Reorganization of Cell Compartmentalization Induced by Stress
Abstract
1. Introduction
2. Eukaryotes
2.1. Nuclear MLOs
2.1.1. Nucleolus
2.1.2. Cajal Bodies
2.1.3. Paraspeckles
2.1.4. Nuclear Speckles
2.1.5. PML-Bodies
2.1.6. NELF-Bodies
2.1.7. Nuclear Stress-Bodies
2.1.8. A-Bodies
2.2. Cytoplasmic MLOs
2.2.1. Stress-Granules
2.2.2. P-Bodies
2.3. MLOs Associated with Membrane-Bound Organelles
2.4. Yeast MLOs
| MLO-Type | Main Components | Stress Factors | Structural Changes in Response to Stress | Main Functions |
|---|---|---|---|---|
| Stress granules | mRNA, Pub1, Pbp1, eIF4GII | Impaired proteostasis, genotoxic stress, temperature, UV irradiation, nutrient deprivation, hypoxia, viral infection, etc. | Assembly of gel-like structures in the cytoplasm. | Storage of capped and polyadenylated mRNAs and their protection from degradation in P-bodies. Regulation of TORC1 signaling |
| P-bodies | mRNA, Dcp2p and Pat1p [147] | Nutrient deprivation, oxidative and osmotic stress | Assembly of liquid droplets in the cytoplasm. Yeast P-bodies mRNA and proteins composition depends on the type of stress. | Translation repression and mRNA turnover: 3′-deadenylation, 5′-decapping, 5′-3′ exonuclease activity, nonsense-mediated decay |
| eIF2B bodies | eIF2B | Glucose deprivation | Formation of eIF2B bodies as a result of eIF2B accumulation in the cytoplasm [147]. | Involved in inhibition of translation initiation |
| Proteasome storage granules | Proteasome 19S and 20S subunits [147] | Glucose deprivation | Relocalization of proteasome subunits and formation of proteasome storage granules in the cytoplasm. | Storage of proteasome subunits |
3. Prokaryotes
| Stress-Linked Organelle | Scaffolding Component | Organism | Structural Changes in Response to Stress | Function |
|---|---|---|---|---|
| SSB condensates | Single-stranded DNA-binding protein (SSB) | Escherichia coli | Disassembled in response to stress that causes DNA damage and accumulation of ssDNA. | Serve as storage capsules for SSB protein and other DNA repairing enzymes. |
| Dps condensates | Dps (DNA-binding protein from starved cells) | Escherichia coli | Transform into denser structures in response to stress. | Compact nucleoid during stress conditions, while preserving transcription of genes. |
| BR bodies (containing RNase E) | RNase E endonuclease | Caulobacter crescentus, Sinorhizobium meli-loti, Agrobacterium tumefacienes, Escherichia coli, and Cyanobacteria | Assembled in bacterial cytoplasm in response to stress. | Isolation of untranslated mRNA during stress. Centers for mRNA decay and degradation. |
| BR bodies (containing RNase Y) [173] | RNase Y endonuclease | Bacillus subtilis | Assembled in bacterial cytoplasm in response to stress. | Isolation of untranslated mRNA during stress. Centers for mRNA decay and degradation. |
| BR bodies (containing RNase J) [174] | RNase J endonuclease | Helicobacter pylori | Assembled in bacterial cytoplasm in response to stress. | Isolation of untranslated mRNA during stress. Centers for mRNA decay and degradation. |
| Granular bodies | IbpA heat shock protein | Acholeplasma laidlawii | Assembled in response to stress. | Regulation of heat shock response. |
| PolyP granules [175] | polyphosphate (polyP) | Pseudomonas aeruginosa | Assembled under nitrogen starvation. | Regulation of bacterial cell cycle exit during starvation survival response. |
4. Factors Regulating Reorganization of MLOs in Stress Response
4.1. “Physical” Factors
4.2. “Biological” Factors
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sekine, Y.; Houston, R.; Sekine, S. Cellular metabolic stress responses via organelles. Exp. Cell Res. 2021, 400, 112515. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Zhou, M.; Chen, S.; Li, D.; Cao, X.; Liu, B. Effects of pH alterations on stress- and aging-induced protein phase separation. Cell. Mol. Life Sci. 2022, 79, 380. [Google Scholar] [CrossRef]
- Dutta, N.; Garcia, G.; Higuchi-Sanabria, R. Hijacking Cellular Stress Responses to Promote Lifespan. Front. Aging 2022, 3, 860404. [Google Scholar] [CrossRef] [PubMed]
- Fulda, S.; Gorman, A.M.; Hori, O.; Samali, A. Cellular stress responses: Cell survival and cell death. Int. J. Cell Biol. 2010, 2010, 214074. [Google Scholar] [CrossRef]
- Antifeeva, I.A.; Fonin, A.V.; Fefilova, A.S.; Stepanenko, O.V.; Povarova, O.I.; Silonov, S.A.; Kuznetsova, I.M.; Uversky, V.N.; Turoverov, K.K. Liquid–liquid phase separation as an organizing principle of intracellular space: Overview of the evolution of the cell compartmentalization concept. Cell. Mol. Life Sci. 2022, 79, 251. [Google Scholar] [CrossRef]
- Fefilova, A.S.; Fonin, A.V.; Vishnyakov, I.E.; Kuznetsova, I.M.; Turoverov, K.K. Stress-Induced Membraneless Organelles in Eukaryotes and Prokaryotes: Bird’s-Eye View. Int J. Mol. Sci. 2022, 23, 5010. [Google Scholar] [CrossRef]
- Turoverov, K.K.; Kuznetsova, I.M.; Fonin, A.V.; Darling, A.L.; Zaslavsky, B.Y.; Uversky, V.N. Stochasticity of Biological Soft Matter: Emerging Concepts in Intrinsically Disordered Proteins and Biological Phase Separation. Trends Biochem. Sci. 2019, 44, 716–728. [Google Scholar] [CrossRef] [PubMed]
- Fonin, A.V.; Darling, A.L.; Kuznetsova, I.M.; Turoverov, K.K.; Uversky, V.N. Intrinsically disordered proteins in crowded milieu: When chaos prevails within the cellular gumbo. Cell. Mol. Life Sci. 2018, 75, 3907–3929. [Google Scholar] [CrossRef]
- Uversky, V.N. Functional roles of transiently and intrinsically disordered regions within proteins. FEBS J. 2015, 282, 1182–1189. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Intrinsically Disordered Proteins and Their “Mysterious” (Meta)Physics. Front. Phys. 2019, 7, 10. [Google Scholar] [CrossRef]
- van Leeuwen, W.; Rabouille, C. Cellular stress leads to the formation of membraneless stress assemblies in eukaryotic cells. Traffic 2019, 20, 623–638. [Google Scholar] [CrossRef]
- Prouteau, M.; Loewith, R. Regulation of Cellular Metabolism through Phase Separation of Enzymes. Biomolecules 2018, 8, 160. [Google Scholar] [CrossRef]
- Hnisz, D.; Shrinivas, K.; Young, R.A.; Chakraborty, A.K.; Sharp, P.A. A Phase Separation Model for Transcriptional Control. Cell 2017, 169, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Sirri, V.; Urcuqui-Inchima, S.; Roussel, P.; Hernandez-Verdun, D. Nucleolus: The fascinating nuclear body. Histochem. Cell Biol. 2008, 129, 13–31. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Yang, J.; Yi, J. Nucleolar Stress: Hallmarks, sensing mechanism and diseases. Cell Stress 2018, 2, 125–140. [Google Scholar] [CrossRef] [PubMed]
- Boulon, S.; Westman, B.J.; Hutten, S.; Boisvert, F.M.; Lamond, A.I. The nucleolus under stress. Mol. Cell 2010, 40, 216–227. [Google Scholar] [CrossRef] [PubMed]
- Al-Baker, E.A.; Oshin, M.; Hutchison, C.J.; Kill, I.R. Analysis of UV-induced damage and repair in young and senescent human dermal fibroblasts using the comet assay. Mech. Ageing Dev. 2005, 126, 664–672. [Google Scholar] [CrossRef]
- Govoni, M.; Farabegoli, F.; Pession, A.; Novello, F. Inhibition of topoisomerase II activity and its effect on nucleolar structure and function. Exp. Cell Res. 1994, 211, 36–41. [Google Scholar] [CrossRef]
- Shav-Tal, Y.; Blechman, J.; Darzacq, X.; Montagna, C.; Dye, B.T.; Patton, J.G.; Singer, R.H.; Zipori, D. Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol. Biol. Cell 2005, 16, 2395–2413. [Google Scholar] [CrossRef]
- David-Pfeuty, T.r.s. Potent inhibitors of cyclin-dependent kinase 2 induce nuclear accumulation of wild-type p53 and nucleolar fragmentation in human untransformed and tumor-derived cells. Oncogene 1999, 18, 7409–7422. [Google Scholar] [CrossRef][Green Version]
- Haaf, T.; Ward, D.C. Inhibition of RNA polymerase II transcription causes chromatin decondensation, loss of nucleolar structure, and dispersion of chromosomal domains. Exp. Cell Res. 1996, 224, 163–173. [Google Scholar] [CrossRef]
- Rubbi, C.P.; Milner, J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 2003, 22, 6068–6077. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lu, H. Signaling to p53: Ribosomal proteins find their way. Cancer Cell 2009, 16, 369–377. [Google Scholar] [CrossRef] [PubMed]
- Lindström, M.S. Emerging functions of ribosomal proteins in gene-specific transcription and translation. Biochem. Biophys. Res. Commun. 2009, 379, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Ofir-Rosenfeld, Y.; Boggs, K.; Michael, D.; Kastan, M.B.; Oren, M. Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Mol. Cell 2008, 32, 180–189. [Google Scholar] [CrossRef]
- Dove, B.K.; You, J.H.; Reed, M.L.; Emmett, S.R.; Brooks, G.; Hiscox, J.A. Changes in nucleolar morphology and proteins during infection with the coronavirus infectious bronchitis virus. Cell. Microbiol. 2006, 8, 1147–1157. [Google Scholar] [CrossRef] [PubMed]
- Eskiw, C.H.; Dellaire, G.; Mymryk, J.S.; Bazett-Jones, D.P. Size, position and dynamic behavior of PML nuclear bodies following cell stress as a paradigm for supramolecular trafficking and assembly. J. Cell Sci. 2003, 116, 4455–4466. [Google Scholar] [CrossRef]
- Chan, P.K.; Aldrich, M.; Busch, H. Alterations in immunolocalization of the phosphoprotein B23 in HeLa cells during serum starvation. Exp. Cell Res. 1985, 161, 101–110. [Google Scholar] [CrossRef]
- Trinkle-Mulcahy, L.; Sleeman, J.E. The Cajal body and the nucleolus: “In a relationship” or “It’s complicated”? RNA Biol. 2017, 14, 739–751. [Google Scholar] [CrossRef]
- Matera, A.G. Nuclear bodies: Multifaceted subdomains of the interchromatin space. Trends Cell Biol. 1999, 9, 302–309. [Google Scholar] [CrossRef]
- Cioce, M.; Lamond, A.I. Cajal bodies: A long history of discovery. Annu. Rev. Cell Dev. Biol. 2005, 21, 105–131. [Google Scholar] [CrossRef]
- Walker, M.P.; Tian, L.; Matera, A.G. Reduced viability, fertility and fecundity in mice lacking the cajal body marker protein, coilin. PLoS ONE 2009, 4, e6171. [Google Scholar] [CrossRef]
- Deryusheva, S.; Gall, J.G. Small Cajal body-specific RNAs of Drosophila function in the absence of Cajal bodies. Mol. Biol. Cell 2009, 20, 5250–5259. [Google Scholar] [CrossRef]
- Collier, S.; Pendle, A.; Boudonck, K.; van Rij, T.; Dolan, L.; Shaw, P. A distant coilin homologue is required for the formation of cajal bodies in Arabidopsis. Mol. Biol. Cell 2006, 17, 2942–2951. [Google Scholar] [CrossRef]
- Tucker, K.E.; Berciano, M.T.; Jacobs, E.Y.; LePage, D.F.; Shpargel, K.B.; Rossire, J.J.; Chan, E.K.; Lafarga, M.; Conlon, R.A.; Matera, A.G. Residual Cajal bodies in coilin knockout mice fail to recruit Sm snRNPs and SMN, the spinal muscular atrophy gene product. J. Cell Biol. 2001, 154, 293–307. [Google Scholar] [CrossRef]
- Strzelecka, M.; Trowitzsch, S.; Weber, G.; Lührmann, R.; Oates, A.C.; Neugebauer, K.M. Coilin-dependent snRNP assembly is essential for zebrafish embryogenesis. Nat. Struct. Mol. Biol. 2010, 17, 403–409. [Google Scholar] [CrossRef]
- Love, A.J.; Yu, C.; Petukhova, N.V.; Kalinina, N.O.; Chen, J.; Taliansky, M.E. Cajal bodies and their role in plant stress and disease responses. RNA Biol. 2017, 14, 779–790. [Google Scholar] [CrossRef]
- Cioce, M.; Boulon, S.; Matera, A.G.; Lamond, A.I. UV-induced fragmentation of Cajal bodies. J. Cell Biol. 2006, 175, 401–413. [Google Scholar] [CrossRef]
- Handwerger, K.E.; Wu, Z.; Murphy, C.; Gall, J.G. Heat shock induces mini-Cajal bodies in the Xenopus germinal vesicle. J. Cell Sci. 2002, 115, 2011–2020. [Google Scholar] [CrossRef]
- Bongiorno-Borbone, L.; De Cola, A.; Barcaroli, D.; Knight, R.A.; Di Ilio, C.; Melino, G.; De Laurenzi, V. FLASH degradation in response to UV-C results in histone locus bodies disruption and cell-cycle arrest. Oncogene 2010, 29, 802–810. [Google Scholar] [CrossRef]
- Andrade, L.E.; Tan, E.M.; Chan, E.K. Immunocytochemical analysis of the coiled body in the cell cycle and during cell proliferation. Proc. Natl. Acad. Sci. USA 1993, 90, 1947–1951. [Google Scholar] [CrossRef]
- Stępiński, D. Cajal body dynamics in soybean root meristem cells under chilling stress and recovery. Environ. Exp. Bot. 2020, 180, 104241. [Google Scholar] [CrossRef]
- Navascues, J.; Bengoechea, R.; Tapia, O.; Casafont, I.; Berciano, M.T.; Lafarga, M. SUMO-1 transiently localizes to Cajal bodies in mammalian neurons. J. Struct. Biol. 2008, 163, 137–146. [Google Scholar] [CrossRef]
- Morency, E.; Sabra, M.; Catez, F.; Texier, P.; Lomonte, P. A novel cell response triggered by interphase centromere structural instability. J. Cell Biol. 2007, 177, 757–768. [Google Scholar] [CrossRef]
- James, N.J.; Howell, G.J.; Walker, J.H.; Blair, G.E. The role of Cajal bodies in the expression of late phase adenovirus proteins. Virology 2010, 399, 299–311. [Google Scholar] [CrossRef]
- Kim, S.H.; Ryabov, E.V.; Kalinina, N.O.; Rakitina, D.V.; Gillespie, T.; MacFarlane, S.; Haupt, S.; Brown, J.W.S.; Taliansky, M. Cajal bodies and the nucleolus are required for a plant virus systemic infection. EMBO J. 2007, 26, 2169–2179. [Google Scholar] [CrossRef]
- Sasaki, Y.T.F.; Ideue, T.; Sano, M.; Mituyama, T.; Hirose, T. MENε/β noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc. Natl. Acad. Sci. USA 2009, 106, 2525–2530. [Google Scholar] [CrossRef]
- Nakagawa, S.; Naganuma, T.; Shioi, G.; Hirose, T. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J. Cell Biol. 2011, 193, 31–39. [Google Scholar] [CrossRef]
- McCluggage, F.; Fox, A.H. Paraspeckle nuclear condensates: Global sensors of cell stress? BioEssays News Rev. Mol. Cell. Dev. Biol. 2021, 43, e2000245. [Google Scholar] [CrossRef]
- Yamazaki, T.; Yamamoto, T.; Yoshino, H.; Souquere, S.; Nakagawa, S.; Pierron, G.; Hirose, T. Paraspeckles are constructed as block copolymer micelles. Embo J. 2021, 40, e107270. [Google Scholar] [CrossRef]
- Nakagawa, S.; Shimada, M.; Yanaka, K.; Mito, M.; Arai, T.; Takahashi, E.; Fujita, Y.; Fujimori, T.; Standaert, L.; Marine, J.C.; et al. The lncRNA Neat1 is required for corpus luteum formation and the establishment of pregnancy in a subpopulation of mice. Development 2014, 141, 4618–4627. [Google Scholar] [CrossRef] [PubMed]
- Choudhry, H.; Albukhari, A.; Morotti, M.; Haider, S.; Moralli, D.; Smythies, J.; Schödel, J.; Green, C.M.; Camps, C.; Buffa, F.; et al. Tumor hypoxia induces nuclear paraspeckle formation through HIF-2α dependent transcriptional activation of NEAT1 leading to cancer cell survival. Oncogene 2015, 34, 4482–4490. [Google Scholar] [CrossRef] [PubMed]
- Lellahi, S.M.; Rosenlund, I.A.; Hedberg, A.; Kiær, L.T.; Mikkola, I.; Knutsen, E.; Perander, M. The long noncoding RNA NEAT1 and nuclear paraspeckles are up-regulated by the transcription factor HSF1 in the heat shock response. J. Biol. Chem. 2018, 293, 18965–18976. [Google Scholar] [CrossRef] [PubMed]
- Todorovski, V.; Fox, A.H.; Choi, Y.S. Matrix stiffness-sensitive long noncoding RNA NEAT1 seeded paraspeckles in cancer cells. Mol. Biol. Cell 2020, 31, 1654–1662. [Google Scholar] [CrossRef]
- Adriaens, C.; Standaert, L.; Barra, J.; Latil, M.; Verfaillie, A.; Kalev, P.; Boeckx, B.; Wijnhoven, P.W.G.; Radaelli, E.; Vermi, W.; et al. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat. Med. 2016, 22, 861–868. [Google Scholar] [CrossRef]
- Wang, Y.; Hu, S.B.; Wang, M.R.; Yao, R.W.; Wu, D.; Yang, L.; Chen, L.L. Genome-wide screening of NEAT1 regulators reveals cross-regulation between paraspeckles and mitochondria. Nat. Cell Biol. 2018, 20, 1145–1158. [Google Scholar] [CrossRef]
- Imamura, K.; Imamachi, N.; Akizuki, G.; Kumakura, M.; Kawaguchi, A.; Nagata, K.; Kato, A.; Kawaguchi, Y.; Sato, H.; Yoneda, M.; et al. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol. Cell 2014, 53, 393–406. [Google Scholar] [CrossRef]
- Beeharry, Y.; Goodrum, G.; Imperiale, C.J.; Pelchat, M. The Hepatitis Delta Virus accumulation requires paraspeckle components and affects NEAT1 level and PSP1 localization. Sci. Rep. 2018, 8, 6031. [Google Scholar] [CrossRef]
- Ma, H.; Han, P.; Ye, W.; Chen, H.; Zheng, X.; Cheng, L.; Zhang, L.; Yu, L.; Wu, X.; Xu, Z.; et al. The Long Noncoding RNA NEAT1 Exerts Antihantaviral Effects by Acting as Positive Feedback for RIG-I Signaling. J. Virol. 2017, 91, e02250–e16. [Google Scholar] [CrossRef]
- Zhang, Q.; Chen, C.Y.; Yedavalli, V.S.; Jeang, K.T. NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression. Mbio 2013, 4, e00596-00512. [Google Scholar] [CrossRef]
- Imamura, K.; Takaya, A.; Ishida, Y.I.; Fukuoka, Y.; Taya, T.; Nakaki, R.; Kakeda, M.; Imamachi, N.; Sato, A.; Yamada, T.; et al. Diminished nuclear RNA decay upon Salmonella infection upregulates antibacterial noncoding RNAs. Embo J. 2018, 37, e97723. [Google Scholar] [CrossRef] [PubMed]
- Spector, D.L.; Lamond, A.I. Nuclear speckles. Cold Spring Harb. Perspect. Biol. 2011, 3, a000646. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, V.; Ellis, J.D.; Shen, Z.; Song, D.Y.; Pan, Q.; Watt, A.T.; Freier, S.M.; Bennett, C.F.; Sharma, A.; Bubulya, P.A.; et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 2010, 39, 925–938. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Arun, G.; Mao, Y.S.; Lazar, Z.; Hung, G.; Bhattacharjee, G.; Xiao, X.; Booth, C.J.; Wu, J.; Zhang, C.; et al. The lncRNA Malat1 Is Dispensable for Mouse Development but Its Transcription Plays a cis-Regulatory Role in the Adult. Cell Rep. 2012, 2, 111–123. [Google Scholar] [CrossRef]
- Engreitz, J.M.; Sirokman, K.; McDonel, P.; Shishkin, A.A.; Surka, C.; Russell, P.; Grossman, S.R.; Chow, A.Y.; Guttman, M.; Lander, E.S. RNA-RNA Interactions Enable Specific Targeting of Noncoding RNAs to Nascent Pre-mRNAs and Chromatin Sites. Cell 2014, 159, 188–199. [Google Scholar] [CrossRef]
- West, J.A.; Davis, C.P.; Sunwoo, H.; Simon, M.D.; Sadreyev, R.I.; Wang, P.I.; Tolstorukov, M.Y.; Kingston, R.E. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell 2014, 55, 791–802. [Google Scholar] [CrossRef]
- Quinodoz, S.A.; Ollikainen, N.; Tabak, B.; Palla, A.; Schmidt, J.M.; Detmar, E.; Lai, M.M.; Shishkin, A.A.; Bhat, P.; Takei, Y.; et al. Higher-Order Inter-chromosomal Hubs Shape 3D Genome Organization in the Nucleus. Cell 2018, 174, 744–757.e724. [Google Scholar] [CrossRef]
- Kim, J.; Venkata, N.C.; Hernandez Gonzalez, G.A.; Khanna, N.; Belmont, A.S. Gene expression amplification by nuclear speckle association. J. Cell Biol. 2019, 219, e201904046. [Google Scholar] [CrossRef]
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef]
- Dutertre, M.; Sanchez, G.; Barbier, J.; Corcos, L.; Auboeuf, D. The emerging role of pre-messenger RNA splicing in stress responses: Sending alternative messages and silent messengers. RNA Biol. 2011, 8, 740–747. [Google Scholar] [CrossRef]
- Biamonti, G.; Caceres, J.F. Cellular stress and RNA splicing. Trends Biochem. Sci. 2009, 34, 146–153. [Google Scholar] [CrossRef]
- Spector, D.L.; Fu, X.D.; Maniatis, T. Associations between distinct pre-mRNA splicing components and the cell nucleus. Embo J. 1991, 10, 3467–3481. [Google Scholar] [CrossRef] [PubMed]
- Melcák, I.; Cermanová, S.; Jirsová, K.; Koberna, K.; Malínský, J.; Raska, I. Nuclear pre-mRNA compartmentalization: Trafficking of released transcripts to splicing factor reservoirs. Mol. Biol. Cell 2000, 11, 497–510. [Google Scholar] [CrossRef]
- Kim, J.; Han, K.Y.; Khanna, N.; Ha, T.; Belmont, A.S. Nuclear speckle fusion via long-range directional motion regulates speckle morphology after transcriptional inhibition. J. Cell Sci. 2019, 132, jcs226563. [Google Scholar] [CrossRef] [PubMed]
- Raina, K.; Rao, B.J. Mammalian nuclear speckles exhibit stable association with chromatin: A biochemical study. Nucleus 2022, 13, 58–73. [Google Scholar] [CrossRef] [PubMed]
- Spector, D.L.; O’Keefe, R.T.; Jiménez-García, L.F. Dynamics of transcription and pre-mRNA splicing within the mammalian cell nucleus. Cold Spring Harb. Symp. Quant. Biol. 1993, 58, 799–805. [Google Scholar] [CrossRef] [PubMed]
- Corpet, A.; Kleijwegt, C.; Roubille, S.; Juillard, F.; Jacquet, K.; Texier, P.; Lomonte, P. PML nuclear bodies and chromatin dynamics: Catch me if you can! Nucleic Acids Res. 2020, 48, 11890–11912. [Google Scholar] [CrossRef] [PubMed]
- Guan, D.; Kao, H.Y. The function, regulation and therapeutic implications of the tumor suppressor protein, PML. Cell Biosci. 2015, 5, 60. [Google Scholar] [CrossRef]
- Hands, K.J.; Cuchet-Lourenco, D.; Everett, R.D.; Hay, R.T. PML isoforms in response to arsenic: High-resolution analysis of PML body structure and degradation. J. Cell Sci. 2014, 127, 365–375. [Google Scholar] [CrossRef] [PubMed]
- Nisole, S.; Maroui, M.A.; Mascle, X.H.; Aubry, M.; Chelbi-Alix, M.K. Differential Roles of PML Isoforms. Front. Oncol. 2013, 3, 125. [Google Scholar] [CrossRef] [PubMed]
- Fonin, A.V.; Silonov, S.A.; Shpironok, O.G.; Antifeeva, I.A.; Petukhov, A.V.; Romanovich, A.E.; Kuznetsova, I.M.; Uversky, V.N.; Turoverov, K.K. The Role of Non-Specific Interactions in Canonical and ALT-Associated PML-Bodies Formation and Dynamics. Int. J. Mol. Sci. 2021, 22, 5821. [Google Scholar] [CrossRef]
- Fonin, A.V.; Silonov, S.A.; Fefilova, A.S.; Stepanenko, O.V.; Gavrilova, A.A.; Petukhov, A.V.; Romanovich, A.E.; Modina, A.L.; Zueva, T.S.; Nedelyaev, E.M.; et al. New Evidence of the Importance of Weak Interactions in the Formation of PML-Bodies. Int. J. Mol. Sci. 2022, 23, 1613. [Google Scholar] [CrossRef]
- Niwa-Kawakita, M.; Wu, H.C.; Thé, H.; Lallemand-Breitenbach, V. PML nuclear bodies, membrane-less domains acting as ROS sensors? Semin. Cell Dev. Biol. 2018, 80, 29–34. [Google Scholar] [CrossRef]
- Rufini, A.; Tucci, P.; Celardo, I.; Melino, G. Senescence and aging: The critical roles of p53. Oncogene 2013, 32, 5129–5143. [Google Scholar] [CrossRef] [PubMed]
- Matt, S.; Hofmann, T.G. Crosstalk between p53 modifiers at PML bodies. Mol. Cell. Oncol. 2018, 5, e1074335. [Google Scholar] [CrossRef]
- Trotman, L.C.; Alimonti, A.; Scaglioni, P.P.; Koutcher, J.A.; Cordon-Cardo, C.; Pandolfi, P.P. Identification of a tumour suppressor network opposing nuclear Akt function. Nature 2006, 441, 523–527. [Google Scholar] [CrossRef] [PubMed]
- Sahin, U.; Ferhi, O.; Jeanne, M.; Benhenda, S.; Berthier, C.; Jollivet, F.; Niwa-Kawakita, M.; Faklaris, O.; Setterblad, N.; de Thé, H.; et al. Oxidative stress–induced assembly of PML nuclear bodies controls sumoylation of partner proteins. J. Cell Biol. 2014, 204, 931–945. [Google Scholar] [CrossRef]
- Sahin, U.; de Thé, H.; Lallemand-Breitenbach, V. PML nuclear bodies: Assembly and oxidative stress-sensitive sumoylation. Nucleus 2014, 5, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Guan, D.; Lim, J.H.; Peng, L.; Liu, Y.; Lam, M.; Seto, E.; Kao, H.Y. Deacetylation of the tumor suppressor protein PML regulates hydrogen peroxide-induced cell death. Cell Death Dis. 2014, 5, e1340. [Google Scholar] [CrossRef]
- Saito, M.; Hess, D.; Eglinger, J.; Fritsch, A.W.; Kreysing, M.; Weinert, B.T.; Choudhary, C.; Matthias, P. Acetylation of intrinsically disordered regions regulates phase separation. Nat. Chem. Biol. 2019, 15, 51–61. [Google Scholar] [CrossRef]
- Lång, A.; Eriksson, J.; Schink, K.O.; Lång, E.; Blicher, P.; Połeć, A.; Brech, A.; Dalhus, B.; Bøe, S.O. Visualization of PML nuclear import complexes reveals FG-repeat nucleoporins at cargo retrieval sites. Nucleus 2017, 8, 404–420. [Google Scholar] [CrossRef] [PubMed]
- Lång, A.; Lång, E.; Bøe, S.O. PML Bodies in Mitosis. Cells 2019, 8, 893. [Google Scholar] [CrossRef]
- Rawat, P.; Boehning, M.; Hummel, B.; Aprile-Garcia, F.; Pandit, A.S.; Eisenhardt, N.; Khavaran, A.; Niskanen, E.; Vos, S.M.; Palvimo, J.J.; et al. Stress-induced nuclear condensation of NELF drives transcriptional downregulation. Mol. Cell 2021, 81, 1013–1026 e1011. [Google Scholar] [CrossRef] [PubMed]
- Ngian, Z.K.; Lin, W.Q.; Ong, C.T. NELF-A controls Drosophila healthspan by regulating heat-shock protein-mediated cellular protection and heterochromatin maintenance. Aging Cell 2021, 20, e13348. [Google Scholar] [CrossRef] [PubMed]
- Goenka, A.; Sengupta, S.; Pandey, R.; Parihar, R.; Mohanta, G.C.; Mukerji, M.; Ganesh, S. Human satellite-III non-coding RNAs modulate heat-shock-induced transcriptional repression. J. Cell Sci. 2016, 129, 3541–3552. [Google Scholar] [CrossRef] [PubMed]
- Jolly, C.; Metz, A.; Govin, J.r.m.; Vigneron, M.; Turner, B.M.; Khochbin, S.; Vourc’h, C. Stress-induced transcription of satellite III repeats. J. Cell Biol. 2003, 164, 25–33. [Google Scholar] [CrossRef]
- Gaglia, G.; Rashid, R.; Yapp, C.; Joshi, G.N.; Li, C.G.; Lindquist, S.L.; Sarosiek, K.A.; Whitesell, L.; Sorger, P.K.; Santagata, S. HSF1 phase transition mediates stress adaptation and cell fate decisions. Nat. Cell Biol. 2020, 22, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Aly, M.K.; Ninomiya, K.; Adachi, S.; Natsume, T.; Hirose, T. Two distinct nuclear stress bodies containing different sets of RNA-binding proteins are formed with HSATIII architectural noncoding RNAs upon thermal stress exposure. Biochem. Biophys. Res. Commun. 2019, 516, 419–423. [Google Scholar] [CrossRef] [PubMed]
- Audas, T.E.; Audas, D.E.; Jacob, M.D.; Ho, J.J.; Khacho, M.; Wang, M.; Perera, J.K.; Gardiner, C.; Bennett, C.A.; Head, T.; et al. Adaptation to Stressors by Systemic Protein Amyloidogenesis. Dev. Cell 2016, 39, 155–168. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Tao, X.; Jacob, M.D.; Bennett, C.A.; Ho, J.J.D.; Gonzalgo, M.L.; Audas, T.E.; Lee, S. Stress-Induced Low Complexity RNA Activates Physiological Amyloidogenesis. Cell Rep. 2018, 24, 1713–1721 e1714. [Google Scholar] [CrossRef] [PubMed]
- Marijan, D.; Tse, R.; Elliott, K.; Chandhok, S.; Luo, M.; Lacroix, E.; Audas, T.E. Stress-specific aggregation of proteins in the amyloid bodies. FEBS Lett. 2019, 593, 3162–3172. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Bokros, M.; Theodoridis, P.R.; Lee, S. Nucleolar Sequestration: Remodeling Nucleoli Into Amyloid Bodies. Front. Genet. 2019, 10, 1179. [Google Scholar] [CrossRef]
- Mahboubi, H.; Stochaj, U. Cytoplasmic stress granules: Dynamic modulators of cell signaling and disease. Biochim. Et Biophys. Acta. Mol. Basis Dis. 2017, 1863, 884–895. [Google Scholar] [CrossRef] [PubMed]
- Khong, A.; Matheny, T.; Jain, S.; Mitchell, S.F.; Wheeler, J.R.; Parker, R. The Stress Granule Transcriptome Reveals Principles of mRNA Accumulation in Stress Granules. Mol. Cell 2017, 68, 808–820.e805. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.; Sumi, S.; Seervi, M. Heat shock proteins-driven stress granule dynamics: Yet another avenue for cell survival. Apoptosis Int. J. Program. Cell Death 2021, 26, 371–384. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, P.; Kedersha, N.; Anderson, P. Stress Granules and Processing Bodies in Translational Control. Cold Spring Harb Perspect. Biol. 2019, 11, a032813. [Google Scholar] [CrossRef]
- Wolozin, B.; Ivanov, P. Stress granules and neurodegeneration. Nat. Rev. Neurosci. 2019, 20, 649–666. [Google Scholar] [CrossRef]
- Asadi, M.R.; Rahmanpour, D.; Moslehian, M.S.; Sabaie, H.; Hassani, M.; Ghafouri-Fard, S.; Taheri, M.; Rezazadeh, M. Stress Granules Involved in Formation, Progression and Metastasis of Cancer: A Scoping Review. Front. Cell Dev. Biol. 2021, 9, 745394. [Google Scholar] [CrossRef]
- Poblete-Durán, N.; Prades-Pérez, Y.; Vera-Otarola, J.; Soto-Rifo, R.; Valiente-Echeverría, F. Who Regulates Whom? An Overview of RNA Granules and Viral Infections. Viruses 2016, 8, 180. [Google Scholar] [CrossRef]
- Protter, D.S.W.; Parker, R. Principles and Properties of Stress Granules. Trends Cell Biol. 2016, 26, 668–679. [Google Scholar] [CrossRef]
- Wheeler, J.R.; Matheny, T.; Jain, S.; Abrisch, R.; Parker, R. Distinct stages in stress granule assembly and disassembly. eLife 2016, 5, e18413. [Google Scholar] [CrossRef] [PubMed]
- Moujaber, O.; Mahboubi, H.; Kodiha, M.; Bouttier, M.; Bednarz, K.; Bakshi, R.; White, J.; Larose, L.; Colmegna, I.; Stochaj, U. Dissecting the molecular mechanisms that impair stress granule formation in aging cells. Biochim. Et Biophys. Acta. Mol. Cell Res. 2017, 1864, 475–486. [Google Scholar] [CrossRef] [PubMed]
- Omer, A.; Patel, D.; Moran, J.L.; Lian, X.J.; Di Marco, S.; Gallouzi, I.E. Autophagy and heat-shock response impair stress granule assembly during cellular senescence. Mech. Ageing Dev. 2020, 192, 111382. [Google Scholar] [CrossRef] [PubMed]
- Maharjan, N.; Künzli, C.; Buthey, K.; Saxena, S. C9ORF72 Regulates Stress Granule Formation and Its Deficiency Impairs Stress Granule Assembly, Hypersensitizing Cells to Stress. Mol. Neurobiol. 2017, 54, 3062–3077. [Google Scholar] [CrossRef]
- Zhao, Y.G.; Codogno, P.; Zhang, H. Machinery, regulation and pathophysiological implications of autophagosome maturation. Nat. Rev. Mol. Cell Biol. 2021, 22, 733–750. [Google Scholar] [CrossRef]
- Cao, X.; Jin, X.; Liu, B. The involvement of stress granules in aging and aging-associated diseases. Aging Cell 2020, 19, e13136. [Google Scholar] [CrossRef]
- Elbaum-Garfinkle, S. Matter over mind: Liquid phase separation and neurodegeneration. J. Biol. Chem. 2019, 294, 7160–7168. [Google Scholar] [CrossRef]
- Namkoong, S.; Ho, A.; Woo, Y.M.; Kwak, H.; Lee, J.H. Systematic Characterization of Stress-Induced RNA Granulation. Mol. Cell 2018, 70, 175–187.e178. [Google Scholar] [CrossRef]
- Ries, R.J.; Pickering, B.F.; Poh, H.X.; Namkoong, S.; Jaffrey, S.R. m6A governs length-dependent enrichment of mRNAs in stress granules. bioRxiv 2022. [Google Scholar] [CrossRef]
- Khong, A.; Matheny, T.; Huynh, T.N.; Babl, V.; Parker, R. Limited effects of m6A modification on mRNA partitioning into stress granules. Nat. Commun. 2022, 13, 3735. [Google Scholar] [CrossRef]
- Poh, H.X.; Mirza, A.H.; Pickering, B.F.; Jaffrey, S.R. Understanding the source of METTL3-independent m6A in mRNA. bioRxiv 2021. [Google Scholar] [CrossRef]
- Riggs, C.L.; Kedersha, N.; Ivanov, P.; Anderson, P. Mammalian stress granules and P bodies at a glance. J. Cell Sci. 2020, 133, jcs242487. [Google Scholar] [CrossRef] [PubMed]
- Kedersha, N.; Anderson, P. Mammalian stress granules and processing bodies. Methods Enzymol. 2007, 431, 61–81. [Google Scholar] [CrossRef]
- Luo, Y.; Na, Z.; Slavoff, S.A. P-Bodies: Composition, Properties, and Functions. Biochemistry 2018, 57, 2424–2431. [Google Scholar] [CrossRef]
- Souquere, S.; Mollet, S.; Kress, M.; Dautry, F.; Pierron, G.; Weil, D. Unravelling the ultrastructure of stress granules and associated P-bodies in human cells. J. Cell Sci. 2009, 122, 3619–3626. [Google Scholar] [CrossRef] [PubMed]
- Ayache, J.; Bénard, M.; Ernoult-Lange, M.; Minshall, N.; Standart, N.; Kress, M.; Weil, D. P-body assembly requires DDX6 repression complexes rather than decay or Ataxin2/2L complexes. Mol. Biol Cell 2015, 26, 2579–2595. [Google Scholar] [CrossRef] [PubMed]
- Hubstenberger, A.; Courel, M.; Bénard, M.; Souquere, S.; Ernoult-Lange, M.; Chouaib, R.; Yi, Z.; Morlot, J.B.; Munier, A.; Fradet, M.; et al. P-Body Purification Reveals the Condensation of Repressed mRNA Regulons. Mol. Cell 2017, 68, 144–157 e145. [Google Scholar] [CrossRef]
- Youn, J.Y.; Dyakov, B.J.A.; Zhang, J.; Knight, J.D.R.; Vernon, R.M.; Forman-Kay, J.D.; Gingras, A.C. Properties of Stress Granule and P-Body Proteomes. Mol. Cell 2019, 76, 286–294. [Google Scholar] [CrossRef] [PubMed]
- Gallois-Montbrun, S.; Kramer, B.; Swanson, C.M.; Byers, H.; Lynham, S.; Ward, M.; Malim, M.H. Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. J. Virol. 2007, 81, 2165–2178. [Google Scholar] [CrossRef]
- Kedersha, N.; Stoecklin, G.; Ayodele, M.; Yacono, P.; Lykke-Andersen, J.; Fritzler, M.J.; Scheuner, D.; Kaufman, R.J.; Golan, D.E.; Anderson, P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 2005, 169, 871–884. [Google Scholar] [CrossRef]
- Weil, T.T.; Parton, R.M.; Herpers, B.; Soetaert, J.; Veenendaal, T.; Xanthakis, D.; Dobbie, I.M.; Halstead, J.M.; Hayashi, R.; Rabouille, C.; et al. Drosophila patterning is established by differential association of mRNAs with P bodies. Nat. Cell Biol. 2012, 14, 1305–1313. [Google Scholar] [CrossRef] [PubMed]
- Buckingham, M.; Liu, J.L. U bodies respond to nutrient stress in Drosophila. Exp. Cell Res. 2011, 317, 2835–2844. [Google Scholar] [CrossRef] [PubMed]
- Buchan, J.R.; Muhlrad, D.; Parker, R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J. Cell Biol. 2008, 183, 441–455. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Schmich, F.; Srivatsa, S.; Weidner, J.; Beerenwinkel, N.; Spang, A. Context-dependent deposition and regulation of mRNAs in P-bodies. eLife 2018, 7, e29815. [Google Scholar] [CrossRef] [PubMed]
- Rieckher, M.; Markaki, M.; Princz, A.; Schumacher, B.; Tavernarakis, N. Maintenance of Proteostasis by P Body-Mediated Regulation of eIF4E Availability during Aging in Caenorhabditis elegans. Cell Rep. 2018, 25, 199–211 e196. [Google Scholar] [CrossRef] [PubMed]
- Ditlev, J.A. Membrane-associated phase separation: Organization and function emerge from a two-dimensional milieu. J. Mol. Cell Biol. 2021, 13, 319–324. [Google Scholar] [CrossRef]
- Zacharogianni, M.; Aguilera-Gomez, A.; Veenendaal, T.; Smout, J.; Rabouille, C. A stress assembly that confers cell viability by preserving ERES components during amino-acid starvation. eLife 2014, 3, e04132. [Google Scholar] [CrossRef]
- Müller, M.; Ahumada-Castro, U.; Sanhueza, M.; Gonzalez-Billault, C.; Court, F.A.; Cárdenas, C. Mitochondria and Calcium Regulation as Basis of Neurodegeneration Associated With Aging. Front. Neurosci. 2018, 12, 470. [Google Scholar] [CrossRef]
- Loncke, J.; Kaasik, A.; Bezprozvanny, I.; Parys, J.B.; Kerkhofs, M.; Bultynck, G. Balancing ER-Mitochondrial Ca(2+) Fluxes in Health and Disease. Trends Cell Biol. 2021, 31, 598–612. [Google Scholar] [CrossRef]
- Barazzuol, L.; Giamogante, F.; Calì, T. Mitochondria Associated Membranes (MAMs): Architecture and physiopathological role. Cell Calcium 2021, 94, 102343. [Google Scholar] [CrossRef]
- Giorgi, C.; Ito, K.; Lin, H.K.; Santangelo, C.; Wieckowski, M.R.; Lebiedzinska, M.; Bononi, A.; Bonora, M.; Duszynski, J.; Bernardi, R.; et al. PML regulates apoptosis at endoplasmic reticulum by modulating calcium release. Science 2010, 330, 1247–1251. [Google Scholar] [CrossRef] [PubMed]
- Carracedo, A.; Ito, K.; Pandolfi, P.P. The nuclear bodies inside out: PML conquers the cytoplasm. Curr. Opin. Cell Biol. 2011, 23, 360–366. [Google Scholar] [CrossRef] [PubMed]
- Missiroli, S.; Bonora, M.; Patergnani, S.; Poletti, F.; Perrone, M.; Gafà, R.; Magri, E.; Raimondi, A.; Lanza, G.; Tacchetti, C.; et al. PML at Mitochondria-Associated Membranes Is Critical for the Repression of Autophagy and Cancer Development. Cell Rep. 2016, 16, 2415–2427. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Li, C.; Yang, S.; Xiao, Y.; Xiong, X.; Chen, W.; Zhao, H.; Zhang, Q.; Han, Y.; Sun, L. Mitochondria-Associated ER Membranes—The Origin Site of Autophagy. Front. Cell Dev. Biol. 2020, 8, 595. [Google Scholar] [CrossRef] [PubMed]
- Belyy, V.; Tran, N.H.; Walter, P. Quantitative microscopy reveals dynamics and fate of clustered IRE1α. Proc. Natl. Acad. Sci. USA 2020, 117, 1533–1542. [Google Scholar] [CrossRef]
- Lee, J.E.; Cathey, P.I.; Wu, H.; Parker, R.; Voeltz, G.K. Endoplasmic reticulum contact sites regulate the dynamics of membraneless organelles. Science 2020, 367, eaay7108. [Google Scholar] [CrossRef] [PubMed]
- Moon, S.L.; Parker, R. Analysis of eIF2B bodies and their relationships with stress granules and P-bodies. Sci Rep. 2018, 8, 12264. [Google Scholar] [CrossRef]
- Mollet, S.; Cougot, N.; Wilczynska, A.; Dautry, F.; Kress, M.; Bertrand, E.; Weil, D. Translationally repressed mRNA transiently cycles through stress granules during stress. Mol. Biol. Cell 2008, 19, 4469–4479. [Google Scholar] [CrossRef]
- Ohn, T.; Kedersha, N.; Hickman, T.; Tisdale, S.; Anderson, P. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat. Cell Biol. 2008, 10, 1224–1231. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, S.F.; Walker, S.E.; Rajagopal, V.; Aitken, C.E.; Lorsch, J.R. Recruiting knotty partners: The roles of translation initiation factors in mRNA recruitment to the eukaryotic ribosome. In Ribosomes: Structure, Function, and Dynamics; Rodnina, M.V., Wintermeyer, W., Green, R., Eds.; Springer Vienna: Vienna, Austria, 2011; pp. 155–169. [Google Scholar]
- Wang, C.Y.; Wen, W.L.; Nilsson, D.; Sunnerhagen, P.; Chang, T.H.; Wang, S.W. Analysis of stress granule assembly in Schizosaccharomyces pombe. RNA (New York N.Y.) 2012, 18, 694–703. [Google Scholar] [CrossRef]
- Ernoult-Lange, M.; Baconnais, S.; Harper, M.; Minshall, N.; Souquere, S.; Boudier, T.; Bénard, M.; Andrey, P.; Pierron, G.; Kress, M.; et al. Multiple binding of repressed mRNAs by the P-body protein Rck/p54. RNA (New York N.Y.) 2012, 18, 1702–1715. [Google Scholar] [CrossRef]
- Teixeira, D.; Parker, R. Analysis of P-body assembly in Saccharomyces cerevisiae. Mol. Biol. Cell 2007, 18, 2274–2287. [Google Scholar] [CrossRef] [PubMed]
- Leung, A.K.; Sharp, P.A. Quantifying Argonaute proteins in and out of GW/P-bodies: Implications in microRNA activities. Adv. Exp. Med. Biol. 2013, 768, 165–182. [Google Scholar] [CrossRef]
- Qi, M.Y.; Wang, Z.Z.; Zhang, Z.; Shao, Q.; Zeng, A.; Li, X.Q.; Li, W.Q.; Wang, C.; Tian, F.J.; Li, Q.; et al. AU-rich-element-dependent translation repression requires the cooperation of tristetraprolin and RCK/P54. Mol. Cell. Biol. 2012, 32, 913–928. [Google Scholar] [CrossRef]
- Laporte, D.; Salin, B.; Daignan-Fornier, B.; Sagot, I. Reversible cytoplasmic localization of the proteasome in quiescent yeast cells. J. Cell Biol. 2008, 181, 737–745. [Google Scholar] [CrossRef] [PubMed]
- Munder, M.C.; Midtvedt, D.; Franzmann, T.; Nüske, E.; Otto, O.; Herbig, M.; Ulbricht, E.; Müller, P.; Taubenberger, A.; Maharana, S.; et al. A pH-driven transition of the cytoplasm from a fluid- to a solid-like state promotes entry into dormancy. eLife 2016, 5, e09347. [Google Scholar] [CrossRef] [PubMed]
- Saad, S.; Cereghetti, G.; Feng, Y.; Picotti, P.; Peter, M.; Dechant, R. Reversible protein aggregation is a protective mechanism to ensure cell cycle restart after stress. Nat. Cell Biol. 2017, 19, 1202–1213. [Google Scholar] [CrossRef] [PubMed]
- Petrovska, I.; Nüske, E.; Munder, M.C.; Kulasegaran, G.; Malinovska, L.; Kroschwald, S.; Richter, D.; Fahmy, K.; Gibson, K.; Verbavatz, J.M.; et al. Filament formation by metabolic enzymes is a specific adaptation to an advanced state of cellular starvation. eLife 2014, 3. [Google Scholar] [CrossRef] [PubMed]
- Cereghetti, G.; Wilson-Zbinden, C.; Kissling, V.M.; Diether, M.; Arm, A.; Yoo, H.; Piazza, I.; Saad, S.; Picotti, P.; Drummond, D.A.; et al. Reversible amyloids of pyruvate kinase couple cell metabolism and stress granule disassembly. Nat. Cell Biol. 2021, 23, 1085–1094, e02409. [Google Scholar] [CrossRef] [PubMed]
- Boor, K.J. Bacterial stress responses: What doesn’t kill them can make then stronger. PLoS Biol. 2006, 4, e23. [Google Scholar] [CrossRef] [PubMed]
- Huo, Y.X.; Rosenthal, A.Z.; Gralla, J.D. General stress response signalling: Unwrapping transcription complexes by DNA relaxation via the sigma38 C-terminal domain. Mol. Microbiol. 2008, 70, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Alba, B.M.; Gross, C.A. Regulation of the Escherichia coli sigma-dependent envelope stress response. Mol. Microbiol. 2004, 52, 613–619. [Google Scholar] [CrossRef]
- Maleki, F.; Khosravi, A.; Nasser, A.; Taghinejad, H.; Azizian, M. Bacterial Heat Shock Protein Activity. J. Clin. Diagn. Res. JCDR 2016, 10, BE01–BE03. [Google Scholar] [CrossRef]
- Keto-Timonen, R.; Hietala, N.; Palonen, E.; Hakakorpi, A.; Lindström, M.; Korkeala, H. Cold Shock Proteins: A Minireview with Special Emphasis on Csp-family of Enteropathogenic Yersinia. Front. Microbiol. 2016, 7, 1151. [Google Scholar] [CrossRef]
- Nandana, V.; Schrader, J.M. Roles of liquid-liquid phase separation in bacterial RNA metabolism. Curr. Opin. Microbiol. 2021, 61, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Azaldegui, C.A.; Vecchiarelli, A.G.; Biteen, J.S. The emergence of phase separation as an organizing principle in bacteria. Biophys. J. 2021, 120, 1123–1138. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Zhang, W.; Chang, R.; Zhang, S.; Yang, G.; Zhao, G. Liquid-Liquid Phase Separation: Unraveling the Enigma of Biomolecular Condensates in Microbial Cells. Front. Microbiol. 2021, 12, 751880. [Google Scholar] [CrossRef] [PubMed]
- Ladouceur, A.M.; Parmar, B.S.; Biedzinski, S.; Wall, J.; Tope, S.G.; Cohn, D.; Kim, A.; Soubry, N.; Reyes-Lamothe, R.; Weber, S.C. Clusters of bacterial RNA polymerase are biomolecular condensates that assemble through liquid-liquid phase separation. Proc. Natl. Acad. Sci. USA 2020, 117, 18540–18549. [Google Scholar] [CrossRef] [PubMed]
- Babl, L.; Giacomelli, G.; Ramm, B.; Gelmroth, A.K.; Bramkamp, M.; Schwille, P. CTP-controlled liquid-liquid phase separation of ParB. J. Mol. Biol. 2022, 434, 167401. [Google Scholar] [CrossRef] [PubMed]
- Lasker, K.; von Diezmann, L.; Zhou, X.; Ahrens, D.G.; Mann, T.H.; Moerner, W.E.; Shapiro, L. Selective sequestration of signalling proteins in a membraneless organelle reinforces the spatial regulation of asymmetry in Caulobacter crescentus. Nat. Microbiol. 2020, 5, 418–429. [Google Scholar] [CrossRef]
- Saurabh, S.; Chong, T.N.; Bayas, C.; Dahlberg, P.D.; Cartwright, H.N.; Moerner, W.E.; Shapiro, L. ATP-responsive biomolecular condensates tune bacterial kinase signaling. Sci. Adv. 2022, 8, eabm6570. [Google Scholar] [CrossRef] [PubMed]
- Hamouche, L.; Billaudeau, C.; Rocca, A.; Chastanet, A.; Ngo, S.; Laalami, S.; Putzer, H. Dynamic Membrane Localization of RNase Y in Bacillus subtilis. mBio 2020, 11, e03337–e19. [Google Scholar] [CrossRef] [PubMed]
- Tejada-Arranz, A.; Galtier, E.; El Mortaji, L.; Turlin, E.; Ershov, D.; De Reuse, H. The RNase J-Based RNA Degradosome Is Compartmentalized in the Gastric Pathogen Helicobacter pylori. mBio 2020, 11, e01173–e20. [Google Scholar] [CrossRef] [PubMed]
- Racki, L.R.; Tocheva, E.I.; Dieterle, M.G.; Sullivan, M.C.; Jensen, G.J.; Newman, D.K. Polyphosphate granule biogenesis is temporally and functionally tied to cell cycle exit during starvation in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2017, 114, E2440–E2449. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Liu, Y.; Wang, Z.; He, R.; Xiang Zhang, J.; Xu, F.; Lei, M.; Deci, M.B.; Nguyen, J.; Bianco, P.R. Super-resolution imaging reveals changes in Escherichia coli SSB localization in response to DNA damage. Genes Cells Devoted Mol. Cell. Mech. 2019, 24, 814–826. [Google Scholar] [CrossRef] [PubMed]
- Harami, G.M.; Kovács, Z.J.; Pancsa, R.; Pálinkás, J.; Baráth, V.; Tárnok, K.; Málnási-Csizmadia, A.; Kovács, M. Phase separation by ssDNA binding protein controlled via protein-protein and protein-DNA interactions. Proc. Natl. Acad. Sci. USA 2020, 117, 26206–26217. [Google Scholar] [CrossRef] [PubMed]
- Janissen, R.; Arens, M.M.A.; Vtyurina, N.N.; Rivai, Z.; Sunday, N.D.; Eslami-Mossallam, B.; Gritsenko, A.A.; Laan, L.; de Ridder, D.; Artsimovitch, I.; et al. Global DNA Compaction in Stationary-Phase Bacteria Does Not Affect Transcription. Cell 2018, 174, 1188–1199 e1114. [Google Scholar] [CrossRef] [PubMed]
- Ceci, P.; Cellai, S.; Falvo, E.; Rivetti, C.; Rossi, G.L.; Chiancone, E. DNA condensation and self-aggregation of Escherichia coli Dps are coupled phenomena related to the properties of the N-terminus. Nucleic Acids Res. 2004, 32, 5935–5944. [Google Scholar] [CrossRef]
- Al-Husini, N.; Tomares, D.T.; Bitar, O.; Childers, W.S.; Schrader, J.M. α-Proteobacterial RNA Degradosomes Assemble Liquid-Liquid Phase-Separated RNP Bodies. Mol. Cell 2018, 71, 1027–1039 e1014. [Google Scholar] [CrossRef] [PubMed]
- Strahl, H.; Turlan, C.; Khalid, S.; Bond, P.J.; Kebalo, J.M.; Peyron, P.; Poljak, L.; Bouvier, M.; Hamoen, L.; Luisi, B.F.; et al. Membrane recognition and dynamics of the RNA degradosome. PLoS Genet. 2015, 11, e1004961. [Google Scholar] [CrossRef] [PubMed]
- Al-Husini, N.; Tomares, D.T.; Pfaffenberger, Z.J.; Muthunayake, N.S.; Samad, M.A.; Zuo, T.; Bitar, O.; Aretakis, J.R.; Bharmal, M.M.; Gega, A.; et al. BR-Bodies Provide Selectively Permeable Condensates that Stimulate mRNA Decay and Prevent Release of Decay Intermediates. Mol. Cell 2020, 78, 670–682 e678. [Google Scholar] [CrossRef] [PubMed]
- Lopez, P.J.; Marchand, I.; Joyce, S.A.; Dreyfus, M. The C-terminal half of RNase E, which organizes the Escherichia coli degradosome, participates in mRNA degradation but not rRNA processing in vivo. Mol. Microbiol. 1999, 33, 188–199. [Google Scholar] [CrossRef] [PubMed]
- Vishnyakov, I.E.; Levitskii, S.A.; Manuvera, V.A.; Lazarev, V.N.; Ayala, J.A.; Ivanov, V.A.; Snigirevskaya, E.S.; Komissarchik, Y.Y.; Borchsenius, S.N. The identification and characterization of IbpA, a novel α-crystallin-type heat shock protein from mycoplasma. Cell Stress Chaperones 2012, 17, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Vishnyakov, I.E.; Levitskii, S.A.; Borchsenius, S.N. The effect of heat shock on phytopathogenic mycoplasma Acholeplasma laidlawii PG-8A. Cell Tissue Biol. 2015, 9, 149–157. [Google Scholar] [CrossRef]
- Vishnyakov, I.E.; Bogachev, M.I.; Salafutdinov, I.; Borchsenius, S.N.; Kayumov, A.R. The Temperature-Dependent Selectivity of Potential Interaction Partners for the Small Heat Shock Protein IbpA from Acholeplasma laidlawii. BioNanoScience 2016, 6, 437–442. [Google Scholar] [CrossRef]
- Strózecka, J.; Chrusciel, E.; Górna, E.; Szymanska, A.; Ziętkiewicz, S.; Liberek, K. Importance of N- and C-terminal regions of IbpA, Escherichia coli small heat shock protein, for chaperone function and oligomerization. J. Biol. Chem. 2012, 287, 2843–2853. [Google Scholar] [CrossRef]
- Lazarev, V.N.; Levitskii, S.A.; Basovskii, Y.I.; Chukin, M.M.; Akopian, T.A.; Vereshchagin, V.V.; Kostrjukova, E.S.; Kovaleva, G.Y.; Kazanov, M.D.; Malko, D.B.; et al. Complete genome and proteome of Acholeplasma laidlawii. J. Bacteriol. 2011, 193, 4943–4953. [Google Scholar] [CrossRef]
- Bright, C.M.; Ellis, D. Intracellular pH changes induced by hypoxia and anoxia in isolated sheep heart Purkinje fibres. Exp. Physiol. 1992, 77, 165–175. [Google Scholar] [CrossRef]
- Dechant, R.; Binda, M.; Lee, S.S.; Pelet, S.; Winderickx, J.; Peter, M. Cytosolic pH is a second messenger for glucose and regulates the PKA pathway through V-ATPase. EMBO J. 2010, 29, 2515–2526. [Google Scholar] [CrossRef]
- Adame-Arana, O.; Weber, C.A.; Zaburdaev, V.; Prost, J.; Jülicher, F. Liquid Phase Separation Controlled by pH. Biophys. J. 2020, 119, 1590–1605. [Google Scholar] [CrossRef]
- Riback, J.A.; Katanski, C.D.; Kear-Scott, J.L.; Pilipenko, E.V.; Rojek, A.E.; Sosnick, T.R.; Drummond, D.A. Stress-Triggered Phase Separation Is an Adaptive, Evolutionarily Tuned Response. Cell 2017, 168, 1028–1040 e1019. [Google Scholar] [CrossRef] [PubMed]
- Hondele, M.; Sachdev, R.; Heinrich, S.; Wang, J.; Vallotton, P.; Fontoura, B.M.A.; Weis, K. DEAD-box ATPases are global regulators of phase-separated organelles. Nature 2019, 573, 144–148. [Google Scholar] [CrossRef] [PubMed]
- Guillén-Boixet, J.; Kopach, A.; Holehouse, A.S.; Wittmann, S.; Jahnel, M.; Schlüßler, R.; Kim, K.; Trussina, I.; Wang, J.; Mateju, D.; et al. RNA-Induced Conformational Switching and Clustering of G3BP Drive Stress Granule Assembly by Condensation. Cell 2020, 181, 346–361 e317. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Yoshizawa, T.; Yamazaki, R.; Fujiwara, A.; Kameda, T.; Kitahara, R. Pressure and Temperature Phase Diagram for Liquid-Liquid Phase Separation of the RNA-Binding Protein Fused in Sarcoma. J. Phys. Chemistry. B 2021, 125, 6821–6829. [Google Scholar] [CrossRef]
- Franzmann, T.M.; Alberti, S. Protein Phase Separation as a Stress Survival Strategy. Cold Spring Harb Perspect. Biol. 2019, 11, a034058. [Google Scholar] [CrossRef]
- Dignon, G.L.; Zheng, W.; Kim, Y.C.; Mittal, J. Temperature-Controlled Liquid-Liquid Phase Separation of Disordered Proteins. ACS Cent. Sci. 2019, 5, 821–830. [Google Scholar] [CrossRef]
- Jalihal, A.P.; Pitchiaya, S.; Xiao, L.; Bawa, P.; Jiang, X.; Bedi, K.; Parolia, A.; Cieslik, M.; Ljungman, M.; Chinnaiyan, A.M.; et al. Multivalent Proteins Rapidly and Reversibly Phase-Separate upon Osmotic Cell Volume Change. Mol. Cell 2020, 79, 978–990.e975. [Google Scholar] [CrossRef]
- Onoguchi-Mizutani, R.; Akimitsu, N. Long noncoding RNA and phase separation in cellular stress response. J. Biochem. 2022, 171, 269–276. [Google Scholar] [CrossRef]
- Snead, W.T.; Gladfelter, A.S. The Control Centers of Biomolecular Phase Separation: How Membrane Surfaces, PTMs, and Active Processes Regulate Condensation. Mol. Cell 2019, 76, 295–305. [Google Scholar] [CrossRef]
- Drino, A.; Schaefer, M.R. RNAs, Phase Separation, and Membrane-Less Organelles: Are Post-Transcriptional Modifications Modulating Organelle Dynamics? BioEssays News Rev. Mol. Cell. Dev. Biol. 2018, 40, e1800085. [Google Scholar] [CrossRef]
- Yang, P.; Mathieu, C.; Kolaitis, R.M.; Zhang, P.; Messing, J.; Yurtsever, U.; Yang, Z.; Wu, J.; Li, Y.; Pan, Q.; et al. G3BP1 Is a Tunable Switch that Triggers Phase Separation to Assemble Stress Granules. Cell 2020, 181, 325–345 e328. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Chen, Y.; Dai, H.; Zhang, H.; Xie, M.; Zhang, H.; Chen, F.; Kang, X.; Bai, X.; Chen, Z. UBAP2L arginine methylation by PRMT1 modulates stress granule assembly. Cell Death Differ. 2020, 27, 227–241. [Google Scholar] [CrossRef]
- Duan, Y.; Du, A.; Gu, J.; Duan, G.; Wang, C.; Gui, X.; Ma, Z.; Qian, B.; Deng, X.; Zhang, K.; et al. PARylation regulates stress granule dynamics, phase separation, and neurotoxicity of disease-related RNA-binding proteins. Cell Res. 2019, 29, 233–247. [Google Scholar] [CrossRef] [PubMed]
- Jongjitwimol, J.; Baldock, R.A.; Morley, S.J.; Watts, F.Z. Sumoylation of eIF4A2 affects stress granule formation. J. Cell Sci. 2016, 129, 2407–2415. [Google Scholar] [CrossRef]
- Carey, J.L.; Guo, L. Liquid-Liquid Phase Separation of TDP-43 and FUS in Physiology and Pathology of Neurodegenerative Diseases. Front. Mol. Biosci. 2022, 9, 826719. [Google Scholar] [CrossRef] [PubMed]
- Hofweber, M.; Dormann, D. Friend or foe-Post-translational modifications as regulators of phase separation and RNP granule dynamics. J. Biol. Chem. 2019, 294, 7137–7150. [Google Scholar] [CrossRef]
- Roth, S.; Khalaila, I. The effect of O-GlcNAcylation on hnRNP A1 translocation and interaction with transportin1. Exp. Cell Res. 2017, 350, 210–217. [Google Scholar] [CrossRef]
- Cheng, X.; Kao, H.Y. Post-translational modifications of PML: Consequences and implications. Front. Oncol. 2012, 2, 210. [Google Scholar] [CrossRef]
- Ohkuni, K.; Pasupala, N.; Peek, J.; Holloway, G.L.; Sclar, G.D.; Levy-Myers, R.; Baker, R.E.; Basrai, M.A.; Kerscher, O. SUMO-Targeted Ubiquitin Ligases (STUbLs) Reduce the Toxicity and Abnormal Transcriptional Activity Associated With a Mutant, Aggregation-Prone Fragment of Huntingtin. Front. Genet. 2018, 9, 379. [Google Scholar] [CrossRef]
- Alberti, S.; Carra, S. Quality Control of Membraneless Organelles. J. Mol. Biol. 2018, 430, 4711–4729. [Google Scholar] [CrossRef]
- Mateju, D.; Franzmann, T.M.; Patel, A.; Kopach, A.; Boczek, E.E.; Maharana, S.; Lee, H.O.; Carra, S.; Hyman, A.A.; Alberti, S. An aberrant phase transition of stress granules triggered by misfolded protein and prevented by chaperone function. EMBO J. 2017, 36, 1669–1687. [Google Scholar] [CrossRef] [PubMed]
- Horne, S.D.; Chowdhury, S.K.; Heng, H.H. Stress, genomic adaptation, and the evolutionary trade-off. Front. Genet. 2014, 5, 92. [Google Scholar] [CrossRef] [PubMed]
- Berchtold, D.; Battich, N.; Pelkmans, L. A Systems-Level Study Reveals Regulators of Membrane-less Organelles in Human Cells. Mol. Cell 2018, 72, 1035–1049 e1035. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Kandi, A.R.; Jayaprakashappa, D.; Thuery, G.; Purohit, D.J.; Huelsmeier, J.; Singh, R.; Pothapragada, S.S.; Ramaswami, M.; Bakthavachalu, B. The transcriptional response to oxidative stress is independent of stress-granule formation. Mol. Biol. Cell 2022, 33, ar25. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Neumann, M.; Stearman, R.; Stauber, R.; Pause, A.; Pavlakis, G.N.; Klausner, R.D. Transcription-dependent nuclear-cytoplasmic trafficking is required for the function of the von Hippel-Lindau tumor suppressor protein. Mol. Cell. Biol. 1999, 19, 1486–1497. [Google Scholar] [CrossRef] [PubMed]
- Marnef, A.; Weil, D.; Standart, N. RNA-related nuclear functions of human Pat1b, the P-body mRNA decay factor. Mol. Biol. Cell 2012, 23, 213–224. [Google Scholar] [CrossRef]
- Borbolis, F.; Syntichaki, P. Cytoplasmic mRNA turnover and ageing. Mech. Ageing Dev. 2015, 152, 32–42. [Google Scholar] [CrossRef]
- Campos-Melo, D.; Hawley, Z.C.E.; Droppelmann, C.A.; Strong, M.J. The Integral Role of RNA in Stress Granule Formation and Function. Front. Cell Dev. Biol. 2021, 9, 621779. [Google Scholar] [CrossRef]
- Sharp, P.A.; Chakraborty, A.K.; Henninger, J.E.; Young, R.A. RNA in formation and regulation of transcriptional condensates. RNA (New York N.Y.) 2022, 28, 52–57. [Google Scholar] [CrossRef]
- Szaflarski, W.; Leśniczak-Staszak, M.; Sowiński, M.; Ojha, S.; Aulas, A.; Dave, D.; Malla, S.; Anderson, P.; Ivanov, P.; Lyons, S.M. Early rRNA processing is a stress-dependent regulatory event whose inhibition maintains nucleolar integrity. Nucleic Acids Res. 2022, 50, 1033–1051. [Google Scholar] [CrossRef]
- Pfister, A.S. Emerging Role of the Nucleolar Stress Response in Autophagy. Front. Cell. Neurosci. 2019, 13, 156. [Google Scholar] [CrossRef]
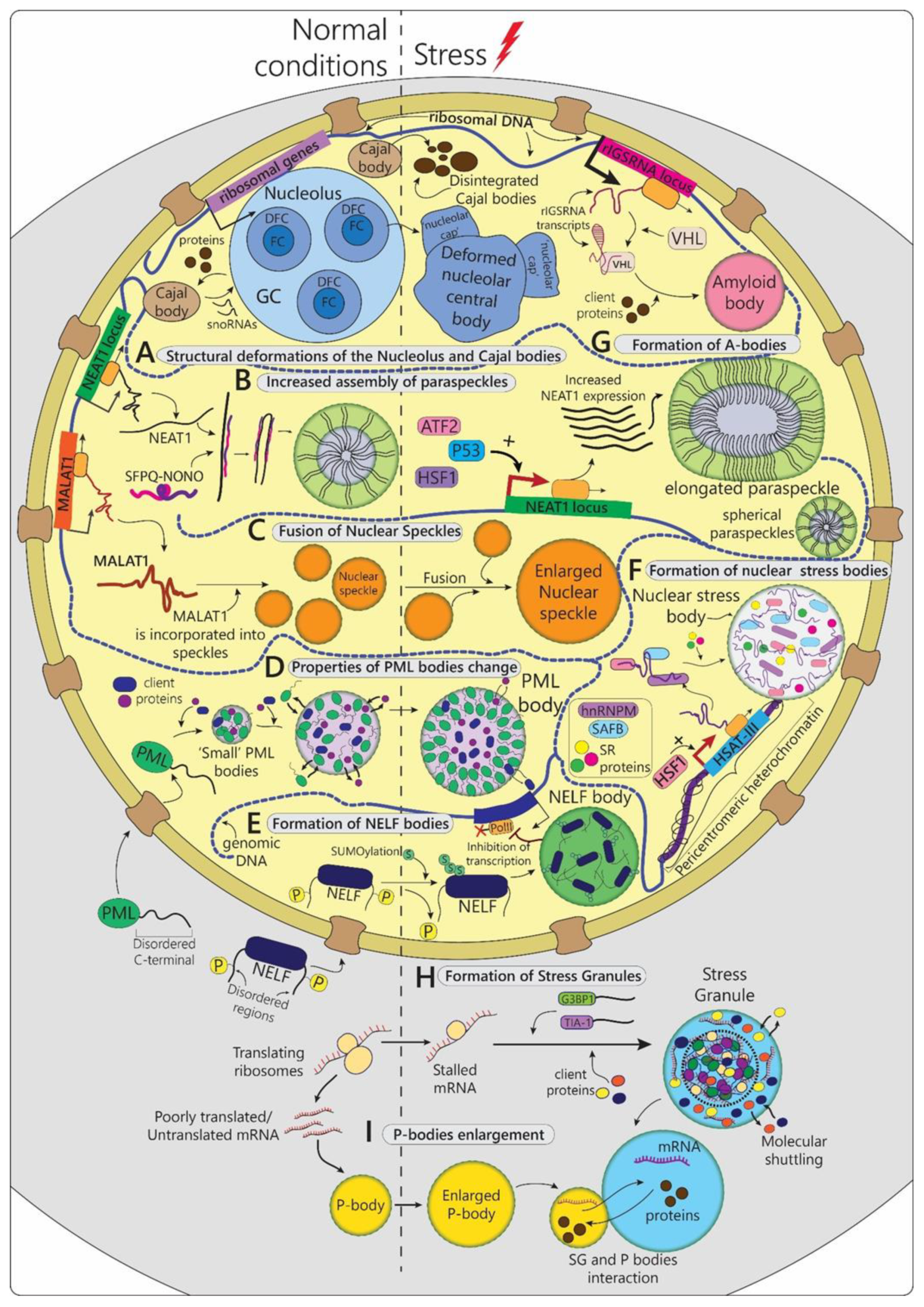

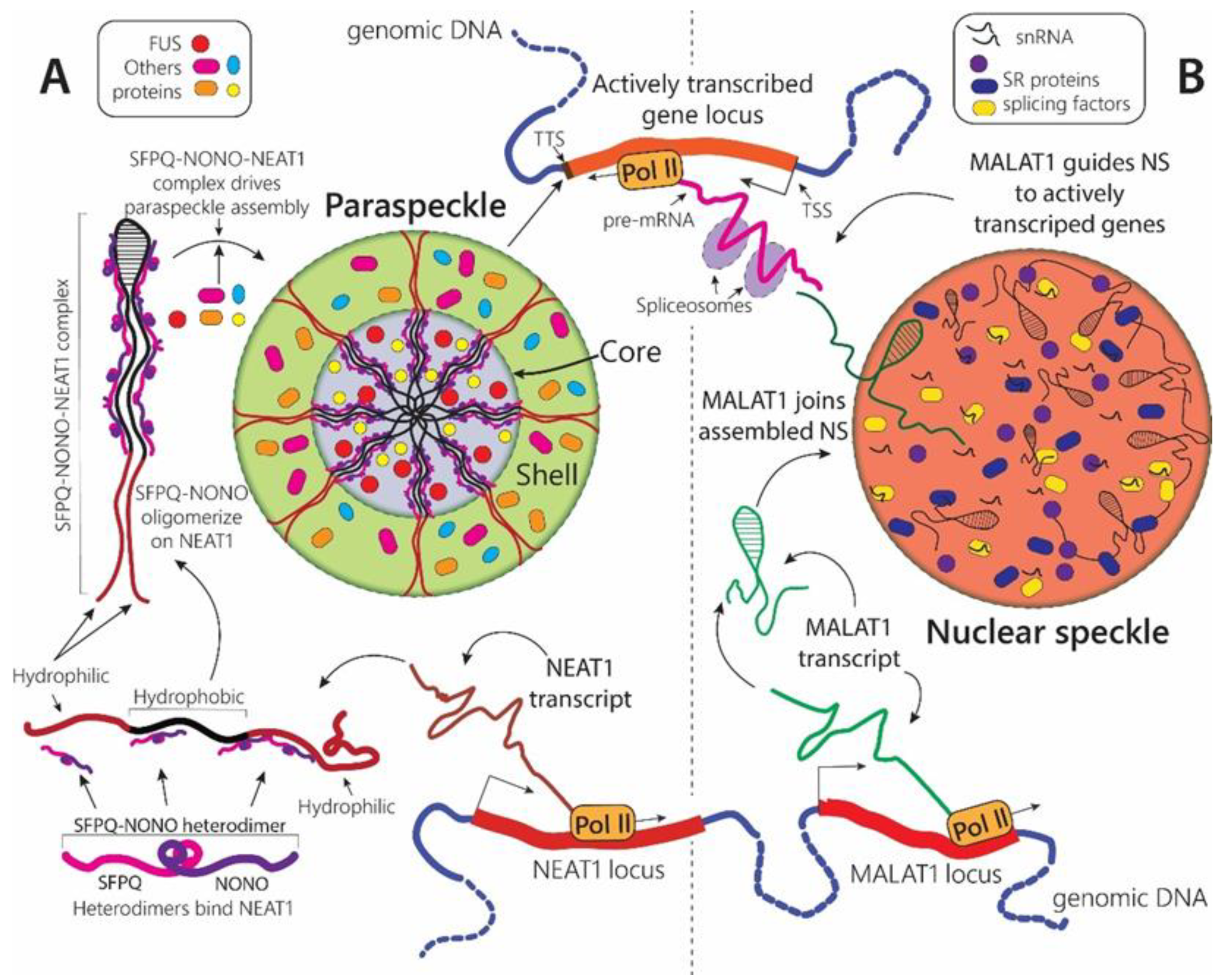
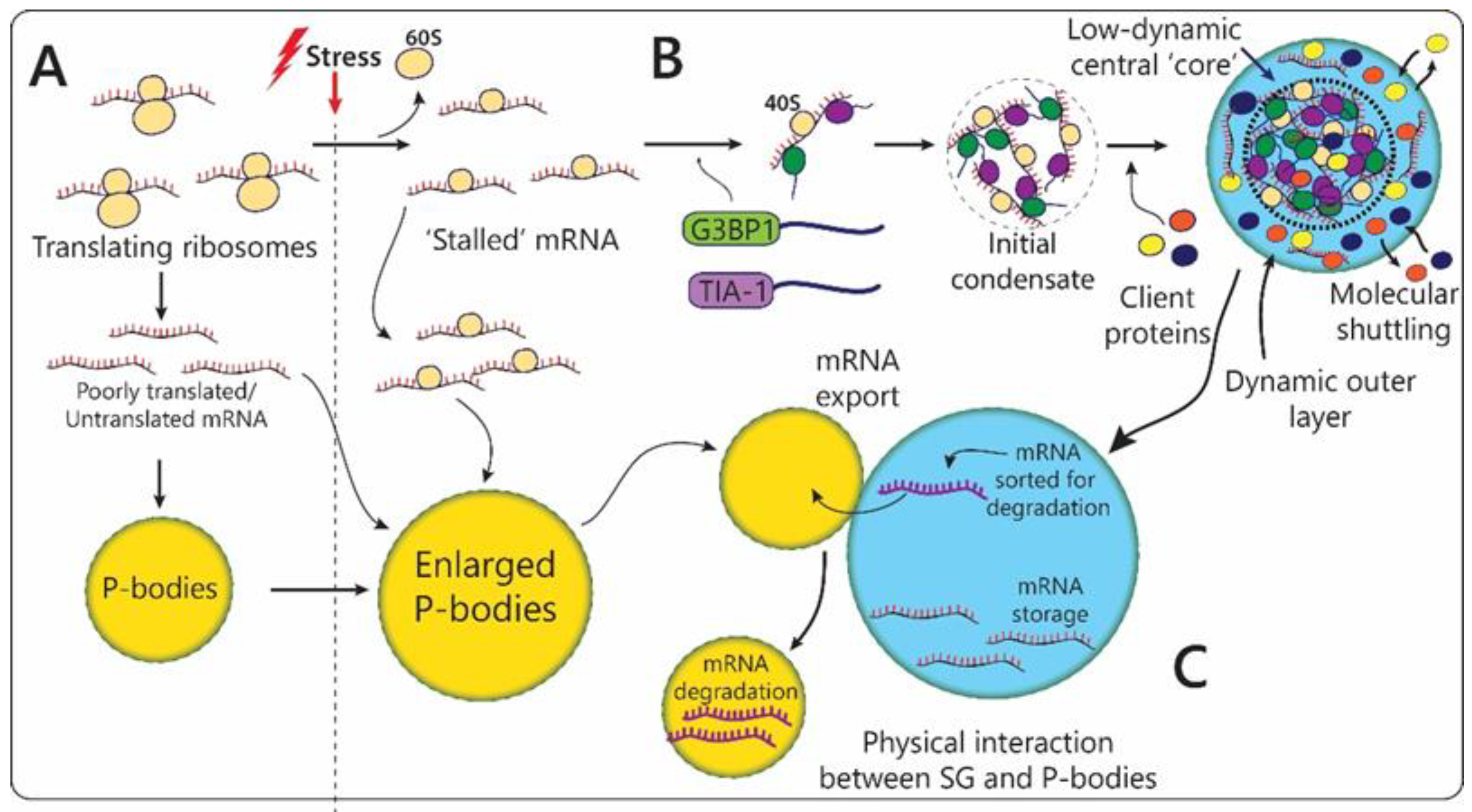
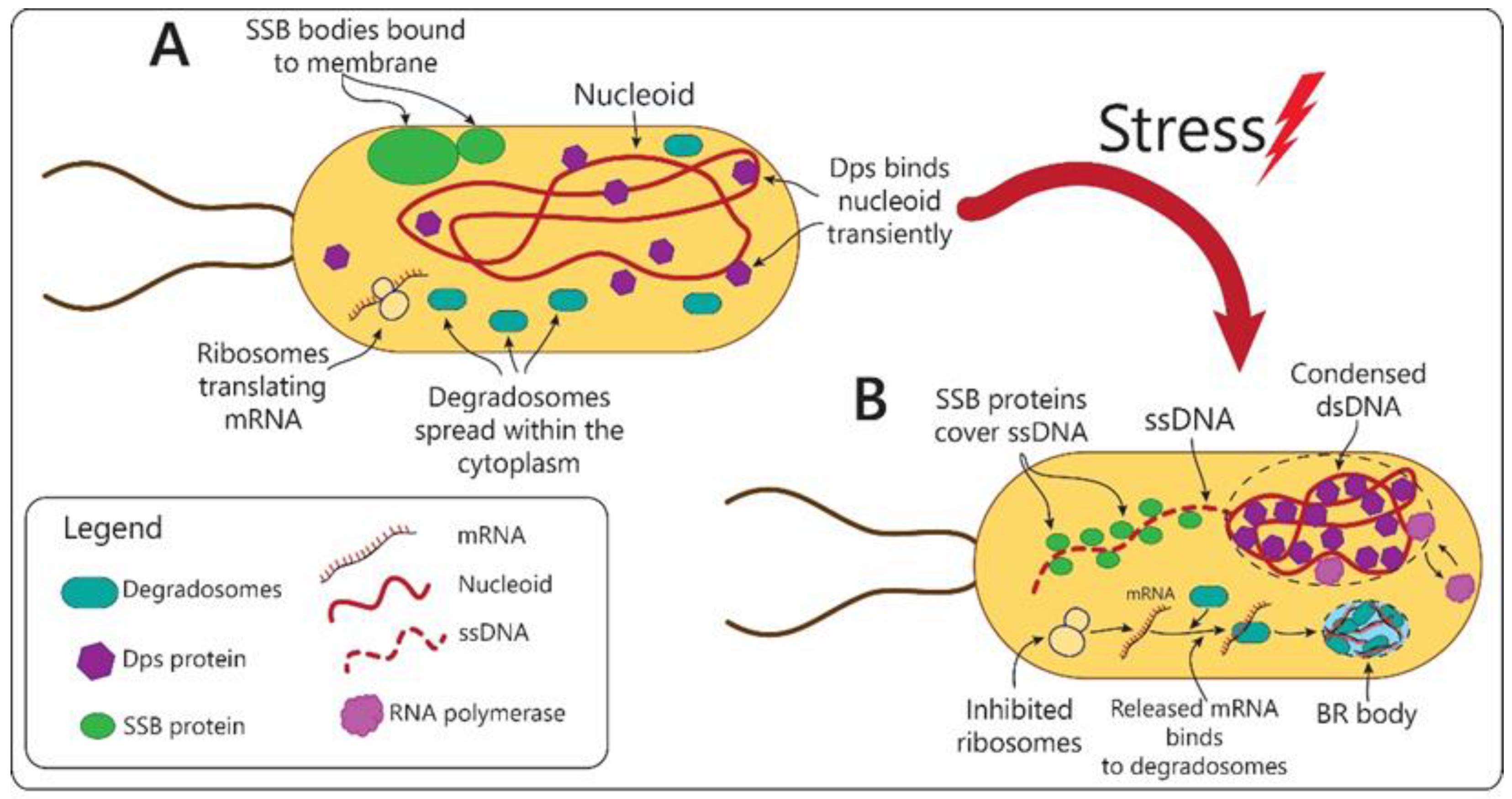
| Stress-Linked Organelle | Main Components | Organism | Structural Changes in Response to Stress | Function | |
|---|---|---|---|---|---|
| Nuclear membrane-less organelles | |||||
| MLOs subject to change and rearrangement in response to stress | Nucleolus | Fibrillarin, nucleophosmin, rRNA, snoRNPs, Nop58, etc. | Eukarya | Release of ribosomal proteins, change in the nucleolar proteome. Nucleolar segregation upon DNA damage or rRNA transcription. Nucleolar fragmentation upon inhibition of RNA Pol II transcription or protein kinases. Nucleolar and FC enlargement upon viral infection | Ribosome biogenesis |
| Cajal bodies | Coilin, SMN1, snRNA, snoRNA, scaRNAs, etc. | Animals and plants | CBs decrease in number and size in response to starvation. CBs undergo disruption and formation of coilin nucleoplasmic microfoci upon UV-C irradiation, osmotic stress, and heat shock. Fusion of transformed CBs with the nucleolus upon GRV infection in plants | Maturation of snoRNA, snRNA, histone mRNA | |
| Paraspeckles | lncRNA Neat1, NONO, SFPQ, FUS, etc. | Mammals | Increase in paraspeckles numbers upon different types of stress: hypoxia, temperature, sulforaphane treatment, softening of the cellular substrate, etc. | Storage of RNAs and proteins involved in the transcription regulation and pre-mRNA processing. | |
| Nuclear speckles | snRNP, SR proteins, lncRNA MALAT1, etc. | Mammals and plants | Enlargement and rounding probably via fusions and reincorporation of splicing factors for temporal storage during stress. | Splicing regulation and storage of proteins | |
| PML-bodies | PML, SUMO-1, Sp100, etc | Mammals Absent in flies, plants and yeasts | Enlargement and decrease in the content mobility upon oxidative stress induced by H2O2. Degradation or cytoplasmic relocalization of the PML isoforms upon oxidative stress induced by As2O3. Decrease in the number and size of PML bodies upon heat stress, heavy metal addition, and expression of adenovirus E1A. | Regulation of the p53-dependent signaling, DNA damage response, DNA repair, telomere homeostasis | |
| Transient assembly in response to stress | NELF bodies | NELF | Human cells | Stress-induced assembly at PolII-active transcription sites driven by NELF protein dephosphorylation and SUMOylation. | Inhibition of RNA Pol II transcription |
| Nuclear stress-bodies | HSF1, HSatIII lncRNA, SAFB, hnRNPM | Primates | Stress-induced formation at sites of HSatIII transcription activated by HSF1 transcription factor. | Protein storage and regulation of mRNA splicing | |
| A-bodies | rIGSRNA, VHL | Mammals, fungi, insects, plants | Assembly and solidification upon the onset of stress at the sites of rIGSRNA transcription. | Temporal storage of amyloidogenic proteins | |
| Cytoplasmic membrane-less organelles | |||||
| Assembly | Stress-granules | G3BP1, TIA-1, FUS, hnRNPA1, untranslated mRNA, etc | Eukaryotic cells | Reversible assembly in response to stress as a result of accumulation of translationally repressed mRNA in the cytoplasm. | mRNA storage and triage, regulation of translation |
| Rearrangement | P-bodies | DDX6, EDC-4, LSM-4, EIF4E-T, poorly translated and untranslated mRNA | Eukaryotic cells | Increase in the number and size of P-bodies under stress conditions. | mRNA translation, processing and degradation |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fefilova, A.S.; Antifeeva, I.A.; Gavrilova, A.A.; Turoverov, K.K.; Kuznetsova, I.M.; Fonin, A.V. Reorganization of Cell Compartmentalization Induced by Stress. Biomolecules 2022, 12, 1441. https://doi.org/10.3390/biom12101441
Fefilova AS, Antifeeva IA, Gavrilova AA, Turoverov KK, Kuznetsova IM, Fonin AV. Reorganization of Cell Compartmentalization Induced by Stress. Biomolecules. 2022; 12(10):1441. https://doi.org/10.3390/biom12101441
Chicago/Turabian StyleFefilova, Anna S., Iuliia A. Antifeeva, Anastasia A. Gavrilova, Konstantin K. Turoverov, Irina M. Kuznetsova, and Alexander V. Fonin. 2022. "Reorganization of Cell Compartmentalization Induced by Stress" Biomolecules 12, no. 10: 1441. https://doi.org/10.3390/biom12101441
APA StyleFefilova, A. S., Antifeeva, I. A., Gavrilova, A. A., Turoverov, K. K., Kuznetsova, I. M., & Fonin, A. V. (2022). Reorganization of Cell Compartmentalization Induced by Stress. Biomolecules, 12(10), 1441. https://doi.org/10.3390/biom12101441












