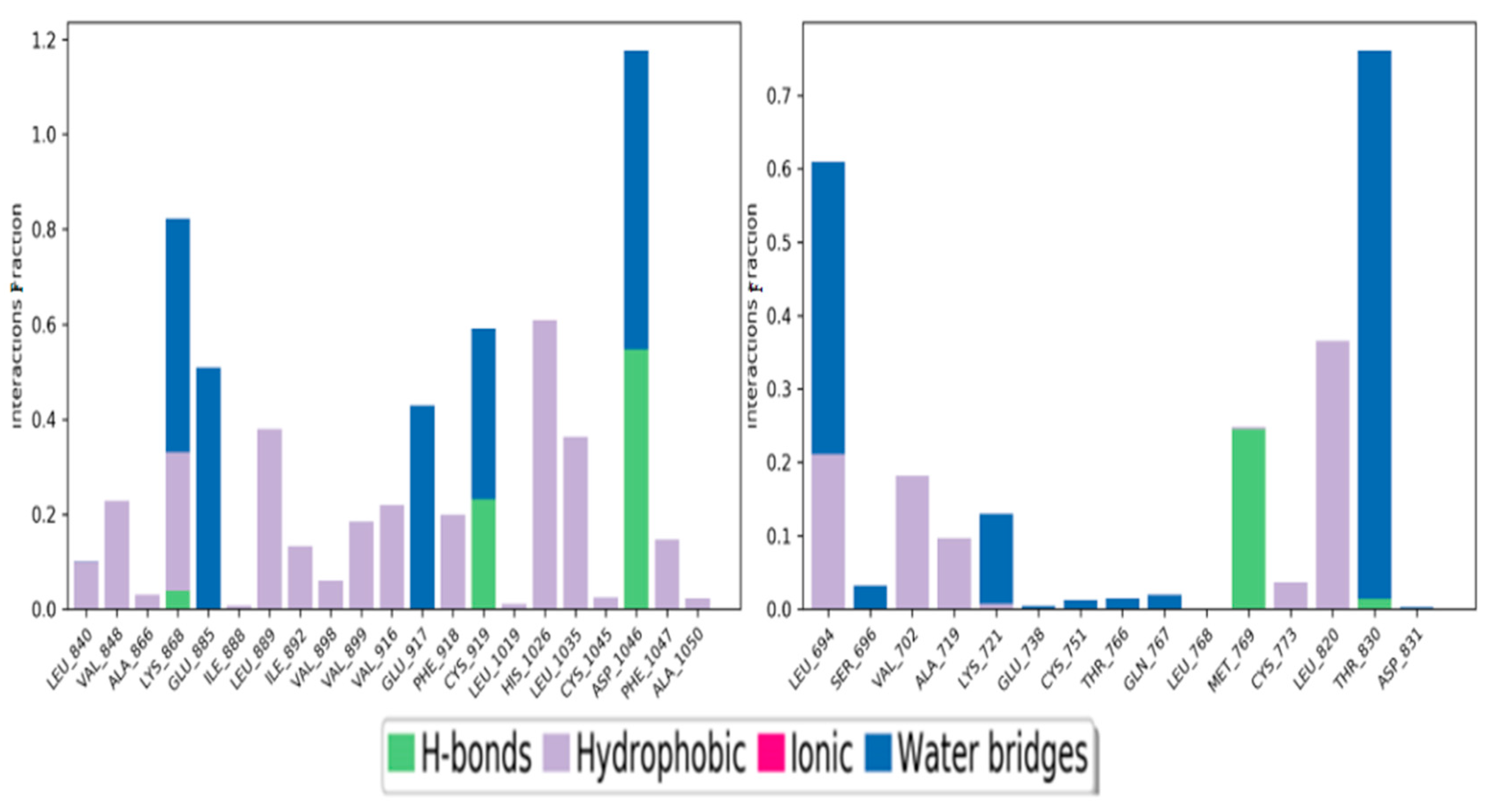
3.3.2. Binding Interaction Studies
Binding Interactions of 1, 3, 4 Oxadiazoles with VEGFR2
Binding Interactions of 1, 3, 4 Oxadiazoles with EGFR
3.4. SeeSAR Analysis
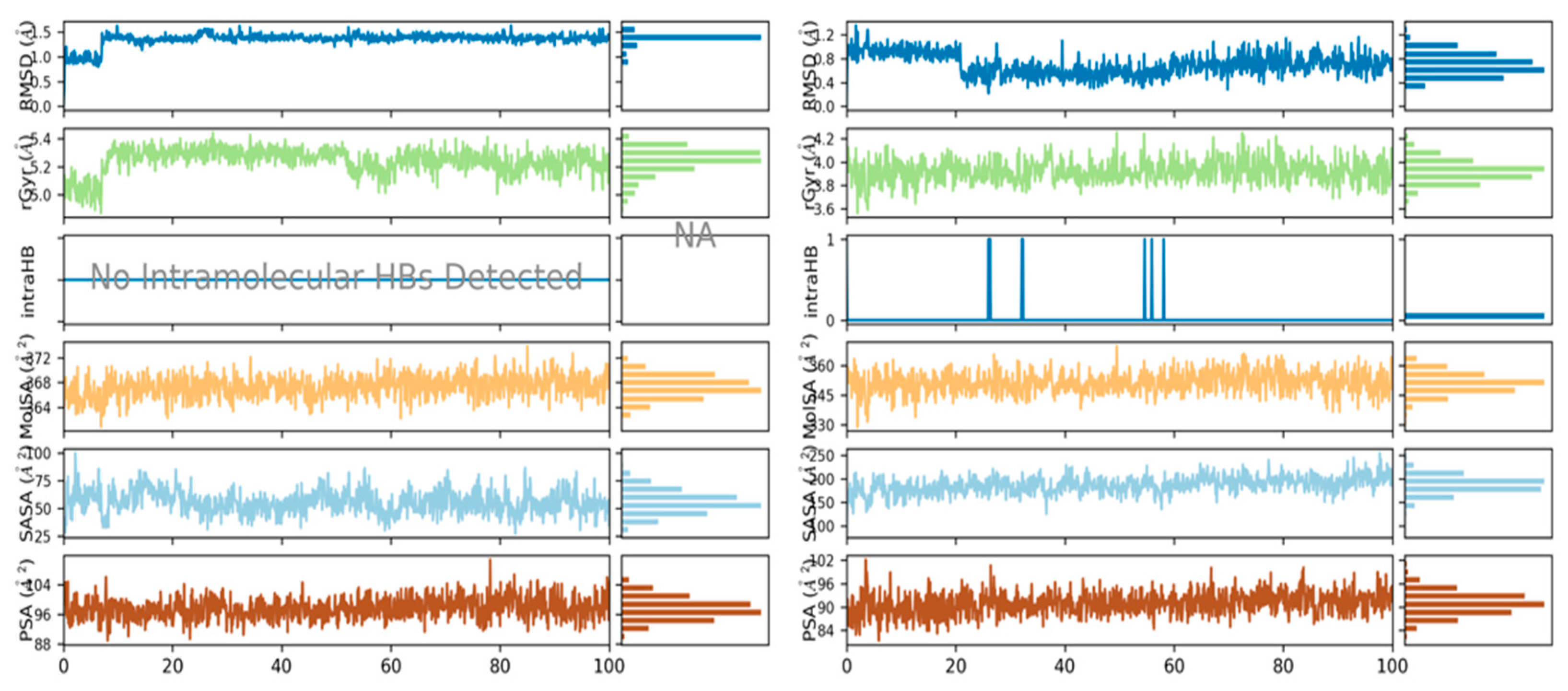
3.5. Molecular Dynamics Simulations
3.6. Cell Viability Assay
3.7. ADMET Properties
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Laversanne, M.; Weiderpass, E.; Soerjomataram, I. The ever-increasing importance of cancer as a leading cause of premature death worldwide. Cancer 2021, 127, 3029–3030. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.L.; Lipscomb, J.; Snyder, C. The burden of illness of cancer: Economic cost and quality of life. Annu. Rev. Public Health 2001, 22, 91. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Leung, D.K.W.; Chan, E.O.T.; Lok, V.; Leung, S.; Wong, I.; Wong, M.C. A global trend analysis of kidney cancer incidence and mortality and their associations with smoking, alcohol consumption, and metabolic syndrome. Eur. Urol. Focus 2022, 8, 200–209. [Google Scholar] [CrossRef]
- Murai, M.; Oya, M. Renal cell carcinoma: Etiology, incidence and epidemiology. Curr. Opin. Urol. 2004, 14, 229–233. [Google Scholar] [CrossRef] [PubMed]
- Roskoski, R., Jr. Src protein-tyrosine kinase structure and regulation. Biochem. Biophys. Res. Commun. 2004, 324, 1155–1164. [Google Scholar] [CrossRef] [PubMed]
- Normanno, N.; De Luca, A.; Bianco, C.; Strizzi, L.; Mancino, M.; Maiello, M.R.; Salomon, D.S. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene 2006, 366, 2–16. [Google Scholar] [CrossRef]
- Raymond, E.; Faivre, S.; Armand, J.P. Epidermal growth factor receptor tyrosine kinase as a target for anticancer therapy. Drugs 2000, 60, 15–23. [Google Scholar] [CrossRef]
- Xu, Y.; Liu, H.; Chen, J.; Zhou, Q. Acquired resistance of lung adenocarcinoma to EGFR-tyrosine kinase inhibitors gefitinib and erlotinib. Cancer Biol. Ther. 2010, 9, 572–582. [Google Scholar] [CrossRef]
- Veikkola, T.; Karkkainen, M.; Claesson-Welsh, L.; Alitalo, K. Regulation of angiogenesis via vascular endothelial growth factor receptors. Cancer Res. 2000, 60, 203–212. [Google Scholar]
- Wise, L.M.; Veikkola, T.; Mercer, A.; Savory, L.J.; Fleming, S.B.; Caesar, C.; Vitali, A.; Makinen, T.; Alitalo, K.; Stacker, S.A. Vascular endothelial growth factor (VEGF)-like protein from orf virus NZ2 binds to VEGFR2 and neuropilin-1. Proc. Natl. Acad. Sci. USA 1999, 96, 3071–3076. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, N.; Han, S.; Wang, D.; Mo, S.; Yu, L.; Chen, J. Dietary compound isoliquiritigenin inhibits breast cancer neoangiogenesis via VEGF/VEGFR-2 signaling pathway. PLoS ONE 2013, 8, e68566. [Google Scholar] [CrossRef]
- Joensuu, H.; Trent, J.C.; Reichardt, P. Practical management of tyrosine kinase inhibitor-associated side effects in GIST. Cancer Treat. Rev. 2011, 37, 75–88. [Google Scholar] [CrossRef] [PubMed]
- Lichtenberger, B.M.; Tan, P.K.; Niederleithner, H.; Ferrara, N.; Petzelbauer, P.; Sibilia, M. Autocrine VEGF Signaling Synergizes with EGFR in Tumor Cells to Promote Epithelial Cancer Development. Cell 2010, 140, 268–279. [Google Scholar] [CrossRef]
- Siwach, A.; Verma, P.K. Therapeutic potential of oxadiazole or furadiazole containing compounds. BMC Chem. 2020, 14, 1–40. [Google Scholar] [CrossRef] [PubMed]
- Dokla, E.M.; Fang, C.-S.; Abouzid, K.A.; Chen, C.S. 1,2,4-Oxadiazole derivatives targeting EGFR and c-Met degradation in TKI resistant NSCLC. Eur. J. Med. Chem. 2019, 182, 111607. [Google Scholar] [CrossRef]
- Iftikhar, M.; Saleem, M.; Riaz, N.; Ahmed, I.; Rahman, J.; Ashraf, M.; Htar, T.T. A novel five-step synthetic route to 1, 3, 4-oxadiazole derivatives with potent α-glucosidase inhibitory potential and their in-silico studies. Arch. Der Pharm. 2019, 352, 1900095. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.-J.; Trucks, G.-W.; Schlegel, H.-B.; Scuseria, G.-E.; Robb, M.-A.; Cheeseman, J.-R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.-A.; et al. Gaussian 09; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Dennington, R.; Keith, T.A.; Millam, J.M. GaussView V 6.1; Semichem Inc.: Shawnee Mission, KS, USA, 2016. [Google Scholar]
- Available online: https//www.rcsb.org/3VHE (accessed on 1 July 2022).
- Available online: https://www.rcsb.org/2GS6 (accessed on 1 July 2022).
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Accelrys Software Inc. Discovery Studio Visualizer, 2; Accelrys Software Inc.: San Diego, CA, USA, 2005. [Google Scholar]
- BioSolveIT-SeeSAR. Available online: https://www.biosolveit.de/SeeSAR/ (accessed on 15 July 2022).
- CambridgeSoft. ChemDraw Ultra 12.0 0 (Copyright) 1986 to 2009; CambridgeSoft Corp.: Cambridge, MA, USA, 2009. [Google Scholar]
- CambridgeSoft. Chem 3D Pro 12.0 (Copyright) 1986 to 2009; CambridgeSoft Corp.: Cambridge, MA, USA, 2009. [Google Scholar]
- Ferreira, L.G.; Dos Santos, R.N.; Oliva, G.; Andricopulo, A.D. Molecular Docking and Structure-Based Drug Design Strategies. Molecules 2015, 20, 13384–13421. [Google Scholar] [CrossRef] [PubMed]
- Zentgraf, M.; Steuber, H.; Koch, C.; La Motta, C.; Sartini, S.; Sotriffer, C.A.; Klebe, G. How reliable are current docking approaches for structure-based drug design? Lessons from aldose reductase. Angew. Chem. Int. Ed. 2007, 46, 3575–3578. [Google Scholar] [CrossRef]
- Yusuf, D.; Davis, A.M.; Kleywegt, G.J.; Schmitt, S. An alternative method for the evaluation of docking performance: RSR vs. RMSD. J. Chem. Inf. Model. 2008, 48, 1411–1422. [Google Scholar] [CrossRef]
- Bowers, K.J.; Chow, D.E.; Xu, H.; Dror, R.O.; Eastwood, M.P.; Gregersen, B.A.; Shaw, D.E. Scalable algorithms for molecular dynamics simulations on commodity clusters. In Proceedings of the SC’06: 2006 ACM/IEEE Conference on Supercomputing, Tampa, FL, USA, 11–17 November 2006; p. 43. [Google Scholar]
- Hildebrand, P.W.; Rose, A.S.; Tiemann, J.K. Bringing Molecular Dynamics Simulation Data into View. Trends Biochem. Sci. 2019, 44, 902–913. [Google Scholar] [CrossRef]
- Rasheed, M.; Iqbal, M.; Saddick, S.; Ali, I.; Khan, F.; Kanwal, S.; Ahmed, D.; Ibrahim, M.; Afzal, U.; Awais, M. Identification of Lead Compounds against Scm (fms10) in Enterococcus faecium Using Computer Aided Drug Designing. Life 2021, 11, 77. [Google Scholar] [CrossRef] [PubMed]
- Shivakumar, D.; Williams, J.; Wu, Y.; Damm, W.; Shelley, J.; Sherman, W. Prediction of Absolute Solvation Free Energies using Molecular Dynamics Free Energy Perturbation and the OPLS Force Field. J. Chem. Theory Comput. 2010, 6, 1509–1519. [Google Scholar] [CrossRef] [PubMed]
- Anderson, P.M.; Wilson, M.R. Developing a force field for simulation of poly (ethylene oxide) based upon ab initio calculations of 1,2-dimethoxyethane. Mol. Phys. 2005, 103, 89–97. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Nikš, M.; Otto, M. Towards an optimized MTT assay. J. Immunol. Methods 1990, 130, 149–151. [Google Scholar] [CrossRef]
- Iqbal, J.; Ejaz, S.A.; Saeed, A.; Al-Rashida, M. Detailed investigation of anticancer activity of sulfamoyl benz (sulfon) amides and 1H-pyrazol-4-yl benzamides: An experimental and computational study. Eur. J. Pharmacol. 2018, 5, 11–24. [Google Scholar] [CrossRef]
- ADMETlab 2.0. Available online: https://admetmesh.scbdd.com/ (accessed on 5 October 2022).
- McTigue, M.A.; Wickersham, J.A.; Pinko, C.; Showalter, R.E.; Parast, C.V.; Tempczyk-Russell, A.; Appelt, K. Crystal structure of the kinase domain of human vascular endothelial growth factor receptor 2: A key enzyme in angiogenesis. Structure 1999, 7, 319–330. [Google Scholar] [CrossRef]
- Zhang, X.; Gureasko, J.; Shen, K.; Cole, P.A.; Kuriyan, J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell 2006, 125, 1137–1149. [Google Scholar] [CrossRef]
- Sargolzaei, M. Effect of nelfinavir stereoisomers on coronavirus main protease: Molecular docking, molecular dynamics simulation and MM/GBSA study. J. Mol. Graph. Model. 2021, 103, 107803. [Google Scholar] [CrossRef]
- Barbosa, L.R.; Ortore, M.G.; Spinozzi, F.; Mariani, P.; Bernstorff, S.; Itri, R. The importance of protein-protein interactions on the pH-induced conformational changes of bovine serum albumin: A small-angle X-ray scattering study. Biophys. J. 2010, 98, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Badieyan, S.; Bevan, D.R.; Zhang, C. Study and design of stability in GH5 cellulases. Biotechnol. Bioeng. 2012, 109, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.M.; Teague, S.J. Hydrogen bonding, hydrophobic interactions, and failure of the rigid receptor hypothesis. Angew. Chem. Int. Ed. 1999, 38, 736–749. [Google Scholar] [CrossRef]
- Rajagopal, S.; Vishveshwara, S. Short hydrogen bonds in proteins. FEBS J. 2005, 272, 1819–1832. [Google Scholar] [CrossRef] [PubMed]
- Aziz, M.; Ejaz, S.A.; Tamam, N.; Siddique, F.; Riaz, N.; Qais, F.A.; Iqbal, J. Identification of potent inhibitors of NEK7 protein using a comprehensive computational approach. Sci. Rep. 2022, 12, 6404. [Google Scholar] [CrossRef]













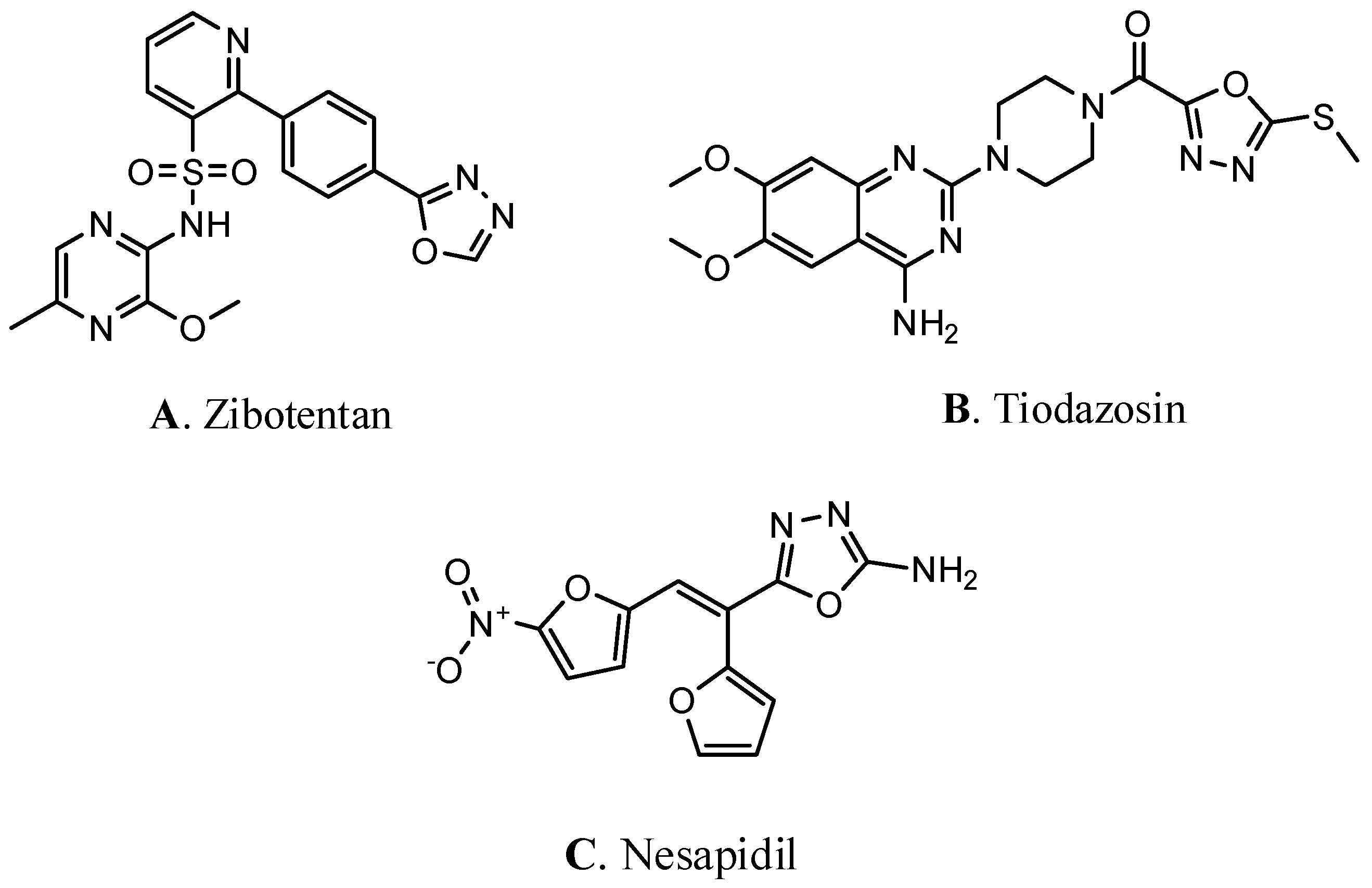
| Codes | Gas | Ethanol | ||
|---|---|---|---|---|
| Optimization Energy (Hatree) | Dipole Moment (Debye) | Optimization Energy (Hatree) | Dipole Moment (Debye) | |
| 7a | −1828.415 | 1.982 | −1828.431 | 4.061 |
| 7b | −1867.705 | 2.210 | −1867.721 | 4.005 |
| 7c | −1867.707 | 2.124 | −1867.722 | 4.471 |
| 7d | −1867.707 | 2.258 | −1867.722 | 4.394 |
| 7e | −1906.995 | 2.371 | −1907.011 | 4.058 |
| 7f | −1906.996 | 2.565 | −1907.012 | 4.316 |
| 7g | −1906.996 | 2.608 | −1907.012 | 5.233 |
| 7h | −1906.997 | 2.404 | −1907.013 | 4.948 |
| 7i | −1906.999 | 2.427 | −1907.014 | 4.821 |
| 7j | −1906.999 | 2.205 | −1907.014 | 4.724 |
| 7k | −1906.994 | 2.903 | −1907.008 | 4.694 |
| 7l | −1906.995 | 2.308 | −1907.011 | 4.211 |
| 7m | −1982.149 | 2.869 | −1982.165 | 4.163 |
| 7n | −1982.151 | 2.933 | −1982.166 | 4.787 |
| Code | EHOMO (eV) | ELUMO (eV) | ∆Egap (eV) | Potential Ionization I (eV) | Affinity A (eV) | Electron Donating Power (ω-) | Electron Accepting Power (ω+) | Electrophilicity (Δω±) |
|---|---|---|---|---|---|---|---|---|
| 7a | −0.2370 | −0.0731 | 0.163 | 0.23707 | 0.0731 | 0.234 | 0.079 | 0.314 |
| 7b | −0.2406 | −0.0713 | 0.169 | 0.24068 | 0.0713 | 0.232 | 0.076 | 0.308 |
| 7c | −0.2337 | −0.0727 | 0.161 | 0.07275 | 0.23379 | 0.079 | 0.232 | 0.312 |
| 7d | −0.2289 | −0.0723 | 0.156 | 0.22892 | 0.07236 | 0.230 | 0.079 | 0.309 |
| 7e | −0.2392 | −0.0703 | 0.168 | 0.23929 | 0.07037 | 0.230 | 0.075 | 0.305 |
| 7f | −0.2378 | −0.0706 | 0.167 | 0.23787 | 0.07069 | 0.230 | 0.076 | 0.306 |
| 7g | −0.2386 | −0.0707 | 0.167 | 0.23866 | 0.07077 | 0.230 | 0.076 | 0.306 |
| 7h | −0.2403 | −0.0712 | 0.169 | 0.24036 | 0.07125 | 0.232 | 0.076 | 0.308 |
| 7i | −0.2255 | −0.0719 | 0.153 | 0.07197 | 0.22553 | 0.079 | 0.228 | 0.307 |
| 7j | −0.2320 | −0.0721 | 0.177 | 0.23009 | 0.07217 | 0.230 | 0.079 | 0.309 |
| 7k | −0.2396 | −0.0707 | 0.168 | 0.23966 | 0.07073 | 0.231 | 0.076 | 0.306 |
| 7l | −0.2294 | −0.0723 | 0.157 | 0.22947 | 0.07231 | 0.230 | 0.036 | 0.199 |
| 7m | −0.2312 | −0.0689 | 0.162 | 0.23125 | 0.06894 | 0.177 | 0.044 | 0.221 |
| 7n | −0.2138 | −0.0716 | 0.142 | 0.21381 | 0.07161 | 0.185 | 0.053 | 0.238 |
| Code | Hardness (η) | Softness (ζ) | Electronegativity (χ) | Chemical Potential (μ) | Electrophilicity Index (ω) |
|---|---|---|---|---|---|
| 7a | 0.082 | 6.099 | 0.155 | −0.155 | 0.147 |
| 7b | 0.085 | 5.904 | 0.156 | −0.156 | 0.144 |
| 7c | −0.081 | −6.210 | 0.153 | −0.153 | −0.146 |
| 7d | 0.078 | 6.387 | 0.151 | −0.151 | 0.145 |
| 7e | 0.084 | 5.920 | 0.155 | −0.155 | 0.142 |
| 7f | 0.084 | 5.982 | 0.154 | −0.154 | 0.142 |
| 7g | 0.084 | 5.956 | 0.155 | −0.155 | 0.143 |
| 7h | 0.085 | 5.913 | 0.156 | −0.156 | 0.144 |
| 7i | −0.077 | −6.512 | 0.149 | −0.149 | −0.144 |
| 7j | 0.085 | 6.332 | 0.151 | −0.151 | 0.145 |
| 7k | 0.084 | 5.920 | 0.155 | −0.155 | 0.143 |
| 7l | 0.079 | 6.363 | 0.151 | −0.151 | 0.145 |
| 7m | 0.081 | 6.161 | 0.150 | −0.150 | 0.139 |
| 7n | 0.071 | 7.032 | 0.143 | −0.143 | 0.143 |
| Code | Vascular Endothelial Growth Factor (VEGFR) | Endothelial Growth Factor Receptor (EGFR) | Selectivity for VEGFR2 | ||
|---|---|---|---|---|---|
| Docking Score (kJ/mole) | Predicted Inhibition Constant (µM) | Docking Score (kJ/mole) | Predicted Inhibition Constant (µM) | ||
| 7a | −41.79 | 0.047 | −32.31 | 2.20 | 46.80 |
| 7b | −42.51 | 0.035 | −32.43 | 2.05 | 58.57 |
| 7c | −44.39 | 0.016 | −32.93 | 1.67 | 104.37 |
| 7d | −43.38 | 0.024 | −33.18 | 1.50 | 62.5 |
| 7e | −44.18 | 0.018 | −34.65 | 0.084 | 4.66 |
| 7f | −40.93 | 3.77 | −31.64 | 2.80 | 0.74 |
| 7g | −46.32 | 0.009 | −31.01 | 3.65 | 405.55 |
| 7h | −44.54 | 0.014 | −31.93 | 2.51 | 179.28 |
| 7i | −44.78 | 0.017 | −33.02 | 0.718 | 42.23 |
| 7j | −48.89 | 0.009 | −33.23 | 1.50 | 166.66 |
| 7k | −43.51 | 0.023 | −31.72 | 2.73 | 118.69 |
| 7l | −45.01 | 0.012 | −34.19 | 1.01 | 84.16 |
| 7m | −42.63 | 0.033 | −27.29 | 6.45 | 195.45 |
| 7n | −42.92 | 0.029 | −32.68 | 1.86 | 64.13 |
| VEGFR2 * | −51.49 | 0.0009 | - | - | - |
| EGFR * | - | - | −23.65 | 71.18 | - |
| Code | % Growth Reduction of HeLa Cells | %Growth Reduction MCF-7 Cells | ||
|---|---|---|---|---|
| After 24 h (µM) | After 48 h (µM) | After 24 h (µM) | After 48 h (µM) | |
| 7a | 30.1 ± 1.23 | 55.6 ± 2.81 | 10.3 ± 0.88 | 49.3 ± 1.66 |
| 7b | 50.1 ± 1.98 | 75.2 ± 3.11 | 54.2 ± 1.78 | 65.1 ± 2.88 |
| 7c | 35.8 ± 0.89 | 51.2 ± 1.98 | 66.1 ± 2.17 | 75.2 ± 1.67 |
| 7d | 60.2 ± 2.89 | 87.4 ± 2.34 | 58.3 ± 1.78 | 83.1 ± 3.11 |
| 7e | 56.8 ± 1.67 | 79.7 ± 2.99 | 69.3 ± 0.76 | 78.9 ± 1.67 |
| 7f | 66.3 ± 4.11 | 72.1 ± 1.78 | 71.2 ± 2.11 | 89.9 ± 3.11 |
| 7g | 70.6 ± 2.98 | 98.1 ± 3.01 | 66.1 ± 1.56 | 96.7 ± 2.89 |
| 7h | 51.4 ± 2.88 | 76.9 ± 1.89 | 28.9 ± 1.67 | 67.5 ± 1.88 |
| 7i | 62.6 ± 2.66 | 82.5 ± 2.04 | 54.7 ± 2.09 | 78.9 ± 2.88 |
| 7j | 67.9 ± 2.78 | 97.8 ± 2.98 | 77.2 ± 1.22 | 89.6 ± 1.98 |
| 7k | 45.1 ± 2.31 | 57.9 ± 2.88 | 81.2 ± 2.12 | 94.3 ± 2.87 |
| 7l | 56.1 ± 2.11 | 89.9 ± 2.65 | 88.8 ± 1.33 | 97.5 ± 3.01 |
| 7m | 43.1 ± 1.99 | 73.1 ± 2.11 | 62.1 ± 1.11 | 88.9 ± 2.11 |
| 7n | 38.9 ± 0.88 | 66.9 ± 1.56 | 57.3 ± 1.58 | 89.3 ± 1.67 |
| Cisplatin | 79.2 ± 2.44 | 89.1 ± 2.44 | 85.2 ± 2.11 | 98.2 ± 1.34 |
| Physicochemical Properties | ||||||||
|---|---|---|---|---|---|---|---|---|
| Molecular Weight | Density | nHA | nHD | TPSA | LogS | LogP | LogD | |
| 7a | 359.05 | 1.064 | 5 | 1 | 68.02 | −5.279 | 4.237 | 4.0 |
| 7b | 373.07 | 1.052 | 5 | 1 | 68.02 | −5.194 | 4.442 | 4.11 |
| 7c | 373.07 | 1.052 | 5 | 1 | 68.02 | −5.572 | 4.69 | 4.176 |
| 7d | 373.07 | 1.052 | 5 | 1 | 68.02 | −5.582 | 4.697 | 4.2 |
| 7e | 387.08 | 1.04 | 5 | 1 | 68.02 | −5.371 | 4.9114 | 4.356 |
| 7f | 387.08 | 1.04 | 5 | 1 | 68.02 | −5.484 | 4.868 | 4.319 |
| 7g | 387.08 | 1.04 | 5 | 1 | 68.02 | −5.462 | 4.914 | 4.343 |
| 7h | 387.08 | 1.04 | 5 | 1 | 68.02 | −5.039 | 4.333 | 4.287 |
| 7i | 387.08 | 1.04 | 5 | 1 | 68.02 | −5.717 | 5.178 | 4.426 |
| 7j | 387.08 | 1.04 | 5 | 1 | 68.02 | −5.865 | 5.135 | 4.265 |
| 7k | 387.08 | 1.04 | 5 | 1 | 68.02 | −5.398 | 4.812 | 4.499 |
| 7l | 387.08 | 1.04 | 5 | 1 | 68.02 | −5.708 | 5.031 | 4.532 |
| 7m | 403.08 | 1.058 | 6 | 1 | 77.25 | −5.38 | 4.439 | 4.269 |
| 7n | 403.08 | 1.058 | 6 | 1 | 77.25 | −5.622 | 4.642 | 4.288 |
| Absorbtion & Distribution Properties | ||||||||
|---|---|---|---|---|---|---|---|---|
| Volume of Distribution (vd) Liters | Human Intestinal Absorption (hia) | Caco-2 Permeability (log) | Blood Brain Barrier (bbb) & Blood-Placenta Barrier (bpb) | Plasma Protein Binding (ppb) % | pgp-Inhibitor | p-Glycoprotein Substrate (pgp-Substrate) | Mdck Permeability (cm/s) | |
| 7a | 2.52 | 0.008 | −4.668 | 0.218 | 99.08 | 0.087 | 0.001 | 1.3 × 10−5 |
| 7b | 2.442 | 0.006 | −4.584 | 0.176 | 99.53 | 0.008 | 0.001 | 1.3 × 10−5 |
| 7c | 2.398 | 0.006 | −4.604 | 0.185 | 99.73 | 0.057 | 0.003 | 1.2 × 10−5 |
| 7d | 2.505 | 0.006 | −4.589 | 0.165 | 99.52 | 0.069 | 0.002 | 1.2 × 10−5 |
| 7e | 2.243 | 0.006 | −4.581 | 0.187 | 96.94 | 0.004 | 0.004 | 1.3 × 10−5 |
| 7f | 2.155 | 0.005 | −4.513 | 0.172 | 98.12 | 0.045 | 0.004 | 1.2 × 10−5 |
| 7g | 2.179 | 0.005 | −4.508 | 0.161 | 100 | 0.056 | 0.005 | 1.2 × 10−5 |
| 7h | 2.817 | 0.005 | −4.548 | 0.159 | 99.99 | 0.007 | 0.002 | 1.3 × 10−5 |
| 7i | 2.418 | 0.006 | −4.611 | 0.16 | 100 | 0.015 | 0.008 | 1.2 × 10−5 |
| 7j | 1.701 | 0.005 | −4.557 | 0.13 | 100 | 0.33 | 0.015 | 1.1 × 10−5 |
| 7k | 2.964 | 0.005 | −4.539 | 0.167 | 99.86 | 0.057 | 0.001 | 1.3 × 10−5 |
| 7l | 2.846 | 0.006 | −4.542 | 0.166 | 99.94 | 0.341 | 0.002 | 1.2 × 10−5 |
| 7m | 2.629 | 0.005 | −4.513 | 0.077 | 99.89 | 0.717 | 0.001 | 1.3 × 10−5 |
| 7n | 2.854 | 0.006 | −4.539 | 0.545 | 100 | 0.627 | 0.001 | 1.2 × 10−5 |
| Metabolism | Excretion | ||||||
|---|---|---|---|---|---|---|---|
| CYP1A2 Inhibitor | CYP2C19 Inhibitor | CYP2C9 Inhibitor | CYP2D6 Inhibitor | CYP3A4 Inhibitor | CL (mL/min.) | T1/2 (h) | |
| 7a | 0.927 | 0.962 | 0.886 | 0.527 | 0.378 | 3.452 | 0.136 |
| 7b | 0.824 | 0.956 | 0.9 | 0.477 | 0.704 | 2.956 | 0.135 |
| 7c | 0.875 | 0.954 | 0.892 | 0.678 | 0.809 | 3.738 | 0.112 |
| 7d | 0.758 | 0.936 | 0.861 | 0.63 | 0.55 | 3.486 | 0.099 |
| 7e | 0.758 | 0.949 | 0.911 | 0.584 | 0.833 | 3.26 | 0.112 |
| 7f | 0.632 | 0.936 | 0.886 | 0.509 | 0.808 | 3.263 | 0.126 |
| 7g | 0.743 | 0.944 | 0.903 | 0.597 | 0.847 | 3.257 | 0.103 |
| 7h | 0.687 | 0.952 | 0.902 | 0.429 | 0.914 | 2.551 | 0.166 |
| 7i | 0.78 | 0.939 | 0.902 | 0.675 | 0.801 | 3.802 | 0.08 |
| 7j | 0.791 | 0.951 | 0.908 | 0.69 | 0.837 | 4.106 | 0.13 |
| 7k | 0.869 | 0.951 | 0.919 | 0.588 | 0.644 | 2.945 | 0.125 |
| 7l | 0.768 | 0.93 | 0.875 | 0.72 | 0.467 | 3.433 | 0.094 |
| 7m | 0.776 | 0.958 | 0.922 | 0.454 | 0.595 | 2.768 | 0.129 |
| 7n | 0.607 | 0.925 | 0.809 | 0.532 | 0.334 | 3.279 | 0.081 |
| Medicinal Properties | Toxicity | ||||||
|---|---|---|---|---|---|---|---|
| Synthetic Accessibility Score | Lipinski Rule | AMES Toxicity | Carcinogenicity | Eye Corrosion | Eye Irritation | Respiratory Toxicity | |
| 7a | 2.331 | None | 0.010 | 0.817 | 0.003 | 0.023 | 0.965 |
| 7b | 2.393 | None | 0.022 | 0.838 | 0.003 | 0.027 | 0.954 |
| 7c | 2.426 | None | 0.014 | 0.846 | 0.003 | 0.018 | 0.952 |
| 7d | 2.379 | None | 0.015 | 0.852 | 0.003 | 0.019 | 0.942 |
| 7e | 2.49 | None | 0.025 | 0.859 | 0.003 | 0.027 | 0.933 |
| 7f | 2.465 | None | 0.016 | 0.866 | 0.003 | 0.022 | 0.933 |
| 7g | 2.477 | None | 0.016 | 0.857 | 0.003 | 0.02 | 0.927 |
| 7h | 2.521 | None | 0.023 | 0.865 | 0.003 | 0.018 | 0.944 |
| 7i | 2.471 | None | 0.012 | 0.881 | 0.003 | 0.021 | 0.926 |
| 7j | 2.547 | None | 0.01 | 0.857 | 0.003 | 0.018 | 0.947 |
| 7k | 2.484 | None | 0.019 | 0.805 | 0.003 | 0.019 | 0.922 |
| 7l | 2.428 | None | 0.013 | 0.846 | 0.003 | 0.017 | 0.89 |
| 7m | 2.476 | None | 0.019 | 0.783 | 0.003 | 0.022 | 0.872 |
| 7n | 2.435 | None | 0.010 | 0.836 | 0.003 | 0.016 | 0.867 |
| TOX21 Pathway | ||||||||
|---|---|---|---|---|---|---|---|---|
| NR-AR | NR-AR-LBD | NR-ER | Antioxidant Response Element | |||||
| Result | Probability | Result | Probability | Result | Probability | Result | Probability | |
| 7a | + | 0.01 | + | 0.156 | - | 0.751 | + | 0.921 |
| 7b | + | 0.013 | + | 0.076 | + | 0.675 | + | 0.91 |
| 7c | + | 0.011 | + | 0.035 | - | 0.745 | + | 0.916 |
| 7d | + | 0.011 | + | 0.042 | - | 0.78 | + | 0.924 |
| 7e | + | 0.021 | + | 0.038 | + | 0.61 | - | 0.912 |
| 7f | + | 0.015 | + | 0.025 | + | 0.693 | + | 0.906 |
| 7g | + | 0.015 | + | 0.024 | + | 0.682 | + | 0.906 |
| 7h | + | 0.023 | + | 0.024 | - | 0.511 | + | 0.88 |
| 7i | + | 0.02 | + | 0.03 | - | 0.732 | + | 0.917 |
| 7j | + | 0.011 | + | 0.014 | - | 0.745 | + | 0.906 |
| 7k | + | 0.012 | + | 0.058 | - | 0.695 | + | 0.908 |
| 7l | + | 0.011 | + | 0.04 | - | 0.801 | + | 0.926 |
| 7m | + | 0.012 | + | 0.271 | - | 0.65 | + | 0.921 |
| 7n | + | 0.004 | + | 0.167 | + | 0.805 | + | 0.936 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bilal, M.S.; Ejaz, S.A.; Zargar, S.; Akhtar, N.; Wani, T.A.; Riaz, N.; Aborode, A.T.; Siddique, F.; Altwaijry, N.; Alkahtani, H.M.; et al. Computational Investigation of 1, 3, 4 Oxadiazole Derivatives as Lead Inhibitors of VEGFR 2 in Comparison with EGFR: Density Functional Theory, Molecular Docking and Molecular Dynamics Simulation Studies. Biomolecules 2022, 12, 1612. https://doi.org/10.3390/biom12111612
Bilal MS, Ejaz SA, Zargar S, Akhtar N, Wani TA, Riaz N, Aborode AT, Siddique F, Altwaijry N, Alkahtani HM, et al. Computational Investigation of 1, 3, 4 Oxadiazole Derivatives as Lead Inhibitors of VEGFR 2 in Comparison with EGFR: Density Functional Theory, Molecular Docking and Molecular Dynamics Simulation Studies. Biomolecules. 2022; 12(11):1612. https://doi.org/10.3390/biom12111612
Chicago/Turabian StyleBilal, Muhammad Sajjad, Syeda Abida Ejaz, Seema Zargar, Naveed Akhtar, Tanveer A. Wani, Naheed Riaz, Adullahi Tunde Aborode, Farhan Siddique, Nojood Altwaijry, Hamad M. Alkahtani, and et al. 2022. "Computational Investigation of 1, 3, 4 Oxadiazole Derivatives as Lead Inhibitors of VEGFR 2 in Comparison with EGFR: Density Functional Theory, Molecular Docking and Molecular Dynamics Simulation Studies" Biomolecules 12, no. 11: 1612. https://doi.org/10.3390/biom12111612
APA StyleBilal, M. S., Ejaz, S. A., Zargar, S., Akhtar, N., Wani, T. A., Riaz, N., Aborode, A. T., Siddique, F., Altwaijry, N., Alkahtani, H. M., & Umar, H. I. (2022). Computational Investigation of 1, 3, 4 Oxadiazole Derivatives as Lead Inhibitors of VEGFR 2 in Comparison with EGFR: Density Functional Theory, Molecular Docking and Molecular Dynamics Simulation Studies. Biomolecules, 12(11), 1612. https://doi.org/10.3390/biom12111612