Label-Free Quantitative Proteomics Analysis of Adriamycin Selected Multidrug Resistant Human Lung Cancer Cells
Abstract
1. Introduction
2. Methodology
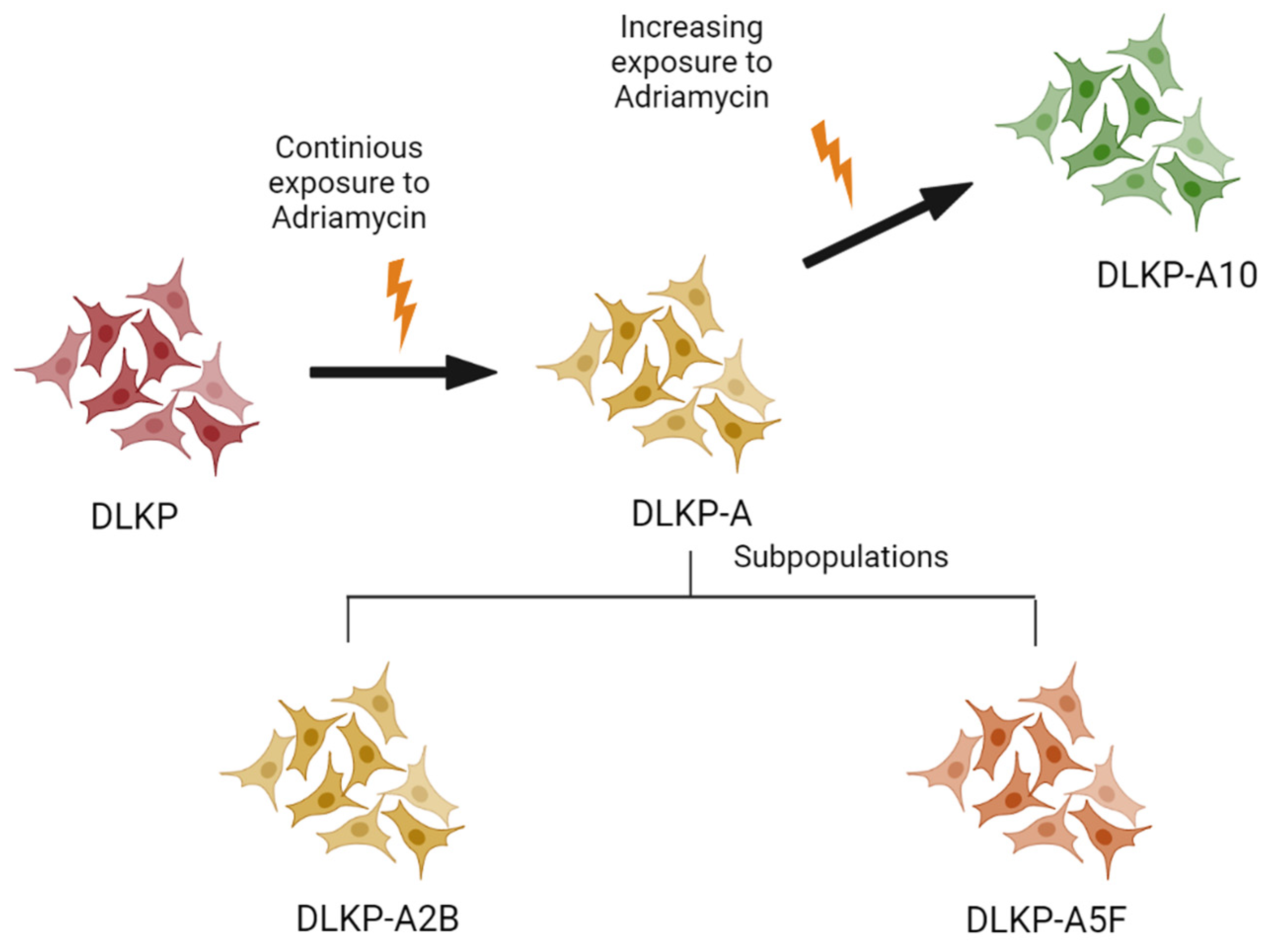
2.1. Cell Culture
2.2. Preparation of Mass Spectrometry Samples
2.3. Mass Spectrometry (the Nano LC-MS/MS)
2.4. Differential Expression (DE) Analysis
2.5. Gene Ontology
2.6. Western Blot for Validation
3. Results and Discussion
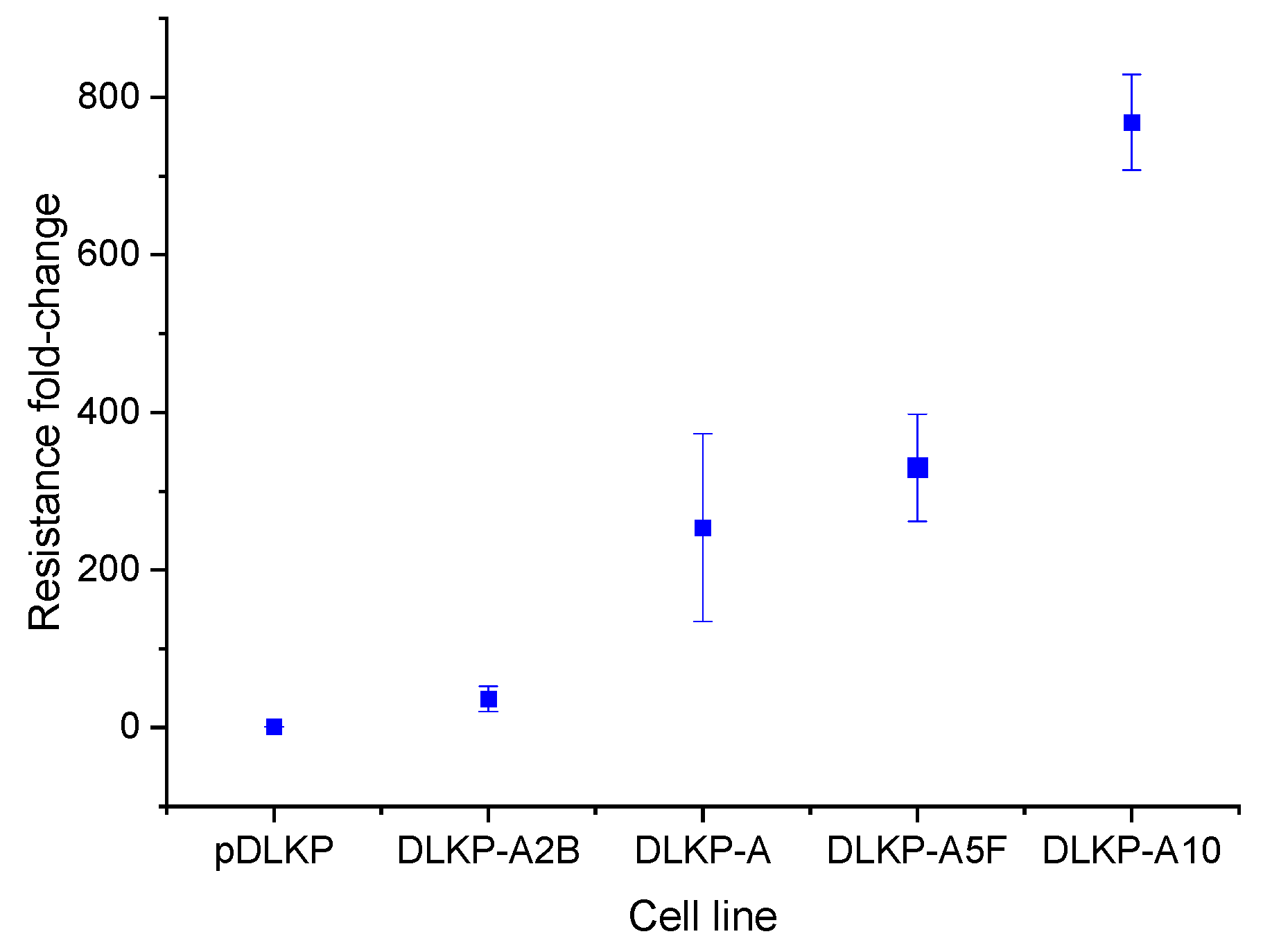
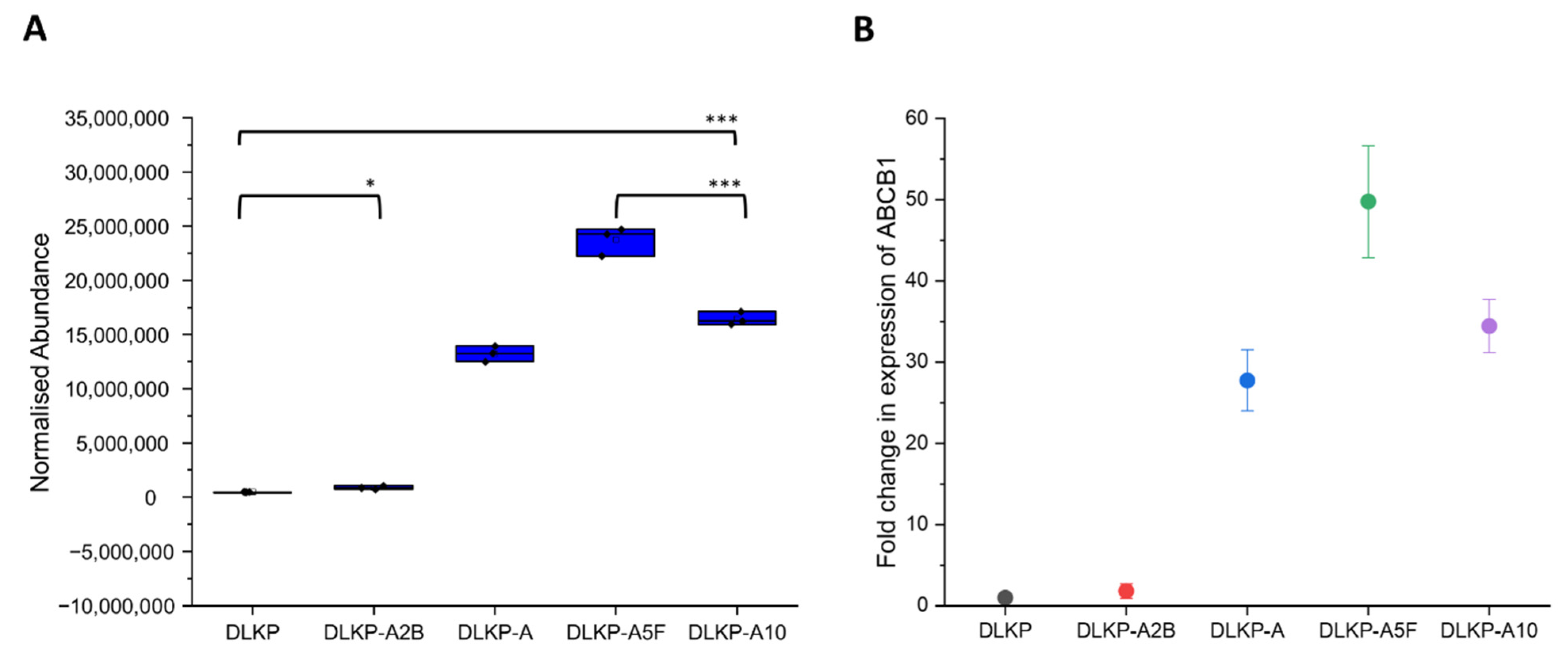
3.1. Drug Resistance in DLKP Subclones and the Role of ABC Transporters: ABCB1 and ABCC1
3.2. Interaction of ABCD1 and ABCD3-Transporter Proteins with Fatty Acid Metabolism and Peroxisome Pathway in Drug Resistance
3.3. Other ABC-Transporters and Their Expression Profiles in Drug-Resistant DLKP Subpopulations
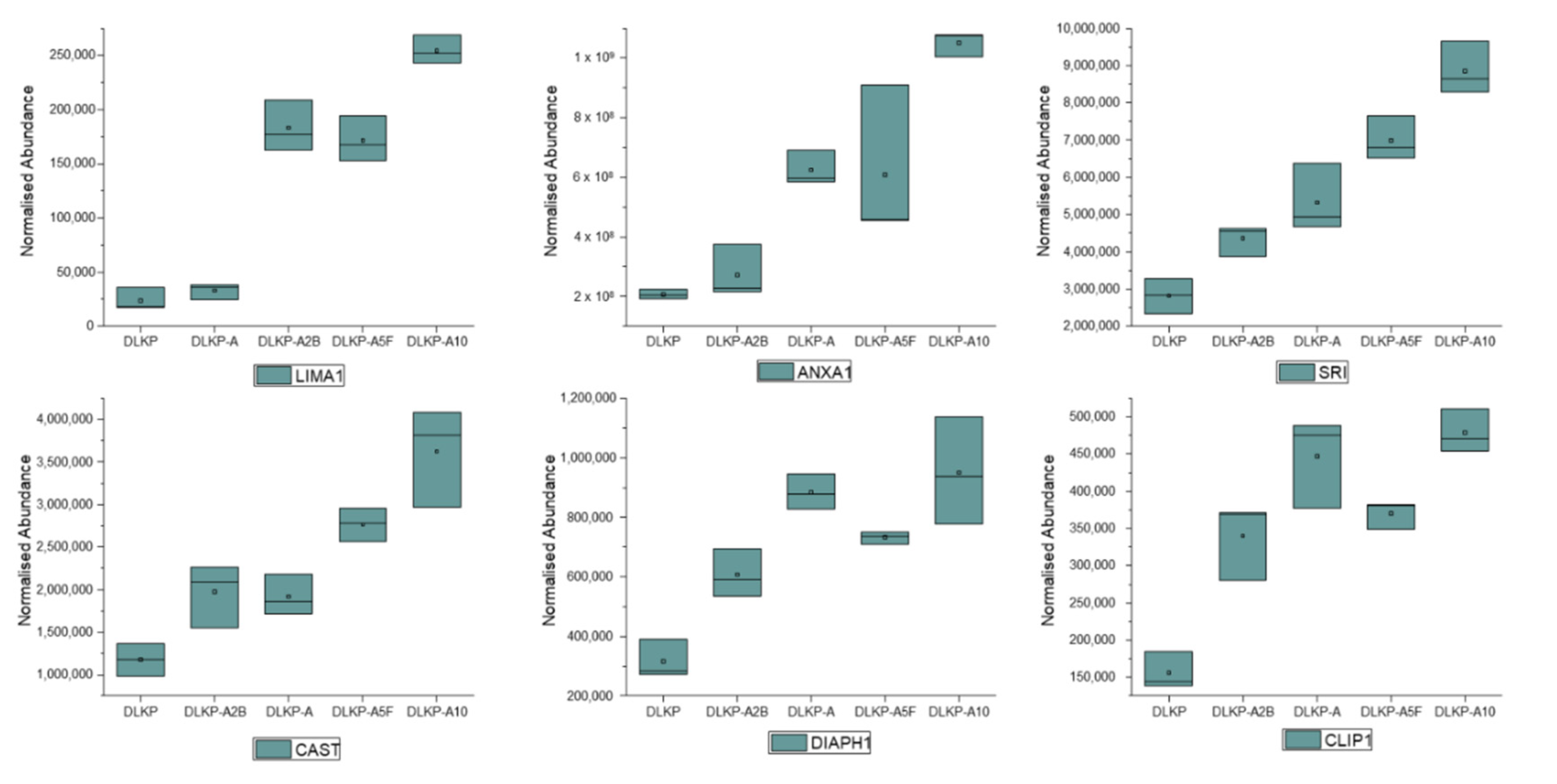
3.4. Expression Levels of Plasma Membrane-Related Proteins in pDLKP and Its Drug-Resistant Subpopulations
3.5. Enriched Pathways and Cellular Proteome Related to Enriched Drug-Resistance Mechanisms Determined by Gene Ontology (GO) Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lung Cancer. Available online: https://www.mariekeating.ie/cancer-information/lung-cancer/ (accessed on 6 June 2022).
- Kumar, S.; Kushwaha, P.P.; Gupta, S. Emerging Targets in Cancer Drug Resistance. Cancer Drug Resist. 2019, 2, 161–177. [Google Scholar] [CrossRef]
- Clynes, M.; Daly, C.; NicAmhlaoibh, R.; Cronin, D.; Elliott, C.; O’Connor, R.; O’Doherty, T.; Connolly, L.; Howlett, A.; Scanlon, K. Recent Developments in Drug Resistance and Apoptosis Research. Crit. Rev. Oncol. Hematol. 1998, 28, 181–205. [Google Scholar] [CrossRef]
- Lai, G.-M.; Moscow, J.A.; Alvarez, M.G.; Fojo, A.T.; Bates, S.E. Contribution of Glutathione and Glutathione-Dependent Enzymes in the Reversal of Adriamycin Resistance in Colon Carcinoma Cell Lines. Int. J. Cancer 1991, 49, 688–695. [Google Scholar] [CrossRef]
- Noguchi, M.; Kabayama, K.; Uemura, S.; Kang, B.; Saito, M.; Igarashi, Y.; Inokuchi, J. Endogenously Produced Ganglioside GM3 Endows Etoposide and Doxorubicin Resistance by Up-Regulating Bcl-2 Expression in 3LL Lewis Lung Carcinoma Cells. Glycobiology 2006, 16, 641–650. [Google Scholar] [CrossRef]
- Clynes, M.; Redmond, A.; Moran, E.; Gilvarry, U. Multiple Drug-Resistance in Variant of a Human Non-Small Cell Lung Carcinoma Cell Line, DLKP-A. Cytotechnology 1992, 10, 75–89. [Google Scholar] [CrossRef] [PubMed]
- Clynes, M. Cellular Models for Multiple Drug Resistance in Cancer. In Vitro Cell. Dev. Biol. 1993, 29, 171–179. [Google Scholar] [CrossRef]
- Law, E.; Gilvarry, U.; Lynch, V.; Gregory, B.; Grant, G.; Clynes, M. Cytogenetic Comparison of Two Poorly Differentiated Human Lung Squamous Cell Carcinoma Lines. Cancer Genet. Cytogenet. 1992, 59, 111–118. [Google Scholar] [CrossRef]
- Keenan, J.; Murphy, L.; Henry, M.; Meleady, P.; Clynes, M. Proteomic Analysis of Multidrug-Resistance Mechanisms in Adriamycin-Resistant Variants of DLKP, a Squamous Lung Cancer Cell Line. Proteomics 2009, 9, 1556–1566. [Google Scholar] [CrossRef] [PubMed]
- Heenan, M.; O’Driscoll, L.; Cleary, I.; Connolly, L.; Clynes, M. Isolation from a Human MDR Lung Cell Line of Multiple Clonal Subpopulations Which Exhibit Significantly Different Drug Resistance. Int. J. Cancer 1997, 71, 907–915. [Google Scholar] [CrossRef]
- Cleary, I.; Doherty, G.; Moran, E.; Clynes, M. The Multidrug-Resistant Human Lung Tumour Cell Line, DLKP-A10, Expresses Novel Drug Accumulation and Sequestration Systems. Biochem. Pharmacol. 1997, 53, 1493–1502. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Dagogo-Jack, I.; Shaw, A.T. Tumour Heterogeneity and Resistance to Cancer Therapies. Nat. Rev. Clin. Oncol. 2018, 15, 81–94. [Google Scholar] [CrossRef]
- Pikor, L.A.; Ramnarine, V.R.; Lam, S.; Lam, W.L. Genetic Alterations Defining NSCLC Subtypes and Their Therapeutic Implications. Lung Cancer 2013, 82, 179–189. [Google Scholar] [CrossRef]
- Robey, R.W.; Pluchino, K.M.; Hall, M.D.; Fojo, A.T.; Bates, S.E.; Gottesman, M.M. Revisiting the Role of ABC Transporters in Multidrug-Resistant Cancer. Nat. Rev. Cancer 2018, 18, 452–464. [Google Scholar] [CrossRef] [PubMed]
- Van der Deen, M.; de Vries, E.G.; Timens, W.; Scheper, R.J.; Timmer-Bosscha, H.; Postma, D.S. ATP-Binding Cassette (ABC) Transporters in Normal and Pathological Lung. Respir. Res. 2005, 6, 59. [Google Scholar] [CrossRef] [PubMed]
- Leonard, G.D.; Fojo, T.; Bates, S.E. The Role of ABC Transporters in Clinical Practice. Oncologist 2003, 8, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Dimanche-Boitrel, M.-T.; Garrido, C.; Chauffert, B. Kinetic Resistance to Anticancer Agents. In Multiple Drug Resistance in Cancer: Cellular, Molecular and Clinical Approaches; Clynes, M., Ed.; Springer: Dordrecht, The Netherlands, 1994; pp. 347–356. ISBN 978-94-011-0826-3. [Google Scholar]
- Choi, C.-H. ABC Transporters as Multidrug Resistance Mechanisms and the Development of Chemosensitizers for Their Reversal. Cancer Cell Int. 2005, 5, 30. [Google Scholar] [CrossRef][Green Version]
- Juliano, R.L.; Ling, V. A Surface Glycoprotein Modulating Drug Permeability in Chinese Hamster Ovary Cell Mutants. Biochim. Biophys. Acta 1976, 455, 152–162. [Google Scholar] [CrossRef]
- Dohse, M.; Robey, R.W.; Brendel, C.; Bates, S.; Neubauer, A.; Scharenberg, C. Efflux of the Tyrosine Kinase Inhibitors Imatinib and Nilotinib (AMN107) Is Mediated by ABCB1 (MDR1)-Type P-Glycoprotein. Blood 2006, 108, 1367. [Google Scholar] [CrossRef]
- Hegedus, C.; Ozvegy-Laczka, C.; Apáti, A.; Magócsi, M.; Német, K.; Orfi, L.; Kéri, G.; Katona, M.; Takáts, Z.; Váradi, A.; et al. Interaction of Nilotinib, Dasatinib and Bosutinib with ABCB1 and ABCG2: Implications for Altered Anti-Cancer Effects and Pharmacological Properties. Br. J. Pharmacol. 2009, 158, 1153–1164. [Google Scholar] [CrossRef]
- Gao, B.; Russell, A.; Beesley, J.; Chen, X.Q.; Healey, S.; Henderson, M.; Wong, M.; Emmanuel, C.; Galletta, L.; Johnatty, S.E.; et al. Paclitaxel Sensitivity in Relation to ABCB1 Expression, Efflux and Single Nucleotide Polymorphisms in Ovarian Cancer. Sci. Rep. 2014, 4, 4669. [Google Scholar] [CrossRef] [PubMed]
- Crawford, R.R.; Potukuchi, P.K.; Schuetz, E.G.; Schuetz, J.D. Beyond Competitive Inhibition: Regulation of ABC Transporters by Kinases and Protein-Protein Interactions as Potential Mechanisms of Drug-Drug Interactions. Drug Metab. Dispos. 2018, 46, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Cole, S.P.; Bhardwaj, G.; Gerlach, J.H.; Mackie, J.E.; Grant, C.E.; Almquist, K.C.; Stewart, A.J.; Kurz, E.U.; Duncan, A.M.; Deeley, R.G. Overexpression of a Transporter Gene in a Multidrug-Resistant Human Lung Cancer Cell Line. Science 1992, 258, 1650–1654. [Google Scholar] [CrossRef] [PubMed]
- Leier, I.; Jedlitschky, G.; Buchholz, U.; Cole, S.P.; Deeley, R.G.; Keppler, D. The MRP Gene Encodes an ATP-Dependent Export Pump for Leukotriene C4 and Structurally Related Conjugates. J. Biol. Chem. 1994, 269, 27807–27810. [Google Scholar] [CrossRef]
- Jedlitschky, G.; Leier, I.; Buchholz, U.; Barnouin, K.; Kurz, G.; Keppler, D. Transport of Glutathione, Glucuronate, and Sulfate Conjugates by the MRP Gene-Encoded Conjugate Export Pump. Cancer Res. 1996, 56, 988–994. [Google Scholar] [PubMed]
- Renes, J.; de Vries, E.G.E.; Jansen, P.L.M.; Müller, M. The (Patho)Physiological Functions of the MRP Family. Drug Resist. Updates 2000, 3, 289–302. [Google Scholar] [CrossRef] [PubMed]
- Morita, M.; Imanaka, T. Peroxisomal ABC Transporters: Structure, Function and Role in Disease. Biochim. Biophys. Acta BBA Mol. Basis Dis. 2012, 1822, 1387–1396. [Google Scholar] [CrossRef]
- Tawbeh, A.; Gondcaille, C.; Trompier, D.; Savary, S. Peroxisomal ABC Transporters: An Update. Int. J. Mol. Sci. 2021, 22, 6093. [Google Scholar] [CrossRef]
- Koundouros, N.; Poulogiannis, G. Reprogramming of Fatty Acid Metabolism in Cancer. Br. J. Cancer 2020, 122, 4–22. [Google Scholar] [CrossRef]
- Hama, K.; Fujiwara, Y.; Hayama, T.; Ozawa, T.; Nozawa, K.; Matsuda, K.; Hashiguchi, Y.; Yokoyama, K. Very Long-Chain Fatty Acids Are Accumulated in Triacylglycerol and Nonesterified Forms in Colorectal Cancer Tissues. Sci. Rep. 2021, 11, 6163. [Google Scholar] [CrossRef]
- Fernández, L.P.; Merino, M.; Colmenarejo, G.; Moreno-Rubio, J.; Sánchez-Martínez, R.; Quijada-Freire, A.; Gómez de Cedrón, M.; Reglero, G.; Casado, E.; Sereno, M.; et al. Metabolic Enzyme ACSL3 Is a Prognostic Biomarker and Correlates with Anticancer Effectiveness of Statins in Non-Small Cell Lung Cancer. Mol. Oncol. 2020, 14, 3135–3152. [Google Scholar] [CrossRef] [PubMed]
- Hillebrand, M.; Gersting, S.W.; Lotz-Havla, A.S.; Schäfer, A.; Rosewich, H.; Valerius, O.; Muntau, A.C.; Gärtner, J. Identification of a New Fatty Acid Synthesis-Transport Machinery at the Peroxisomal Membrane. J. Biol. Chem. 2012, 287, 210–221. [Google Scholar] [CrossRef] [PubMed]
- Hoppel, C. The Role of Carnitine in Normal and Altered Fatty Acid Metabolism. Am. J. Kidney Dis. 2003, 41, S4–S12. [Google Scholar] [CrossRef]
- Longo, N.; Frigeni, M.; Pasquali, M. Carnitine transport and fatty acid oxidation. Biochim. Biophys. Acta 2016, 1863, 2422–2435. [Google Scholar] [CrossRef]
- Drury, J.; Rychahou, P.G.; He, D.; Jafari, N.; Wang, C.; Lee, E.Y.; Weiss, H.L.; Evers, B.M.; Zaytseva, Y.Y. Inhibition of Fatty Acid Synthase Upregulates Expression of CD36 to Sustain Proliferation of Colorectal Cancer Cells. Front. Oncol. 2020, 10, 1185. [Google Scholar] [CrossRef]
- Xu, S.; Chen, T.; Dong, L.; Li, T.; Xue, H.; Gao, B.; Ding, X.; Wang, H.; Li, H. Fatty Acid Synthase Promotes Breast Cancer Metastasis by Mediating Changes in Fatty Acid Metabolism. Oncol. Lett. 2021, 21, 27. [Google Scholar] [CrossRef]
- Fhu, C.W.; Ali, A. Fatty Acid Synthase: An Emerging Target in Cancer. Molecules 2020, 25, 3935. [Google Scholar] [CrossRef]
- Menendez, J.A.; Lupu, R. Fatty Acid Synthase and the Lipogenic Phenotype in Cancer Pathogenesis. Nat. Rev. Cancer 2007, 7, 763–777. [Google Scholar] [CrossRef]
- Pizer, E.S.; Wood, F.D.; Heine, H.S.; Romantsev, F.E.; Pasternack, G.R.; Kuhajda, F.P. Inhibition of Fatty Acid Synthesis Delays Disease Progression in a Xenograft Model of Ovarian Cancer. Cancer Res. 1996, 56, 1189–1193. [Google Scholar]
- Bao, L.; Wu, J.; Dodson, M.; Rojo de la Vega, E.M.; Ning, Y.; Zhang, Z.; Yao, M.; Zhang, D.D.; Xu, C.; Yi, X. ABCF2, an Nrf2 Target Gene, Contributes to Cisplatin Resistance in Ovarian Cancer Cells. Mol. Carcinog. 2017, 56, 1543–1553. [Google Scholar] [CrossRef]
- Kara, G.; Tuncer, S.; Türk, M.; Denkbaş, E.B. Downregulation of ABCE1 via SiRNA Affects the Sensitivity of A549 Cells against Chemotherapeutic Agents. Med. Oncol. 2015, 32, 103. [Google Scholar] [CrossRef]
- Jiang, W.G.; Martin, T.A.; Lewis-Russell, J.M.; Douglas-Jones, A.; Ye, L.; Mansel, R.E. Eplin-Alpha Expression in Human Breast Cancer, the Impact on Cellular Migration and Clinical Outcome. Mol. Cancer 2008, 7, 71. [Google Scholar] [CrossRef]
- Sanders, A.J.; Martin, T.A.; Ye, L.; Mason, M.D.; Jiang, W.G. EPLIN Is a Negative Regulator of Prostate Cancer Growth and Invasion. J. Urol. 2011, 186, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Onozawa, H.; Saito, M.; Saito, K.; Kanke, Y.; Watanabe, Y.; Hayase, S.; Sakamoto, W.; Ishigame, T.; Momma, T.; Ohki, S.; et al. Annexin A1 Is Involved in Resistance to 5-FU in Colon Cancer Cells. Oncol. Rep. 2017, 37, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Serfass, L.; Roy, M.O.; Wong, J.; Bonneau, A.M.; Georges, E. Annexin-I Expression Modulates Drug Resistance in Tumor Cells. Biochem. Biophys. Res. Commun. 2004, 314, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Oshi, M.; Tokumaru, Y.; Mukhopadhyay, S.; Yan, L.; Matsuyama, R.; Endo, I.; Takabe, K. Annexin A1 Expression Is Associated with Epithelial-Mesenchymal Transition (EMT), Cell Proliferation, Prognosis, and Drug Response in Pancreatic Cancer. Cells 2021, 10, 653. [Google Scholar] [CrossRef] [PubMed]
- Belvedere, R.; Bizzarro, V.; Forte, G.; Dal Piaz, F.; Parente, L.; Petrella, A. Annexin A1 Contributes to Pancreatic Cancer Cell Phenotype, Behaviour and Metastatic Potential Independently of Formyl Peptide Receptor Pathway. Sci. Rep. 2016, 6, 29660. [Google Scholar] [CrossRef]
- Battista, T.; Fiorillo, A.; Chiarini, V.; Genovese, I.; Ilari, A.; Colotti, G. Roles of Sorcin in Drug Resistance in Cancer: One Protein, Many Mechanisms, for a Novel Potential Anticancer Drug Target. Cancers 2020, 12, 887. [Google Scholar] [CrossRef] [PubMed]
- Wendt, A.; Thompson, V.F.; Goll, D.E. Interaction of Calpastatin with Calpain: A Review. Biol. Chem. 2004, 385, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Fu, X.; Wang, Y.; Wang, X.; Wang, H.; Wen, J.; Kang, N. Protein Diaphanous Homolog 1 (Diaph1) Promotes Myofibroblastic Activation of Hepatic Stellate Cells by Regulating Rab5a Activity and TGFβ Receptor Endocytosis. FASEB J. 2020, 34, 7345–7359. [Google Scholar] [CrossRef]
- Miao, S.; Schäfer, P.; Nojszewski, J.; Meyer, F.; Windhorst, S. DIAPH1 Regulates Chromosomal Instability of Cancer Cells by Controlling Microtubule Dynamics. Eur. J. Cell Biol. 2021, 100, 151156. [Google Scholar] [CrossRef] [PubMed]
- Kita, K.; Thakkar, P.V.; Galletti, G.; Madhukar, N.; Navarro, E.V.; Barasoain, I.; Goodson, H.V.; Sackett, D.; Díaz, J.F.; Elemento, O.; et al. Imatinib Overrides Taxane Resistance by Selective Inhibition of Novel CLIP1 Variant Obstructing the Microtubule Pore. bioRxiv 2019, 838334v1. [Google Scholar] [CrossRef]
- CLIP1-LTK Fusion: A Novel Druggable Gene Fusion in NSCLC, Identified Using LC-SCRUM-Asia Genomic Screening Platform—A Step Further to Precision Medicine. Available online: https://www.amed.go.jp/en/news/20211129-01.html (accessed on 30 May 2022).
- Della Fazia, M.A.; Castelli, M.; Bartoli, D.; Pieroni, S.; Pettirossi, V.; Piobbico, D.; Viola-Magni, M.; Servillo, G. HOPS: A Novel CAMP-Dependent Shuttling Protein Involved in Protein Synthesis Regulation. J. Cell Sci. 2005, 118, 3185–3194. [Google Scholar] [CrossRef][Green Version]
- Fu, H.; Xu, J.; Chen, J.; Li, G.; Zhao, X.; Chen, P. Microarray Analysis Reveals Tmub1 as a Cell Cycle-Associated Protein in Rat Hepatocytes. Mol. Med. Rep. 2018, 17, 4337–4344. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Zhang, Y.; Chen, J.; Zhou, B.; Chen, G.; Chen, P. Tmub1 Suppresses Hepatocellular Carcinoma by Promoting the Ubiquitination of ΔNp63 Isoforms. Mol. Ther. Oncolytics 2020, 18, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Haxho, F.; Neufeld, R.J.; Szewczuk, M.R. Neuraminidase-1: A Novel Therapeutic Target in Multistage Tumorigenesis. Oncotarget 2016, 7, 40860–40881. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yang, S.; Jakoncic, J.; Zhang, J.J.; Huang, X.-Y. Migrastatin Analogues Target Fascin to Block Tumour Metastasis. Nature 2010, 464, 1062–1066. [Google Scholar] [CrossRef] [PubMed]
- Zhe, N.; Wang, J.; Chen, S.; Lin, X.; Chai, Q.; Zhang, Y.; Zhao, J.; Fang, Q. Heme Oxygenase-1 Plays a Crucial Role in Chemoresistance in Acute Myeloid Leukemia. Hematology 2015, 20, 384–391. [Google Scholar] [CrossRef]
- Heasman, S.-A.; Zaitseva, L.; Bowles, K.M.; Rushworth, S.A.; Macewan, D.J. Protection of Acute Myeloid Leukaemia Cells from Apoptosis Induced by Front-Line Chemotherapeutics Is Mediated by Haem Oxygenase-1. Oncotarget 2011, 2, 658–668. [Google Scholar] [CrossRef] [PubMed]
- Barbazan, J.; Dunkel, Y.; Li, H.; Nitsche, U.; Janssen, K.-P.; Messer, K.; Ghosh, P. Prognostic Impact of Modulators of G Proteins in Circulating Tumor Cells from Patients with Metastatic Colorectal Cancer. Sci. Rep. 2016, 6, 22112. [Google Scholar] [CrossRef] [PubMed]
- Hua, Y.-Q.; Zhang, K.; Sheng, J.; Ning, Z.-Y.; Li, Y.; Shi, W.; Liu, L.-M. NUCB1 Suppresses Growth and Shows Additive Effects With Gemcitabine in Pancreatic Ductal Adenocarcinoma via the Unfolded Protein Response. Front. Cell Dev. Biol. 2021, 9, 641836. [Google Scholar] [CrossRef] [PubMed]
- Oliva, C.R.; Markert, T.; Gillespie, G.Y.; Griguer, C.E. Nuclear-Encoded Cytochrome c Oxidase Subunit 4 Regulates BMI1 Expression and Determines Proliferative Capacity of High-Grade Gliomas. Oncotarget 2015, 6, 4330–4344. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Guccini, I.; Di Mitri, D.; Brina, D.; Revandkar, A.; Sarti, M.; Pasquini, E.; Alajati, A.; Pinton, S.; Losa, M.; et al. Compartmentalized Activities of the Pyruvate Dehydrogenase Complex Sustain Lipogenesis in Prostate Cancer. Nat. Genet. 2018, 50, 219–228. [Google Scholar] [CrossRef]
- Cevatemre, B.; Ulukaya, E.; Dere, E.; Dilege, S.; Acilan, C. Pyruvate Dehydrogenase Contributes to Drug Resistance of Lung Cancer Cells Through Epithelial Mesenchymal Transition. Front. Cell Dev. Biol. 2022, 9, 3745. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Cao, J.; Zhao, J.; Li, X.; Suo, Z.; Li, H. PDHA1 Gene Knockout In Human Esophageal Squamous Cancer Cells Resulted In Greater Warburg Effect And Aggressive Features In Vitro And In Vivo. OncoTargets Ther. 2019, 12, 9899–9913. [Google Scholar] [CrossRef]
- Lin, S.-R.; Wen, Y.-C.; Yeh, H.-L.; Jiang, K.-C.; Chen, W.-H.; Mokgautsi, N.; Huang, J.; Chen, W.-Y.; Liu, Y.-N. EGFR-Upregulated LIFR Promotes SUCLG2-Dependent Castration Resistance and Neuroendocrine Differentiation of Prostate Cancer. Oncogene 2020, 39, 6757–6775. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Zhou, J.; Zhang, K.; Li, L.; Xu, Y.; Ma, K.; Xie, H.; Cai, L.; Gong, Y.; Gong, K. Identification and Validation of the Clinical Roles of the VHL-Related LncRNAs in Clear Cell Renal Cell Carcinoma. J. Cancer 2021, 12, 2702–2714. [Google Scholar] [CrossRef] [PubMed]
- Xing, C.; Cai, Z.; Gong, J.; Zhou, J.; Xu, J.; Guo, F. Identification of Potential Biomarkers Involved in Gastric Cancer Through Integrated Analysis of Non-Coding RNA Associated Competing Endogenous RNAs Network. Clin. Lab. 2018, 64, 1661–1669. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Cai, Y.; Zhao, G.; Li, M. A Ten N6-Methyladenosine-Related Long Non-Coding RNAs Signature Predicts Prognosis of Triple-Negative Breast Cancer. J. Clin. Lab. Anal. 2021, 35, e23779. [Google Scholar] [CrossRef]
- Lee, I.-N.; Yang, J.-T.; Huang, C.; Huang, H.-C.; Wu, Y.-P.; Chen, J.-C. Elevated XRCC5 Expression Level Can Promote Temozolomide Resistance and Predict Poor Prognosis in Glioblastoma. Oncol. Lett. 2021, 21, 443. [Google Scholar] [CrossRef] [PubMed]
- Piskareva, O.; Harvey, H.; Nolan, J.; Conlon, R.; Alcock, L.; Buckley, P.; Dowling, P.; Henry, M.; O’Sullivan, F.; Bray, I.; et al. The Development of Cisplatin Resistance in Neuroblastoma Is Accompanied by Epithelial to Mesenchymal Transition in Vitro. Cancer Lett. 2015, 364, 142–155. [Google Scholar] [CrossRef] [PubMed]
- Bau, D.-T.; Tsai, C.-W.; Wu, C.-N. Role of the XRCC5/XRCC6 Dimer in Carcinogenesis and Pharmacogenomics. Pharmacogenomics 2011, 12, 515–534. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; García-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D.J.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2021, 50, D543–D552. [Google Scholar] [CrossRef] [PubMed]

| Term ID | Term Description | Observed Gene Count | Background Gene Count | False Discovery Rate |
|---|---|---|---|---|
| Enriched Biological Processes | ||||
| GO:0017144 | drug metabolic process | 112 | 622 | 8.45 × 10−18 |
| GO:0042493 | response to drug | 100 | 900 | 1.17 × 10−5 |
| GO:2001038 | regulation of cellular response to drug | 9 | 28 | 0.0035 |
| GO:2001024 | negative regulation of response to drug | 8 | 29 | 0.0144 |
| GO:0035690 | cellular response to drug | 33 | 310 | 0.0455 |
| Enriched Molecular Functions | ||||
| GO:0008144 | drug binding | 226 | 1710 | 7.03 × 10−21 |
| Accession | Description | Anova (p) | A2B | A | A5F | A10 |
|---|---|---|---|---|---|---|
| Differentially expressed proteins in drug related biological processes | ||||||
| P09601 | HMOX1 | 0.000186 | −3.2 | −7.9 | −34.5 | −56.1 |
| P34913 | EPHX2 | 0.000329 | 1.2 | 2.5 | 3.9 | 7.4 |
| Q02818 | NUCB1 | 4.23 × 10−5 | −1.2 | −1.9 | −2.0 | −2.8 |
| P22695 | UQCRC2 | 6.41 × 10−5 | 2.0 | 2.3 | 2.5 | 2.8 |
| P36957 | DLST | 8.11 × 10−6 | 1.6 | 1.8 | 1.9 | 2.3 |
| Q96I99 | SUCLG2 | 9.31 × 10−5 | 1.3 | 1.5 | 1.5 | 1.9 |
| P08559 | PDHA1 | 0.001339 | 1.3 | 1.5 | 1.6 | 1.7 |
| P13073 | COX4I1 | 0.000831 | −1.2 | −1.5 | −1.5 | −1.6 |
| P13010 | XRCC5 | 0.000174 | 1.0 | 1.3 | 1.4 | 1.5 |
| Differentially expressed proteins in drug related molecular functions | ||||||
| P34932 | HSPA4 | 0.000191948 | 1.3 | 1.5 | 1.6 | 1.9 |
| Q9P2J5 | LARS1 | 0.000250465 | 1.1 | 1.3 | 1.7 | 1.9 |
| O95757 | HSPA4L | 3.60 × 10−5 | 1.2 | 1.4 | 1.5 | 1.8 |
| Q08211 | DHX9 | 2.72 × 10−5 | 1.2 | 1.3 | 1.4 | 1.6 |
| P50991 | CCT4 | 0.000158646 | 1.0 | 1.3 | 1.4 | 1.6 |
| P11216 | PYGB | 0.004613192 | 1.1 | 1.2 | 1.3 | 1.7 |
| Q9UHJ6 | SHPK | 3.91 × 10−6 | 2.0 | 6.1 | 6.9 | 10.2 |
| Q16658 | FSCN1 | 7.52 × 10−7 | −1.5 | −1.5 | −3.2 | −4.7 |
| Biological/Molecular Assignment | Gene Name | DLKP vs. Resistant Max. Fold Change * | SK-MES-1 vs. Resistant Max. Fold Change | COR-23 vs. Resistant Max. Fold Change |
|---|---|---|---|---|
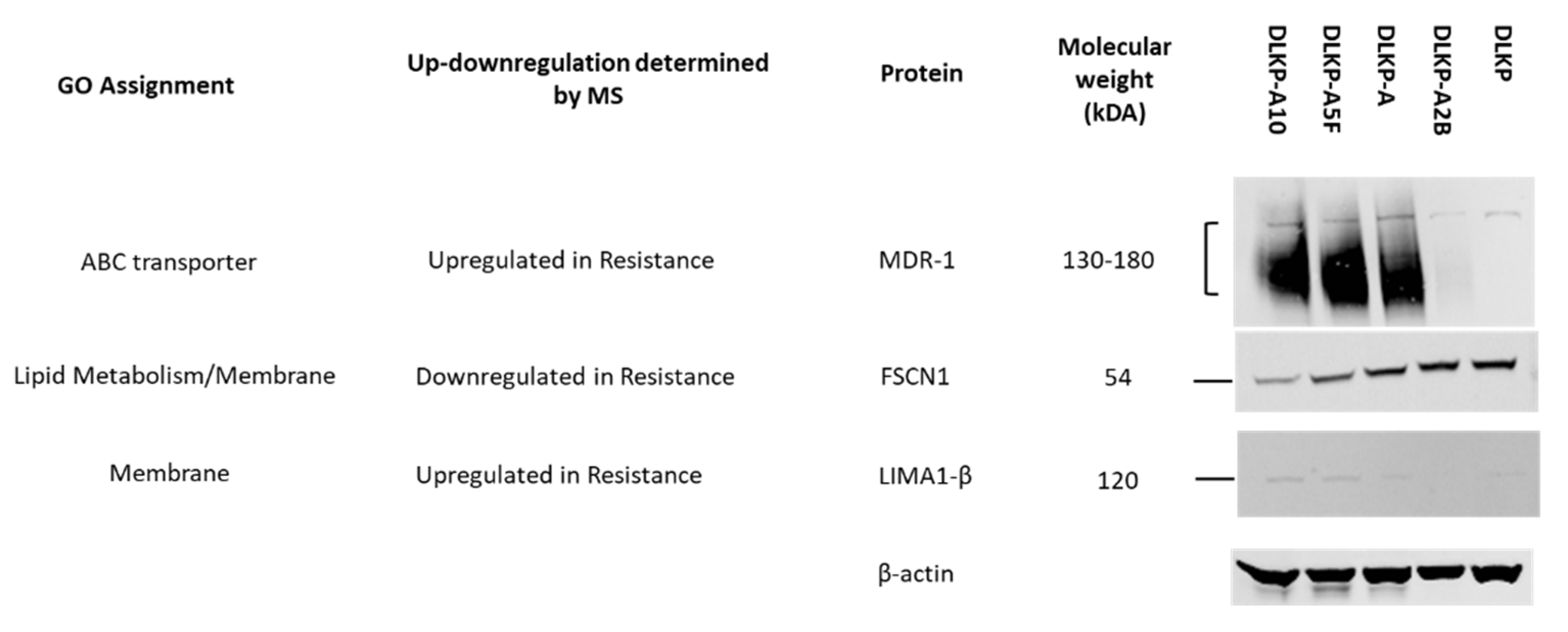
| Drug transport | ABCB1 | +(49.8-fold) | +(13.5-fold) | - |
| ABCC1 | +(18.8-fold) | - | +(15.1-fold) | |
| Lipid metabolism | ABCD1 | −(131.1-fold) | - | - |
| ABCD3 | +(2.0-fold) | +(4.1-fold) | - | |
| ALDH1B1 | +(2.6-fold) | - | - | |
| ALDH3A2 | +(2.6-fold) | +(4.9-fold) | - | |
| GCDH | +(4.4-fold) | - | - | |
| FASN | +(1.6-fold) | - | - | |
| ACACA | +(2-fold) | +(2.7-fold) | −(2.1-fold) | |
| CROT | +(52.8-fold) | - | - | |
| Membrane proteins | LIMA1 | +(10.7-fold) | - | −(1.5-fold) |
| SRI | +(3.1-fold) | +(4.8-fold) | +(1.6-fold) | |
| ANXA1 | +(5.1-fold) | - | +(4.6-fold) | |
| CAST | +(3.1-fold) | +(2.1-fold) | - | |
| DIAPH1 | +(3.0-fold) | −(1.2-fold) | −(3.3-fold) | |
| CLIP1 | +(3.1-fold) | +(5.2-fold) | +(1.9-fold) | |
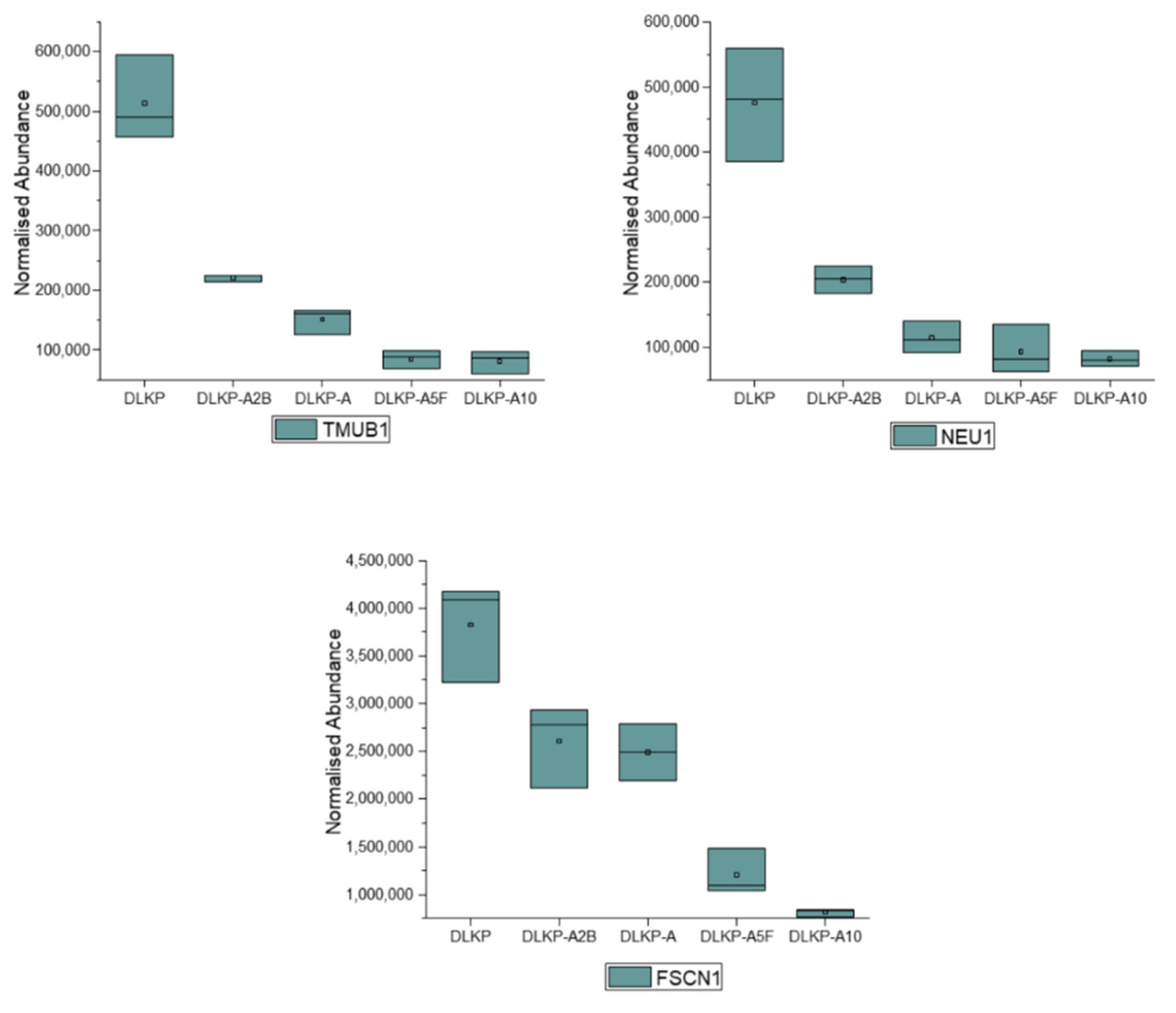
| TMUB1 | −(6.3-fold) | - | - | |
| NEU1 | −(5.8-fold) | +(2.8-fold) | - | |
| FSCN1 | −(4.7-fold) | - | −(1.3-fold) | |
| Gene Ontology-drug metabolism related DE proteins | HMOX1 | −(56.1-fold) | - | - |
| EPHX2 | +(7.3-fold) | - | - | |
| NUCB1 | −(2.8-fold) | - | −(1.3-fold) | |
| UQCRC2 | +(2.8-fold) | +(1.2-fold) | - | |
| DLST | +(2.3-fold) | - | −(1.5-fold) | |
| SUCLG2 | +(1.9-fold) | - | - | |
| PDHA1 | +(1.7-fold) | +(2.2-fold) | - | |
| COX4I1 | −(1.6-fold) | - | +(1.3-fold) | |
| XRCC5 | +(1.5-fold) | - | - | |
| HSPA4 | +(1.9-fold) | - | - | |
| LARS1 | +(1.8-fold) | - | +(1.6-fold) | |
| HSPA4L | +(1.8-fold) | −(2.1-fold) | - | |
| DHX9 | +(1.6-fold) | - | +(1.5-fold) | |
| CCT4 | +(1.6-fold) | - | +(1.6-fold) | |
| PYGB | +(1.7-fold) | - | +(1.6-fold) | |
| SHPK | +(10.3-fold) | - | −(2.1-fold) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Efeoglu, E.; Henry, M.; Clynes, M.; Meleady, P. Label-Free Quantitative Proteomics Analysis of Adriamycin Selected Multidrug Resistant Human Lung Cancer Cells. Biomolecules 2022, 12, 1401. https://doi.org/10.3390/biom12101401
Efeoglu E, Henry M, Clynes M, Meleady P. Label-Free Quantitative Proteomics Analysis of Adriamycin Selected Multidrug Resistant Human Lung Cancer Cells. Biomolecules. 2022; 12(10):1401. https://doi.org/10.3390/biom12101401
Chicago/Turabian StyleEfeoglu, Esen, Michael Henry, Martin Clynes, and Paula Meleady. 2022. "Label-Free Quantitative Proteomics Analysis of Adriamycin Selected Multidrug Resistant Human Lung Cancer Cells" Biomolecules 12, no. 10: 1401. https://doi.org/10.3390/biom12101401
APA StyleEfeoglu, E., Henry, M., Clynes, M., & Meleady, P. (2022). Label-Free Quantitative Proteomics Analysis of Adriamycin Selected Multidrug Resistant Human Lung Cancer Cells. Biomolecules, 12(10), 1401. https://doi.org/10.3390/biom12101401






