Identification of Proteins Responsible for the Neuroprotective Effect of the Secretome Derived from Blood Cells of Remote Ischaemic Conditioned Rats
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Experimental Design
2.3. Preparation of the Blood Cell-Derived Secretome
2.4. Bioactivity Testing
2.4.1. Native PAGE
2.4.2. In Vitro Assays
Cell Culture
Cell Toxicity and Viability Assays
Cell Death Detection
2.4.3. Proteomic Analysis of Blood Cell-Derived Secretomes
Labelling Proteins with Fluorescent Dyes
SDS-PAGE and Sample Processing for Mass Spectrometric Analysis
Mass Spectrometric Analysis and Data Processing
2.5. Data Analysis and Statistics
3. Results
3.1. Neuroprotective Effects of Blood Cell-Derived Secretomes
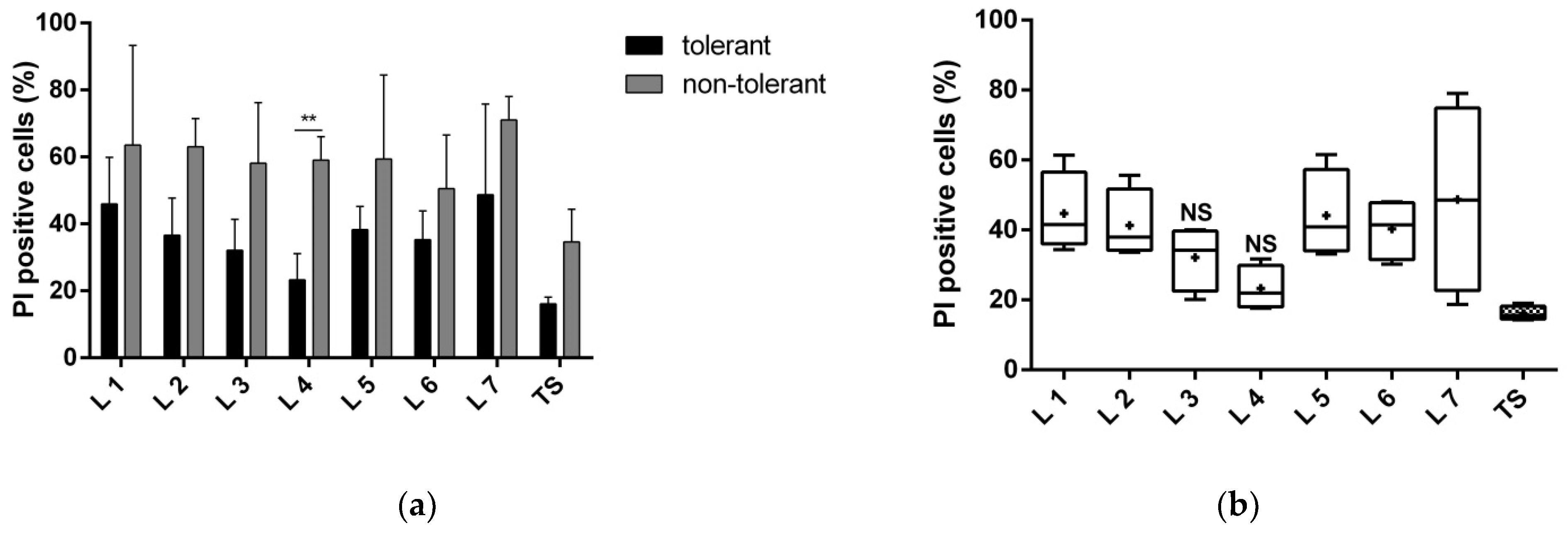
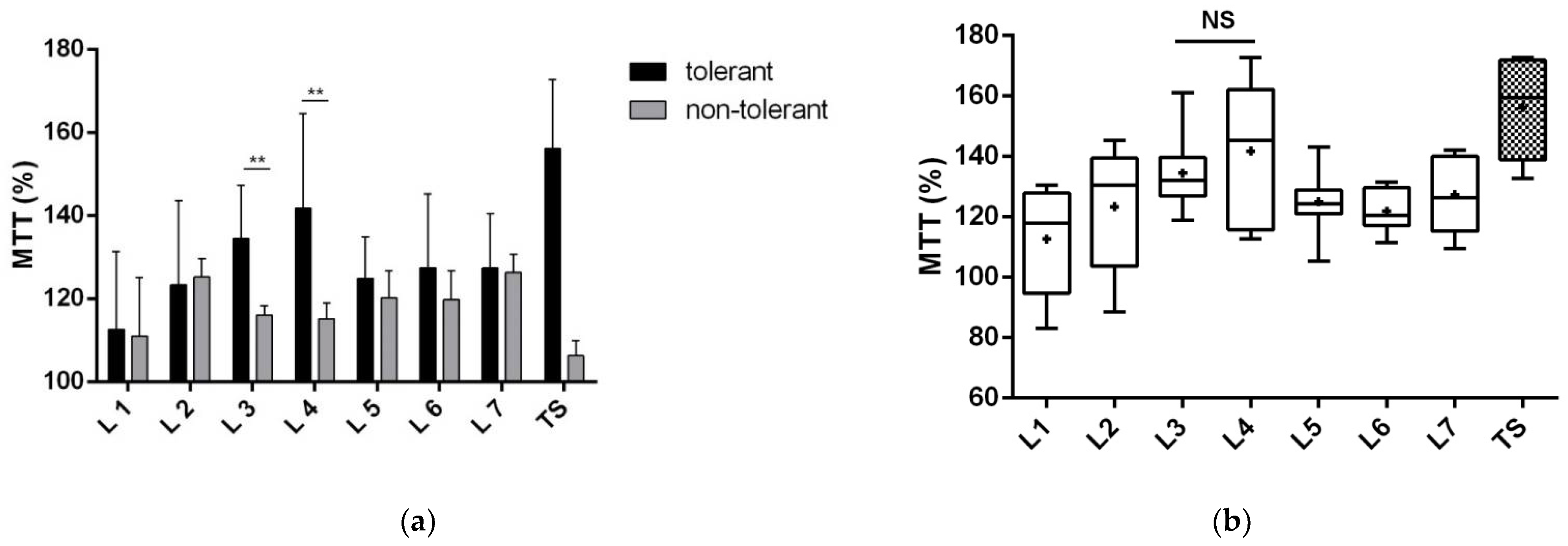
3.1.1. Proteins of Fractions 3 and 4 Reduce Neuronal Death
3.1.2. Proteins of Fractions 3 and 4 Reduce the Toxic Effect of Glutamate and Improve Neuronal Viability
3.2. RIC-Mediated Changes in Proteins of the Blood Cell-Derived Secretome
3.3. RIC Alters the Functional Interaction of Bioactive Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marshall, R.S. Progress in Intravenous Thrombolytic Therapy for Acute Stroke. JAMA Neurol. 2015, 72, 928–934. [Google Scholar] [CrossRef] [PubMed]
- Baig, S.; Moyle, B.; Nair, K.P.S.; Redgrave, J.; Majid, A.; Ali, A. Remote ischaemic conditioning for stroke: Unanswered questions and future directions. Stroke Vasc. Neurol. 2021, 6, 298–309. [Google Scholar] [CrossRef] [PubMed]
- Che, R.; Zhao, W.; Ma, Q.; Jiang, F.; Wu, L.; Yu, Z.; Zhang, Q.; Dong, K.; Song, H.; Huang, X.; et al. rt-PA with remote ischemic postconditioning for acute ischemic stroke. Ann. Clin. Transl. Neurol. 2019, 6, 364–372. [Google Scholar] [CrossRef] [PubMed]
- Bonova, P.; Gottlieb, M. Blood as the carrier of ischemic tolerance in rat brain. J. Neurosci. Res. 2015, 93, 1250–1257. [Google Scholar] [CrossRef]
- Bonova, P.; Nemethova, M.; Matiasova, M.; Bona, M.; Gottlieb, M. Blood cells serve as a source of factor-inducing rapid ischemic tolerance in brain. Eur. J. Neurosci. 2016, 44, 2958–2965. [Google Scholar] [CrossRef]
- Bonova, P.; Jachova, J.; Nemethova, M.; Macakova, L.; Bona, M.; Gottlieb, M. Rapid remote conditioning mediates modulation of blood cell paracrine activity and leads to the production of a secretome with neuroprotective features. J. Neurochem. 2020, 154, 99–111. [Google Scholar] [CrossRef]
- Hu, S.; Dong, H.L.; Zhang, H.P.; Wang, S.Q.; Hou, L.C.; Chen, S.Y.; Zhang, J.S.; Xiong, L.Z. Noninvasive limb remote ischemic preconditioning contributes neuroprotective effects via activation of adenosine A1 receptor and redox status after transient focal cerebral ischemia in rats. Brain Res. 2012, 1459, 81–90. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Brewer, G.J.; Torricelli, J.R. Isolation and culture of adult neurons and neurospheres. Nat. Protoc. 2007, 2, 1490–1498. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic. Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Thangaraju, K.; Neerukonda, S.N.; Katneni, U.; Buehler, P.W. Extracellular Vesicles from Red Blood Cells and Their Evolving Roles in Health, Coagulopathy and Therapy. Int. J. Mol. Sci. 2021, 22, 153. [Google Scholar] [CrossRef] [PubMed]
- Mildner, M.; Hacker, S.; Haider, T.; Gschwandtner, M.; Werba, G.; Barresi, C.; Zimmermann, M.; Golabi, B.; Tschachler, E.; Ankersmit, H.J. Secretome of peripheral blood mononuclear cells enhances wound healing. PLoS ONE 2013, 8, e60103. [Google Scholar] [CrossRef] [PubMed]
- Beer, L.; Zimmermann, M.; Mitterbauer, A.; Ellinger, A.; Gruber, F.; Narzt, M.S.; Zellner, M.; Gyongyosi, M.; Madlener, S.; Simader, E.; et al. Analysis of the Secretome of Apoptotic Peripheral Blood Mononuclear Cells: Impact of Released Proteins and Exosomes for Tissue Regeneration. Sci. Rep. 2015, 5, 16662. [Google Scholar] [CrossRef] [PubMed]
- Altmann, P.; Mildner, M.; Haider, T.; Traxler, D.; Beer, L.; Ristl, R.; Golabi, B.; Gabriel, C.; Leutmezer, F.; Ankersmit, H.J. Secretomes of apoptotic mononuclear cells ameliorate neurological damage in rats with focal ischemia. F1000Research 2014, 3, 131. [Google Scholar] [CrossRef]
- Hepponstall, M.; Ignjatovic, V.; Binos, S.; Monagle, P.; Jones, B.; Cheung, M.H.; d’Udekem, Y.; Konstantinov, I.E. Remote ischemic preconditioning (RIPC) modifies plasma proteome in humans. PLoS ONE 2012, 7, e48284. [Google Scholar] [CrossRef]
- Van Vlierberghe, H.; Langlois, M.; Delanghe, J. Haptoglobin polymorphisms and iron homeostasis in health and in disease. Clin. Chim. Acta Int. J. Clin. Chem. 2004, 345, 35–42. [Google Scholar] [CrossRef]
- Tseng, C.F.; Lin, C.C.; Huang, H.Y.; Liu, H.C.; Mao, S.J. Antioxidant role of human haptoglobin. Proteomics 2004, 4, 2221–2228. [Google Scholar] [CrossRef]
- Theilgaard-Monch, K.; Jacobsen, L.C.; Nielsen, M.J.; Rasmussen, T.; Udby, L.; Gharib, M.; Arkwright, P.D.; Gombart, A.F.; Calafat, J.; Moestrup, S.K.; et al. Haptoglobin is synthesized during granulocyte differentiation, stored in specific granules, and released by neutrophils in response to activation. Blood 2006, 108, 353–361. [Google Scholar] [CrossRef]
- Huntoon, K.M.; Wang, Y.; Eppolito, C.A.; Barbour, K.W.; Berger, F.G.; Shrikant, P.A.; Baumann, H. The acute phase protein haptoglobin regulates host immunity. J. Leukoc. Biol. 2008, 84, 170–181. [Google Scholar] [CrossRef]
- Huntoon, K.M.; Russell, L.; Tracy, E.; Barbour, K.W.; Li, Q.; Shrikant, P.A.; Berger, F.G.; Garrett-Sinha, L.A.; Baumann, H. A unique form of haptoglobin produced by murine hematopoietic cells supports B-cell survival, differentiation and immune response. Mol. Immunol. 2013, 55, 345–354. [Google Scholar] [CrossRef]
- Oh, S.K.; Pavlotsky, N.; Tauber, A.I. Specific binding of haptoglobin to human neutrophils and its functional consequences. J. Leukoc. Biol. 1990, 47, 142–148. [Google Scholar] [CrossRef]
- Morimoto, M.; Nakano, T.; Egashira, S.; Irie, K.; Matsuyama, K.; Wada, M.; Nakamura, Y.; Shigemori, Y.; Ishikura, H.; Yamashita, Y.; et al. Haptoglobin Regulates Macrophage/Microglia-Induced Inflammation and Prevents Ischemic Brain Damage Via Binding to HMGB1. J. Am. Heart Assoc. 2022, 11, e024424. [Google Scholar] [CrossRef]
- Yang, F.; Naylor, S.L.; Lum, J.B.; Cutshaw, S.; McCombs, J.L.; Naberhaus, K.H.; McGill, J.R.; Adrian, G.S.; Moore, C.M.; Barnett, D.R.; et al. Characterization, mapping, and expression of the human ceruloplasmin gene. Proc. Natl. Acad. Sci. USA 1986, 83, 3257–3261. [Google Scholar] [CrossRef]
- Williams, D.M.; Lee, G.R.; Cartwright, G.E. Ferroxidase activity of rat ceruloplasmin. Am. J. Physiol. 1974, 227, 1094–1097. [Google Scholar] [CrossRef][Green Version]
- Klebanoff, S.J. Bactericidal effect of Fe2+, ceruloplasmin, and phosphate. Arch. Biochem. Biophys. 1992, 295, 302–308. [Google Scholar] [CrossRef]
- Chalise, U.; Daseke, M.J., 2nd; Kalusche, W.J.; Konfrst, S.R.; Rodriguez-Paar, J.R.; Flynn, E.R.; Cook, L.M.; Becirovic-Agic, M.; Lindsey, M.L. Macrophages secrete murinoglobulin-1 and galectin-3 to regulate neutrophil degranulation after myocardial infarction. Mol. Omics 2022, 18, 186–195. [Google Scholar] [CrossRef]
- Mollet, I.; Martins, C.; Angelo-Dias, M.; Carvalho, A.S.; Aloria, K.; Matthiesen, R.; Viana-Baptista, M.; Borrego, L.M.; Vieira, H.L.A. Pilot study in human healthy volunteers on the mechanisms underlying remote ischemic conditioning (RIC)-Targeting circulating immune cells and immune-related proteins. J. Neuroimmunol. 2022, 367, 577847. [Google Scholar] [CrossRef]
- Pang, T.; Zhao, Y.; Zhang, N.R.; Jin, S.Q.; Pan, S.Q. Transient limb ischemia alters serum protein expression in healthy volunteers: Complement C3 and vitronectin may be involved in organ protection induced by remote ischemic preconditioning. Oxidative Med. Cell. Longev. 2013, 2013, 859056. [Google Scholar] [CrossRef]
- Mocco, J.; Mack, W.J.; Ducruet, A.F.; Sosunov, S.A.; Sughrue, M.E.; Hassid, B.G.; Nair, M.N.; Laufer, I.; Komotar, R.J.; Claire, M.; et al. Complement component C3 mediates inflammatory injury following focal cerebral ischemia. Circ. Res. 2006, 99, 209–217. [Google Scholar] [CrossRef]
- Alper, C.A.; Johnson, A.M.; Birtch, A.G.; Moore, F.D. Human C’3: Evidence for the liver as the primary site of synthesis. Science 1969, 163, 286–288. [Google Scholar] [CrossRef]
- Lubbers, R.; van Essen, M.F.; van Kooten, C.; Trouw, L.A. Production of complement components by cells of the immune system. Clin. Exp. Immunol. 2017, 188, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Botto, M.; Lissandrini, D.; Sorio, C.; Walport, M.J. Biosynthesis and secretion of complement component (C3) by activated human polymorphonuclear leukocytes. J. Immunol. 1992, 149, 1348–1355. [Google Scholar]
- Maas, C.; Oschatz, C.; Renne, T. The plasma contact system 2.0. Semin. Thromb. Hemost. 2011, 37, 375–381. [Google Scholar] [CrossRef]
- Kleinschnitz, C.; Stoll, G.; Bendszus, M.; Schuh, K.; Pauer, H.U.; Burfeind, P.; Renne, C.; Gailani, D.; Nieswandt, B.; Renne, T. Targeting coagulation factor XII provides protection from pathological thrombosis in cerebral ischemia without interfering with hemostasis. J. Exp. Med. 2006, 203, 513–518. [Google Scholar] [CrossRef]
- Penna, C.; Mancardi, D.; Rastaldo, R.; Losano, G.; Pagliaro, P. Intermittent activation of bradykinin B2 receptors and mitochondrial KATP channels trigger cardiac postconditioning through redox signaling. Cardiovasc. Res. 2007, 75, 168–177. [Google Scholar] [CrossRef] [PubMed]
- Danielisova, V.; Burda, J.; Nemethova, M.; Gottlieb, M.; Burda, R. An effective combination of two different methods of postconditioning. Neurochem. Res. 2012, 37, 2085–2091. [Google Scholar] [CrossRef] [PubMed]
- Danielisova, V.; Gottlieb, M.; Bonova, P.; Nemethova, M.; Burda, J. Bradykinin postconditioning ameliorates focal cerebral ischemia in the rat. Neurochem. Int. 2014, 72, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Mechirova, E.; Danielisova, V.; Domorakova, I.; Dankova, M.; Stebnicky, M.; Mickova, H.; Burda, J. Bradykinin preconditioning affects the number of degenerated neurons and the level of antioxidant enzymes in spinal cord ischemia in rabbits. Acta Histochem. 2014, 116, 252–257. [Google Scholar] [CrossRef]
- Boehm-Cagan, A.; Michaelson, D.M. Reversal of apoE4-driven brain pathology and behavioral deficits by bexarotene. J. Neurosci. Off. J. Soc. Neurosci. 2014, 34, 7293–7301. [Google Scholar] [CrossRef] [PubMed]
- Cramer, P.E.; Cirrito, J.R.; Wesson, D.W.; Lee, C.Y.; Karlo, J.C.; Zinn, A.E.; Casali, B.T.; Restivo, J.L.; Goebel, W.D.; James, M.J.; et al. ApoE-directed therapeutics rapidly clear beta-amyloid and reverse deficits in AD mouse models. Science 2012, 335, 1503–1506. [Google Scholar] [CrossRef]
- Liu, S.; Liu, J.; Weng, R.; Gu, X.; Zhong, Z. Apolipoprotein E gene polymorphism and the risk of cardiovascular disease and type 2 diabetes. BMC Cardiovasc. Disord. 2019, 19, 213. [Google Scholar] [CrossRef]
- Mabuchi, T.; Kitagawa, K.; Ohtsuki, T.; Kuwabara, K.; Yagita, Y.; Yanagihara, T.; Hori, M.; Matsumoto, M. Contribution of microglia/macrophages to expansion of infarction and response of oligodendrocytes after focal cerebral ischemia in rats. Stroke 2000, 31, 1735–1743. [Google Scholar] [CrossRef]
- Kitagawa, K.; Matsumoto, M.; Kuwabara, K.; Ohtsuki, T.; Hori, M. Delayed, but marked, expression of apolipoprotein E is involved in tissue clearance after cerebral infarction. J. Cereb. Blood Flow Metab. Off. J. Int. Soc. Cereb. Blood Flow Metab. 2001, 21, 1199–1207. [Google Scholar] [CrossRef]
- Ng, M.W.; Angerosa, J.; Konstantinov, I.E.; Cheung, M.M.; Pepe, S. Remote ischaemic preconditioning modifies serum apolipoprotein D, met-enkephalin, adenosine, and nitric oxide in healthy young adults. Clin. Exp. Pharmacol. Physiol. 2019, 46, 995–1000. [Google Scholar] [CrossRef]


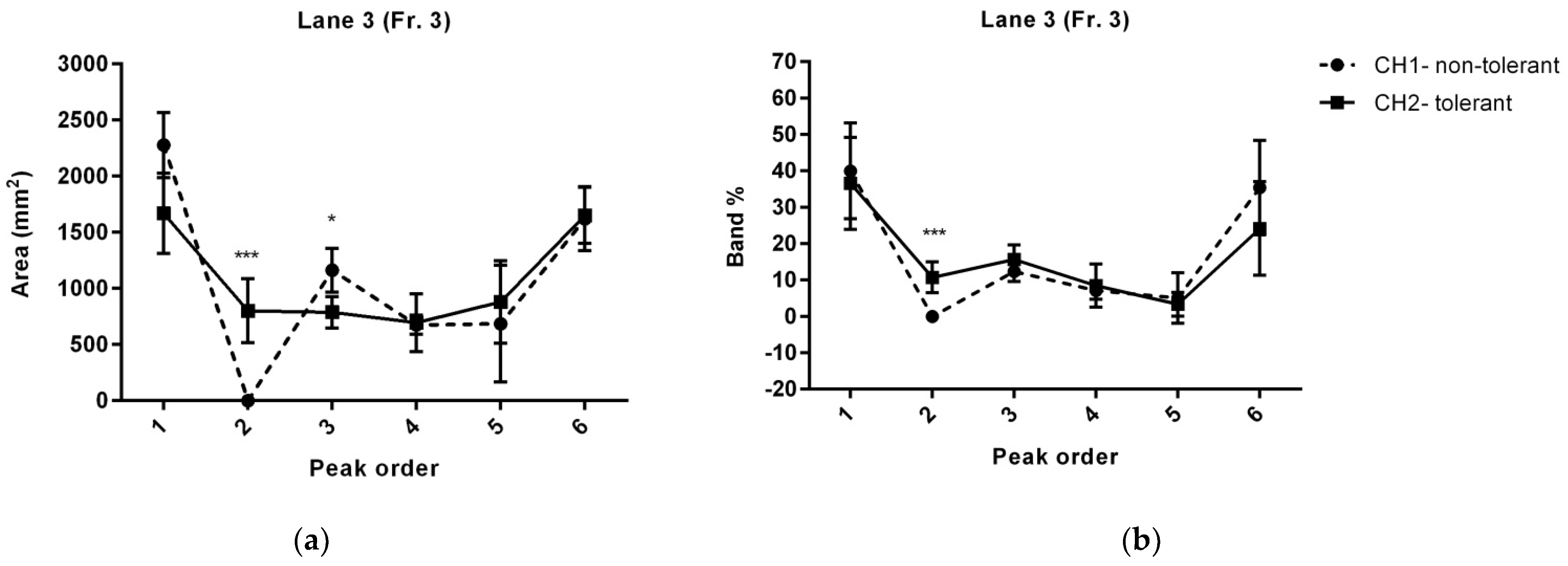
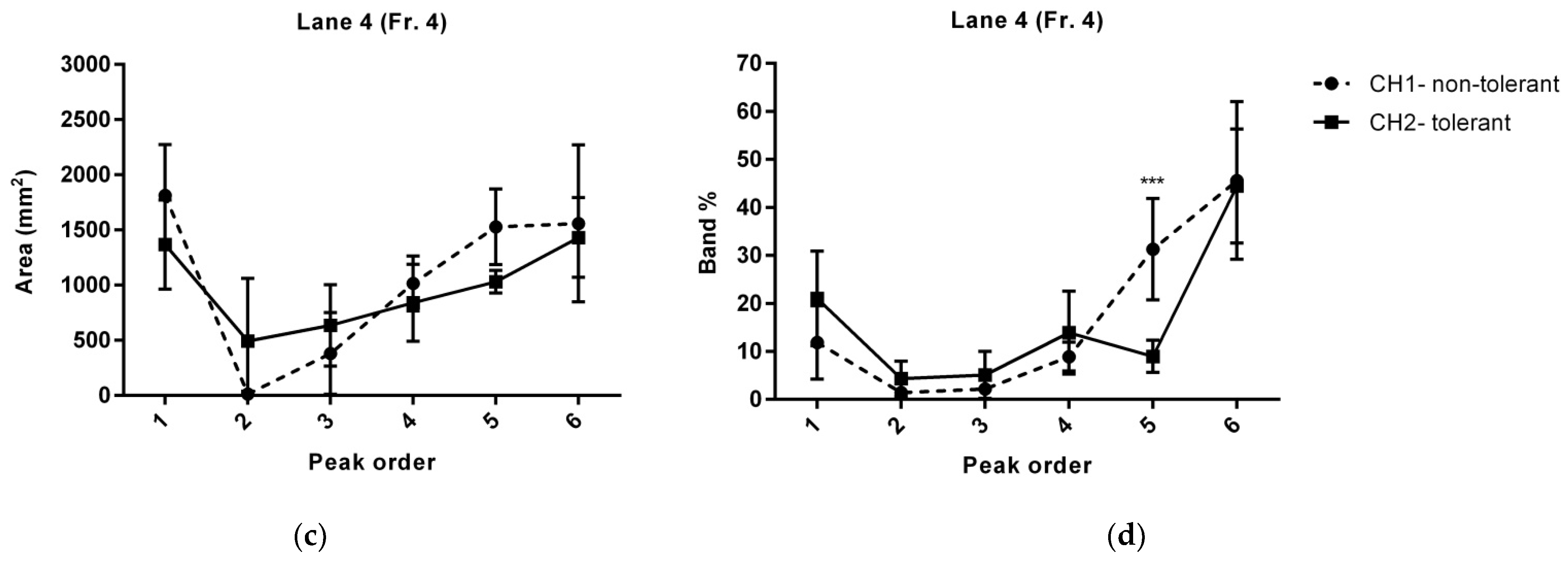

| Distance (mm) | Peak Sequence | Area (mm2) | Area (p-Value) | Band (%) | Band% (p-Value) | Protein | ||
|---|---|---|---|---|---|---|---|---|
| Non-Tolerant (CH1) | Tolerant (CH2) | Non-Tolerant | Tolerant | |||||
| Line 3 | ||||||||
| 5 | peak 1 | 2277.53 ± 144 | 1669.68 ± 205 | 0.0540651 | 39.99 ± 6 | 36.57 ± 5.7 | 0.685846 | Murinoglobulin-2 |
| 10 | Alpha-1-inhibitor 3 | |||||||
| Complement C3 | ||||||||
| 15 | Complement C3 | |||||||
| Guanine nucleotide-binding protein subunit alpha-13 | ||||||||
| 20 | peak 2 | X | 802.17 ± 142 | 0.00135494 * | X | 10.74 ± 1.9 | 0.000471288 * | Haptoglobin |
| 25 | peak 3 | 1162.79 ± 99 | 789.13 ± 70 | 0.0211761 * | 12.34 ± 1.2 | 15.68 ± 1.8 | 0.163233 | Centrosomal protein of 70 kDa |
| Glycerol-3-phosphate dehydrogenase, mitochondrial | ||||||||
| 3-hydroxy-3-methylglutaryl-coenzyme A reductase | ||||||||
| Kininogen-1 OS = Rattus norvegicus | ||||||||
| Thyroid hormone receptor alpha | ||||||||
| 30 | peak 4 | 675.5 ± 43 | 696.3 ± 129 | 0.882889 | 7.11 ± 1.1 | 8.47 ± 2.7 | 0.64914 | Fetuin-B OS = Rattus norvegicus |
| 35 | ||||||||
| 40 | peak 5 | 685.5 ± 260 | 880.08 ± 185 | 0.564531 | 5.06 ± 3.1 | 3.34 ± 1.5 | 0.628697 | Apolipoprotein E |
| 45 | peak 6 | 1620.67 ± 142 | 1650.91 ± 144 | 0.889648 | 35.4 ± 5.8 | 24.16 ± 5.8 | 0.206228 | Collagen alpha-2(I) chain |
| POU domain, class 4, transcription factor 2 | ||||||||
| Line 4 | ||||||||
| 5 | peak1 | 1814.12 ± 206 | 1368.27 ± 181 | 0.141957 | 11.88 ± 3.1 | 21.03 ± 4.1 | 0.102411 | |
| 10 | Murinoglobulin-1 | |||||||
| 15 | Ceruloplasmin | |||||||
| 20 | peak 2 | X | 493.21 ± 284 | 0.0885759 | X | 4.32 ± 1.5 | 0.0652243 | Haptoglobin |
| 25 | peak 3 | 381.37 ± 165 | 635.92 ± 165 | 0.307385 | 2.18 ± 1.2 | 5.12 ± 2 | 0.237078 | Alpha-2-HS-glycoprotein |
| 30 | peak 4 | 1015.21 ± 111 | 840.07 ± 156 | 0.387032 | 8.92 ± 1.2 | 13.94 ± 3.5 | 0.208264 | Alpha-2-HS-glycoprotein |
| 35 | ||||||||
| 40 | peak 5 | 1529.37 ± 171 | 1030.58 ± 59 | 0.058 | 31.3 ± 4.3 | 8.97 ± 1.4 | 0.000596955 * | Apolipoprotein E |
| 45 | peak 6 | 1559.96 ± 318 | 1432.38 ± 161 | 0.7299 | 45.61 ± 6.7 | 44.45 ± 4.8 | 0.891795 | Protein polyglycylase TTLL10 |
| #Term ID | Term Description | Matching Proteins |
|---|---|---|
| GO:0044092 | Negative regulation of molecular function | Thra, Pou4f2, Mug1, Hmgcr, A1i3, Hp, Apoe, Fetub, Ahsg, Mug2, Kng2, C3 |
| GO:0043086 | Negative regulation of catalytic activity | Mug1, Hmgcr, A1i3, Hp, Apoe, Fetub, Ahsg, Mug2, Kng2, C3 |
| GO:0032269 | Negative regulation of cellular protein metabolic process | Mug1, Hmgcr, A1i3, Apoe, Fetub, Ahsg, Mug2, Kng2, C3 |
| GO:0030162 | Regulation of proteolysis | Mug1, A1i3, Apoe, Fetub, Ahsg, Mug2, Kng2, C3 |
| GO:0031324 | Negative regulation of cellular metabolic process | Thra, Pou4f2, Mug1, Hmgcr, A1i3, Hp, Apoe, Fetub, Ahsg, Mug2, Kng2, C3 |
| GO:0050790 | Regulation of catalytic activity | Mug1, Hmgcr, A1i3, Hp, Apoe, Fetub, Gna13, Ahsg, Mug2, Kng2, C3 |
| GO:0002526 | Acute inflammatory response | Mug1, Hp, Ahsg, Kng2 |
| GO:0051172 | Negative regulation of nitrogen compound metabolic process | Thra, Pou4f2, Mug1, Hmgcr, A1i3, Apoe, Fetub, Ahsg, Mug2, Kng2, C3 |
| GO:0010605 | Negative regulation of macromolecule metabolic process | Thra, Pou4f2, Mug1, Hmgcr, A1i3, Apoe, Fetub, Ahsg, Mug2, Kng2, C3 |
| GO:0051336 | Regulation of hydrolase activity | Mug1, A1i3, Fetub, Gna13, Ahsg, Mug2, Kng2, C3 |
| GO:0006953 | Acute-phase response | Mug1, Hp, Ahsg |
| GO:0065009 | Regulation of molecular function | Thra, Pou4f2, Mug1, Hmgcr, A1i3, Hp, Apoe, Fetub, Gna13, Ahsg, Mug2, Kng2, C3 |
| GO:0009725 | Response to hormone | Thra, Pou4f2, Col1a2, Hp, Apoe, Ahsg, C3 |
| GO:0033993 | Response to lipid | Thra, Pou4f2, Col1a2, Hmgcr, Hp, Apoe, C3 |
| GO:0061045 | Negative regulation of wound healing | Hmgcr, Apoe, Kng2 |
| GO:0006469 | Negative regulation of protein kinase activity | Hmgcr, Apoe, Ahsg, Kng2 |
| GO:0014070 | Response to organic cyclic compound | Thra, Pou4f2, Col1a2, Hmgcr, Hp, Apoe, C3 |
| GO:0043407 | Negative regulation of map kinase activity | Hmgcr, Apoe, Kng2 |
| GO:0006954 | Inflammatory response | Mug1, Hp, Ahsg, Kng2, C3 |
| GO:0032101 | Regulation of response to external stimulus | Pou4f2, Hmgcr, Apoe, Ahsg, Kng2, C3 |
| GO:1900221 | Regulation of amyloid-beta clearance | Hmgcr, Apoe |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bonova, P.; Koncekova, J.; Nemethova, M.; Petrova, K.; Bona, M.; Gottlieb, M. Identification of Proteins Responsible for the Neuroprotective Effect of the Secretome Derived from Blood Cells of Remote Ischaemic Conditioned Rats. Biomolecules 2022, 12, 1423. https://doi.org/10.3390/biom12101423
Bonova P, Koncekova J, Nemethova M, Petrova K, Bona M, Gottlieb M. Identification of Proteins Responsible for the Neuroprotective Effect of the Secretome Derived from Blood Cells of Remote Ischaemic Conditioned Rats. Biomolecules. 2022; 12(10):1423. https://doi.org/10.3390/biom12101423
Chicago/Turabian StyleBonova, Petra, Jana Koncekova, Miroslava Nemethova, Klaudia Petrova, Martin Bona, and Miroslav Gottlieb. 2022. "Identification of Proteins Responsible for the Neuroprotective Effect of the Secretome Derived from Blood Cells of Remote Ischaemic Conditioned Rats" Biomolecules 12, no. 10: 1423. https://doi.org/10.3390/biom12101423
APA StyleBonova, P., Koncekova, J., Nemethova, M., Petrova, K., Bona, M., & Gottlieb, M. (2022). Identification of Proteins Responsible for the Neuroprotective Effect of the Secretome Derived from Blood Cells of Remote Ischaemic Conditioned Rats. Biomolecules, 12(10), 1423. https://doi.org/10.3390/biom12101423






