Prostate Cancer-Associated miRNAs in Saliva: First Steps to an Easily Accessible and Reliable Screening Tool
Abstract
1. Introduction
1.1. Background
1.2. Literature Review
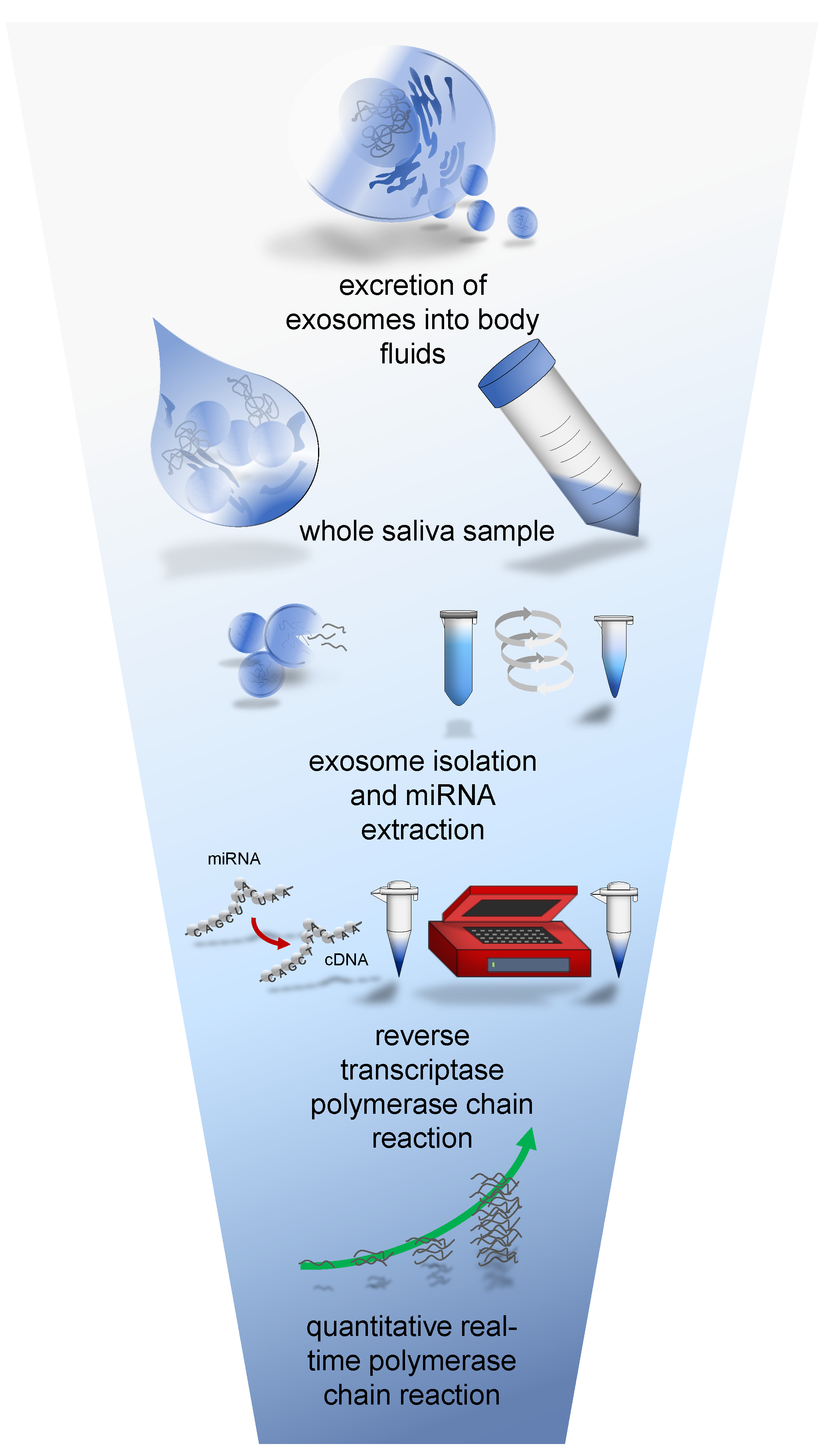
1.3. Specimen Collection and Methodological Adaptions
2. Materials & Methods
2.1. Research Subjects
2.2. qRT-PCR
2.3. Statistical Analysis
| Participant Collective | Cancer Group | Control Group | Total | |
|---|---|---|---|---|
| [N] (n/total) | 43 (58%) | 31 (42%) | 74 (100%) | |
| Age (Years) | ||||
| MW/SD | 69.32/8.82 | 66.96/9.33 | 68.34/9.05 | |
| MDN | 70.00 | 69.00 | 70.00 | |
| Range | 41.00 | 39.00 | 42.00 | |
| PSA (ng/mL) | ||||
| MW/SD | 30.84/70.49 | 11.20/17.43 | 22.61/55.49 | |
| MDN | 8.63 | 6.80 | 8.40 | |
| Range | 429.24 | 99.16 | 429.48 | |
| fPSA (Unbound PSA) (ng/mL) | ||||
| MW/SD | 5.73/16.19 | 1.96/2.74 | 4.15/12.55 | |
| MDN | 1.26 | 1.29 | 1.27 | |
| Range | 98.75 | 15.32 | 98.75 | |
| PSA Ratio (fPSA/PSA) | ||||
| MW/SD | 0.16/0.07 | 0.20/0.09 | 0.17/0.08 | |
| MDN | 0.16 | 0.18 | 0.16 | |
| Range | 0.29 | 0.38 | 0.42 | |
| Prostate Volume (mL) * | ||||
| MW/SD | 43.09/22.90 | 69.45/40.57 | 54.13/33.91 | |
| MDN | 37.00 | 52.00 | 45.00 | |
| Range | 98.00 | 139.00 | 148.00 | |
| Urine Culture with bacterial Growth [N] (n/Group) | ||||
| Positive | 10 (23%) | 13 (42%) | 25 (31%) | |
| Negative | 33 (77%) | 18 (58%) | 51 (69%) | |
| Gleason Score [N] (n/Cancer Group) | ||||
| 6 | 7a | 7b | 8 | 9 |
| 11 (26%) | 14 (33%) | 13 (30%) | 1 (2%) | 4 (9%) |
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PSA | prostate-specific antigen |
| qRT-PCR | quantitative real time polymerase chain reaction |
| ROC | receiver operating characteristic |
| RT | Reverse transcriptase |
| snRNA | small nuclear ribonucleic acid |
| CT | cycle threshold |
| Bmi-1 | B cell-specific Moloney murine leukemia virus integration site 1 |
| EMT | epithelial mesenchymal transition |
| ERBB | receptor tyrosine kinases |
| DOHH | deoxyhypusine hydroxylase |
| HuR | human antigen R |
| miRNA | microRNA |
References
- Ferlay, J.; Colombet, M.; Soerjomoataram, I.; Parkin, D.M.; Pineros, M.; Znaor, A.; Bray, F. Cancer statistics for the year 2020: An overview. Int. J. Cancer 2021, 149, 778–789. [Google Scholar] [CrossRef]
- Leitlinienprogramm Onkologie (Deutsche Krebsgesellschaft, D.K., AWMF). Interdisziplinäre Leitlinie der Qualität S3 zur Früherkennung, Diagnose und Therapie der Verschiedenen Stadien des Prostatakarzinoms. 2018. Available online: https://www.awmf.org/leitlinien/detail/ll/043-022OL.html (accessed on 13 June 2021).
- Adhyam, M.; Gupta, A. A Review on the Clinical Utility of PSA in Cancer Prostate. Indian J. Surg. Oncol. 2012, 3, 120–129. [Google Scholar] [CrossRef] [PubMed]
- Ilic, D.; Djulbegovic, M.; Jung, J.H.; Hwang, E.C.; Zhou, Q.; Cleves, A.; Agoritsas, T.; Dahm, P. Prostate cancer screening with prostate-specific antigen (PSA) test: A systematic review and meta-analysis. BMJ 2018, 362, k3519. [Google Scholar] [CrossRef]
- Hoogendam, A.; Buntinx, F.; de Vet, H.C. The diagnostic value of digital rectal examination in primary care screening for prostate cancer: A meta-analysis. Fam. Pract. 1999, 16, 621–626. [Google Scholar] [CrossRef] [PubMed]
- Maricic, A.; Valencic, M.; Sotosek, S.; Oguic, R.; Ivancic, A.; Ahel, J. Transrectal sonography in prostate cancer detection--our 25 years experience of implementation. Coll. Antropol. 2010, 34, 239–242. [Google Scholar]
- Oesterling, J. Prostate specific antigen: A critical assessment of the most useful tumor marker for adenocarcinoma of the prostate. J. Urol. 1991, 145, 907–923. [Google Scholar] [CrossRef]
- Sapre, N.; Selth, L.A. Circulating MicroRNAs as Biomarkers of Prostate Cancer: The State of Play. Hindawi Publ. Corp. Prostate Cancer 2013, 2013, 539680. [Google Scholar] [CrossRef] [PubMed]
- Kroh, E.; Parkin, R.; Mitchell, P.; Tewari, M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods 2010, 50, 298–301. [Google Scholar] [CrossRef] [PubMed]
- Sita-Lumsden, A.; Dart, D.A.; Waxman, J.; Bevan, C.L. Circulating microRNAs as potential new biomarkers for prostate cancer. Br. J. Cancer 2013, 108, 1925–1930. [Google Scholar] [CrossRef] [PubMed]
- Markert, L.; Holdmann, J.; Klinger, C.; Kaufmann, M.; Schork, K.; Turewicz, M.; Eisenacher, M.; Savelsbergh, A. Small RNAs as biomarkers to differentiate benign and malign prostate diseases: An alternative for transrectal punch biopsy of the prostate? PLoS ONE 2021, 16, e0247930. [Google Scholar] [CrossRef] [PubMed]
- Vanacore, D.; Boccellino, M.; Rossetti, S.; Cavaliere, C.; D’Aniello, C.; Di Franco, R.; Romano, F.J.; Montanari, M.; La Mantia, E.; Piscitelli, R. Micrornas in prostate cancer: An overview. Oncotarget 2017, 8, 50240–50251. [Google Scholar] [CrossRef]
- Galvao-Lima, L.J.; Moaris, A.H.F.; Valentim, R.A.M.; Barreto, E. miRNAs as biomarkers for early cancer detection and their application in the development of new diagnostic tools. BioMed. Eng. OnLine 2021, 20, 21. [Google Scholar] [CrossRef] [PubMed]
- Carthew, R.W.; Sontheimer, E.J. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, J.; Hasnain, S.; Siddiqui, M.; Ahamed, M.; Musarrat, J.; Al-Khedhairy, A. MicroRNA in carcinogenesis & cancer diagnostics: A new paradigm. Indian J. Med. Res. 2013, 137, 680–694. [Google Scholar] [PubMed Central]
- Gallo, A.; Tandon, M.; Alevizos, I.; Illei, G.G. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS ONE 2012, 7, e30679. [Google Scholar] [CrossRef]
- Lawrie, C.H.; Gal, S.; Dunlop, H.M.; Pushkaran, B.; Liggins, A.P.; Pulford, K.; Banham, A.H.; Pezzella, F.; Boultwood, J.; Wainscoat, J.S.; et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br. J. Haematol. 2008, 141, 672–675. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef]
- Chen, C.-Z.; Zhang, G.-L.; Li, H.-R.; Luo, J.-D.; Li, Z.-X.; Chen, G.-M.; Yang, J. A panel of five circulating microRNAs as potential biomarkers for prostate cancer. Prostate 2012, 72, 1443–1452. [Google Scholar] [CrossRef]
- Volinia, S.; Calin, G.A.; Liu, C.G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M.; et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef] [PubMed]
- Kachakova, D.; Mitkova, A.; Popov, E.; Popov, I.; Vlahova, A.; Dikov, T.; Christova, S.; Mitev, V.; Slavov, C.; Kaneva, R. Combinations of serum prostate-specific antigen and plasma expression levels of let-7c, miR-30c, miR-141, and miR-375 as potential better diagnostic biomarkers for prostate cancer. DNA Cell Biol. 2015, 34, 189–200. [Google Scholar] [CrossRef]
- Haj-Ahmad, T.A.; Abdalla, M.A.; Haj-Ahmad, Y. Potential Urinary miRNA Biomarker Candidates for the Accurate Detection of Prostate Cancer among Benign Prostatic Hyperplasia Patients. J. Cancer 2014, 5, 182–191. [Google Scholar] [CrossRef] [PubMed]
- Fabris, L.; Ceder, Y.; Chinnaiyan, A.; Jenster, G.; Sorensen, K.; Tomlins, S.; Visakorpi, T.; Calin, G. The Potential of MicroRNAs as Prostate Cancer Biomarkers. Eur. J. Urol. 2016, 70, 312–322. [Google Scholar] [CrossRef] [PubMed]
- Bhagirath, D.; Liston, M.; Patel, N.; Akoto, T.; Lui, B.; Yang, T.L.; To, D.M.; Majid, S.; Dahiya, R.; Tabatabai, Z.L.; et al. MicroRNA determinants of neuroendocrine differentiation in metastatic castration-resistant prostate cancer. Oncogene 2020, 39, 7209–7223. [Google Scholar] [CrossRef]
- Pfaffe, T.; Cooper-White, J.; Beyerlein, P.; Kostner, K.; Punyadeera, C. Diagnostic potential of saliva: Current state and future applications. Clin. Chem. 2011, 57, 675–687. [Google Scholar] [CrossRef]
- Mittal, S.; Bansal, V.; Garg, S.; Atreja, G.; Bansal, S. The diagnostic role of saliva—A Review. J. Clin. Exp. Dent. 2010, 3, e314–e320. [Google Scholar] [CrossRef]
- Sembler-Moller, M.; Belstrom, D.; Locht, H.; Pedersen, A. Distinct microRNA expression profiles in saliva and salivary gland tissue differentiate patients with primary Sjögren’s syndrome from non-Sjögren’s sicca patients. Oral. Pathol. Med. 2020, 49, 1044–1052. [Google Scholar] [CrossRef]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. BMJ 2009, 339, b2535. [Google Scholar] [CrossRef]
- Bryant, R.J.; Pawlowski, T.; Catto, J.W.F.; Marsden, G.; Vessella, R.L.; Rhees, B.; Kuslich, C.; Visakorpi, T.; Hamdy, F.C. Changes in circulating microRNA levels associated with prostate cancer. Br. J. Cancer 2012, 106, 768–774. [Google Scholar] [CrossRef]
- Brase, J.C.; Johannes, M.; Schlomm, T.; Fälth, M.; Haese, A.; Steuber, T.; Beissbarth, T.; Kuner, R.; Sültmann, H. Circulating miRNAs are correlated with tumor progression in prostate cancer. Int. J. Cancer 2011, 128, 608–616. [Google Scholar] [CrossRef]
- Moltzahn, F.; Olshen, A.B.; Baehner, L.; Peek, A.; Fong, L.; Stoppler, H.; Simko, J.; Hilton, J.F.; Carroll, P.; Blelloch, R. Microfluidic-based multiplex qRT-PCR identifies diagnostic and prognostic microRNA signatures in the sera of prostate cancer patients. Cancer Res. 2011, 71, 550–560. [Google Scholar] [CrossRef]
- Wiegand, C.; Heusser, P.; Klinger, C.; Cysarz, D.; Büssing, A.; Ostermann, T.; Savelsbergh, A. Stress-associated changes in salivary microRNAs can be detected in response to the Trier Social Stress Test: An exploratory study. Sci. Rep. 2018, 8, 7112. [Google Scholar] [CrossRef] [PubMed]
- Nadler, R.B.; Humphrey, P.A.; Smith, D.S.; Catalona, W.J.; Ratliff, T.L. Effect of Inflammation and Benign Prostatic Hyperplasia on Elevated Serum Prostate Specific Antigen Levels. J. Urol. 1995, 154, 407–413. [Google Scholar] [CrossRef]
- De Sarkar, N.; Roy, R.; Mitra, J.K.; Ghose, S.; Chakraborty, A.; Paul, R.R.; Mukhopadhyay, I.; Roy, B. A quest for miRNA bio-marker: A track back approach from gingivo buccal cancer to two different types of precancers. PLoS ONE 2014, 9, e104839. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.L.; Liu, X.; Jin, Z.; Gou, C.; Liang, M.; Cui, L.; Zhao, X. A three miRNAs signature for predicting the transformation of oralleukoplakia to oral squamous cell carcinoma. Cancer Res. 2018, 8, 1403–1413. [Google Scholar]
- Kao, Y.Y.; Tu, H.F.; Kao, S.Y.; Chang, K.W.; Lin, S.C. The increase of oncogenic miRNA expression in tongue carcinogenesis of a mouse model. Oral Oncol. 2015, 51, 1103–1112. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Mann, H.B.; Whitney, D.R. On a Test of Whether one of Two Random Variables is Stochastically Larger than the Other. Ann. Math. Stat. 1947, 18, 50–60. [Google Scholar] [CrossRef]
- Zweig, M.; Campbell, G. Receiver-operating characteristic (ROC) plots: A fundamental evaluation tool in clinical medicine. Clin. Chem. 1993, 39, 561–577. [Google Scholar] [CrossRef]
- Ruopp, M.; Perkins, N.; Whitcomb, B.; Schisterman, E. Youden Index and optimal cut-point estimated from observations affected by a lower limit of detection. Biom. J. 2008, 50, 419–430. [Google Scholar] [CrossRef]
- Akobeng, A. Understanding diagnostic tests 1: Sensitivity, specificity and predictive values. Acta Paed. 2007, 96, 338–341. [Google Scholar] [CrossRef]
- Vezyraki, P.; Vlachaki, A.; Baltogiannis, D.; Batistatou, A.; Tsampalas, S.; Simos, Y.; Kaltsas, A.; Pappas, P.; Dounousi, E.; Ragos, V.; et al. Impact of total PSA and percent free PSA in the differentiation of prostate disease: A retrospective comparative study implicating neoplastic and non-neoplastic entities. J. BUON. 2019, 24, 2107–2113. [Google Scholar] [PubMed]
- Katz, B.; Reis, S.; Viana, N.; Morais, D.; Moura, C.; Dip, N.; Silva, I.; Iscaife, A.; Srougi, M.; Leite, K. Comprehensive Study of Gene and microRNA Expression Related to Epithelial-Mesenchymal Transition in Prostate Cancer. PLoS ONE 2014, 9, e113700. [Google Scholar] [CrossRef] [PubMed]
- Epis, M.R.; Barker, A.; Giles, K.M.; Beveridge, D.J.; Leedman, P.J. The RNA-binding Protein HuR Opposes the Repression of ERBB-2 Gene Expression by MicroRNA miR-331-3p in Prostate Cancer Cells. J. Biol. Chem. 2011, 286, 41442–41454. [Google Scholar] [CrossRef] [PubMed]
- Epis, M.R.; Giles, K.M.; Barker, A.; Kendrick, T.S.; Leedman, P.J. miR-331-3p regulates ERBB-2 expression and androgen receptor signaling in prostate cancer. J. Biol. Chem. 2009, 284, 24696–24704. [Google Scholar] [CrossRef]
- Wang, L.; Tang, H.; Thayanithy, V.; Subramanian, S.; Oberg, A.; Cunningham, J.; Cerhan, J.; Steer, C.; Thibodeau, S. Gene networks and microRNAs implicated in aggressive prostate cancer. Cancer Res. 2009, 69, 9490–9497. [Google Scholar] [CrossRef]
- Yu, J.; Lu, Y.; Cui, D.; Li, E.; Zhu, Y.; Zhao, Y.; Zhao, F.; Xia, S. miR-200b suppresses cell proliferation, migration and enhances chemosensitivity in prostate cancer by regulating Bmi-1. Oncol. Rep. 2014, 31, 910–918. [Google Scholar] [CrossRef]
- Bracken, C.; Gregory, P.; Kolesnikkoff, N.; Bert, A.; Wang, J.; Shannon, M.; Goodall, G. A Double-Negative Feedback Loop between ZEB1-SIP1 and the microRNA-200 Family Regulates Epithelial-Mesenchymal Transition. Cancer Res. 2008, 68, 7846–7854. [Google Scholar] [CrossRef]
- Hugo, H.; Ackland, M.; Blick, T.; Lawrence, M.; Clements, J.; Williams, E.; Thompson, E. Epithelial--mesenchymal and mesenchymal--epithelial transitions in carcinoma progression. J. Cell. Physiol. 2007, 213, 374–383. [Google Scholar] [CrossRef]
- Peinado, H.; Olmeda, D.; Cano, A. Snail, Zeb and bHLH factors in tumour progression: An alliance against the epithelial phenotype? Nat. Rev. Cancer 2007, 7, 415–428. [Google Scholar] [CrossRef]
- Fujii, T.; Shimada, K.; Tatsumi, Y.; Tanaka, N.; Fujimoto, K.; Konishi, N. Syndecan-1 up-regulates microRNA-331-3p and mediates epithelial-to-mesenchymal transition in prostate cancer. Mol. Carcinog. 2016, 55, 1378–1386. [Google Scholar] [CrossRef]
- Shee, S.M.E.; Koh, R.Y.; Voon, K.G.L.; Chye, S.M.; Othman, I.; Ng, K.Y. The roles of microRNA-331 Family in Cancers. J. Cancer Res. Pract. 2019, 6, 1–6. [Google Scholar] [CrossRef]
- Souza, M.; Kuasen, H.; Camargo Barros-Filho, M.; Cliliao, H.; Marchi, F.; Fuganti, P.; Rossi Paschoal, A.; Rogatto, S.; Syllos Colus, I. Circulating mRNAs and miRNAs as candidate markers for the diagnosis and prognosis of prostate cancer. PLoS ONE 2017, 12, e0184094. [Google Scholar] [CrossRef]
- Godoy, P.; Bhakta, N.; Barczak, A.; Cakmak, H.; Fisher, S.; MacKenzie, T.; Patel, T.; Price, R.; Smith, J.; Woodruff, P.; et al. Large Differences in Small RNA Composition Between Human Biofluids. Cell Rep. 2018, 25, 1346–1358. [Google Scholar] [CrossRef] [PubMed]
- Endzelins, E.; Berger, A.; Melne, V.; Bajo-Santos, C.; Sobolevska, K.; Abols, A.; Rodriguez, M.; Santare, D.; Rudnickiha, A.; Lietuvietis, V.; et al. Detection of circulating miRNAs: Comparative analysis of extracellular vesicle-incorporated miRNAs and cell-free miRNAs in whole plasma of prostate cancer patients. BMC Cancer 2017, 17, 730. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Li, Z.; Bai, F.; Ji, N.; Zheng, Y.; Li, Y.; Chen, J.; Mao, X. MicroRNA expression profiles in benign prostatic hyperplasia. Mol. Med. Rep. 2018, 17, 3853–3858. [Google Scholar] [CrossRef] [PubMed]
- Pezaro, C.; Woo, H.H.; Davis, I.D. Prostate cancer: Measuring PSA. Intern. Med. J. 2014, 44, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Sari Motlagh, R.; Yanagisawa, T.; Kawada, T.; Laukhtina, E.; Rajwa, P.; Aydh, A.; König, F.; Pallauf, M.; Huebner, N.A.; Baltzer, P.A.; et al. Accuracy of SelectMDx compared to mpMRI in the diagnosis of prostate cancer: A systematic review and diagnostic meta-analysis. Prostate Cancer Prostatic Dis. 2022, 25, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Crowley, E.; Di Nicolantonio, F.; Loupakis, F.; Bardelli, A. Liquid biopsy: Monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 2013, 10, 472–484. [Google Scholar] [CrossRef] [PubMed]


| Source of Literature | miRNA |
|---|---|
| Chen et al. (2012) [19] | hsa-mir: 622 |
| Bryant et al. (2012) [29] | hsa-mir: 574-3p, 625, 331-3p, 141, 130b, 432, 484, 375, 107, 181a, 2110, 301a, 326 |
| Brase et al. (2011) [30] | hsa-mir: 200b, 141, 375 |
| Moltzahn et al. (2011) [31] | hsa-mir: 106a |
| Equipment | |
|---|---|
| PCR tower | Jena Bioscience, Jena, Germany |
| Sorvall MGX-120 Ultracentrifuge | Thermo Fisher Scientific, Waltham, MA, USA |
| Chemicals | |
| TRIzol™ Reagent | Thermo Fisher Scientific, Waltham, MA, USA |
| Chloroform | Merck, Darmstadt, Germany |
| Isopropanol C3H8O | Merck, Darmstadt, Germany |
| RNAse free H2O | Thermo Fisher Scientific, Waltham, MA, USA |
| Ethanol (75%) | Merck, Darmstadt, Germany |
| Software | |
| qPCR Soft | Thermo Fisher Scientific, Waltham, MA, USA |
| Microsoft Office | Microsoft, Redmond, WA, USA |
| SPSS | IBM, Armonk, NY, USA |
| Endnote | Thomson Reuters, Toronto, ON, Canada |
| microRNA | Mature | Accession | Sequence |
|---|---|---|---|
| MIR106A | hsa-miR-106a-5p | MIMAT0000103 | 13-AAAAGUGCUUACAGUGCAGGUAG-35 |
| MIR130B | hsa-miR-130b-5p | MIMAT0004680 | 13-ACUCUUUCCCUGUUGCACUAC-33 |
| MIR301A | hsa-miR-301a-5p | MIMAT0022696 | 14-GCUCUGACUUUAUUGCACUACU-35 |
| MIR331 | hsa-miR-331 | MIMAT0000760 | 61-GCCCCUGGGCCUAUCCUAGAA-81 |
| MIR326 | hsa-miR-326 | MIMAT0000756 | 60-CCUCUGGGCCCUUCCUCCAG-79 |
| MIR375 | hsa-miR-375-3p | MIMAT0000728 | 40-UUUGUUCGUUCGGCUCGCGUGA-61 |
| MIR484 | hsa-miR-484 | MIMAT0002174 | 8-UCAGGCUCAGUCCCCUCCCGAU-29 |
| MIR2110 | hsa-miR-2110 | MIMAT0010133 | 8-UUGGGGAAACGGCCGCUGAGUG-29 |
| MIR107 | hsa-mir-107 | MIMAT0000104 | 50-AGCAGCAUUGUACAGGGCUAUCA-72 |
| MIR622 | hsa-mir-622 | MIMAT0003291 | 61-ACAGUCUGCUGAGGUUGGAGG-81 |
| MIR141 | hsa-mir-141 | MIMAT0000432 | 5ß-UAACACUGUCUGGUAAAGAUGG-38 |
| MIR432 | hsa-mir-432 | MIMAT0002814 | 14-UCUUGGAGUAGGUCAUUGGGUGG-36 |
| MIR574 | hsa-mir-574-3p | MIMAT0003239 | 61-CACGCUCAUGCACACACCCACA-82 |
| MIR625 | hsa-mir-625 | MIMAT0003294 | 15-AGGGGGAAAGUUCUAUAGUCC-35 |
| MIR181A | hsa-mir-181a-2-3p | MIMAT0004558 | 77-ACCACUGACCGUUGACUGUACC-98 |
| MIR200b | hsa-mir-200b | MIMAT0000318 | 57-UAAUACUGCCUGGUAAUGAUGA-78 |
| PCa-Specific microRNA | ∆CT Cancer Group (Mean/SD) | ∆CT Control Group (Mean/SD) | Foldchange (Control Group—Cancer Group) | p-Value |
|---|---|---|---|---|
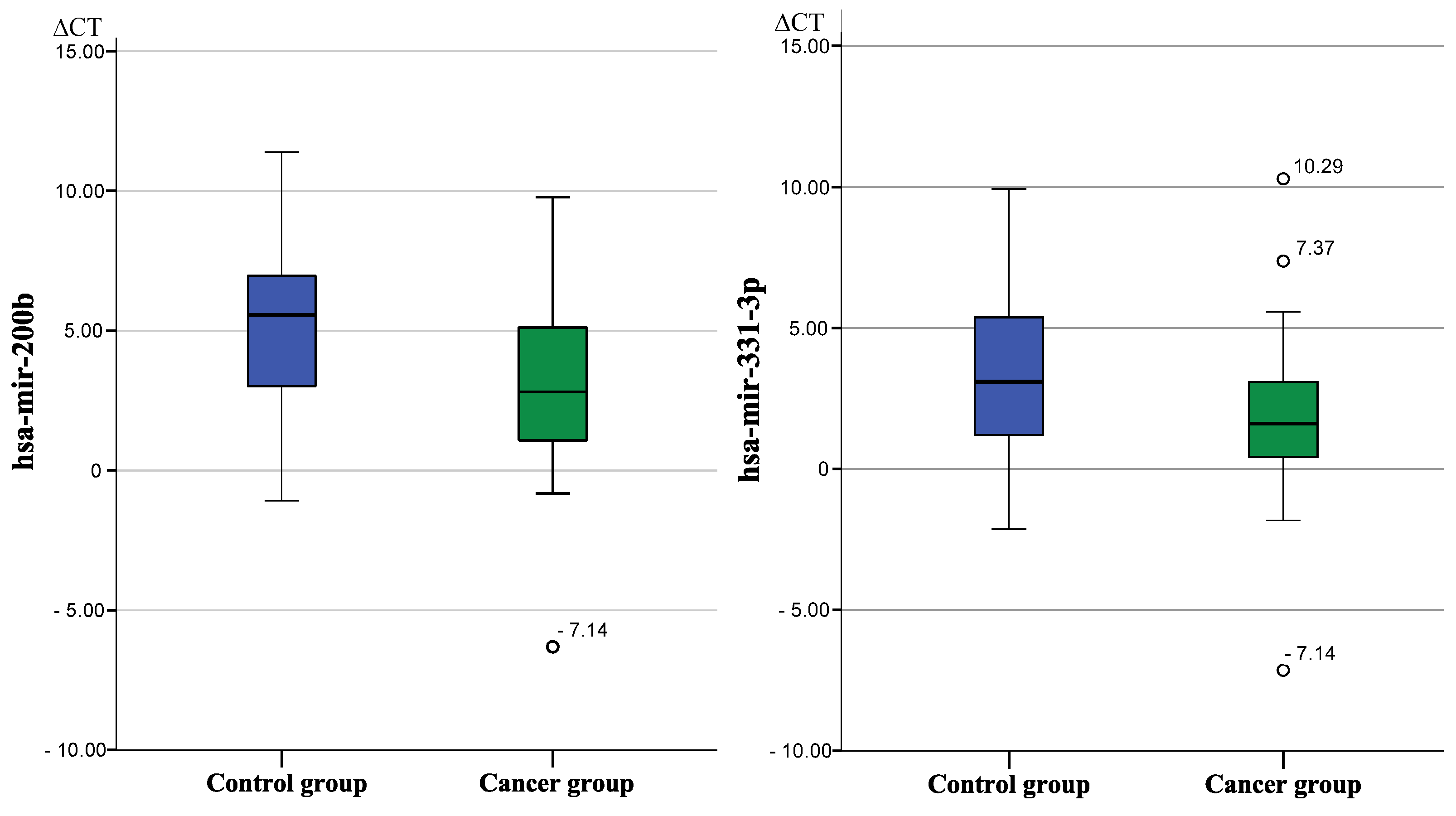
| hsa-mir-200b * | 3.27/3.11 | 5.12/3.23 | −3.60 | 0.017 |
| hsa-mir-331-3p * | 1.78/2.81 | 3.17/2.84 | −2.64 | 0.031 |
| hsa-mir-107 | 1.41/3.38 | 2.34/3.84 | No significant difference | 0.290 |
| hsa-mir-141 | 0.83/2.66 | 1.47/2.95 | No significant difference | 0.224 |
| hsa-mir-432 | 2.87/2.71 | 3.46/3.05 | No significant difference | 0.446 |
| hsa-mir-574 | 5.83/3.40 | 6.00/2.36 | No significant difference | 0.874 |
| hsa-mir-625 | 1.30/3.00 | 2.41/3.45 | No significant difference | 0.174 |
| hsa-mir-181 | 1.78/3.81 | 1.99/3.19 | No significant difference | 0.657 |
| hsa-mir-622 | 1.79/2.81 | 3.17/2.84 | No significant difference | 0.890 |
| hsa-mir-375 | 1.56/4.47 | 0.97/4.40 | No significant difference | 0.806 |
| hsa-mir-484 | 3.22/3.90 | 2.80/3.10 | No significant difference | 0.766 |
| hsa-mir-2110 | 1.55/4.49 | 2.47/2.70 | No significant difference | 0.433 |
| hsa-mir-130b | 1.39/3.66 | 1.82/2.87 | No significant difference | 0.552 |
| hsa-mir-301a | 3.27/3.55 | 2.98/2.67 | No significant difference | 0.739 |
| hsa-mir-326 | 6.12/3.29 | 5.87/2.47 | No significant difference | 0.959 |
| hsa-mir-106a | 5.00/3.33 | 5.19/2.77 | No significant difference | 0.782 |
| hsa-mir-200b, Cut off ∆CT = 5.5 | Tested Positive (X ≤ 5.5) | Tested Negative (X ≥ 5.5) | |
|---|---|---|---|
| cancer group [N] = 43 | 35 | 8 | sensitivity0.814 |
| control group [N] = 31 | 14 | 17 | specificity0.548 |
| Predictive value | 0.714 | 0.680 | |
| hsa-mir-331, cut off ∆CT = 2.87 | tested positive (X ≤ 2.87) | tested negative (X ≥ 2.87) | |
| cancer group [N] = 43 | 32 | 11 | sensitivity0.744 |
| control group [N] = 31 | 13 | 18 | specificity0.581 |
| predictive value | 0.711 | 0.462 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luedemann, C.; Reinersmann, J.-L.; Klinger, C.; Degener, S.; Dreger, N.M.; Roth, S.; Kaufmann, M.; Savelsbergh, A. Prostate Cancer-Associated miRNAs in Saliva: First Steps to an Easily Accessible and Reliable Screening Tool. Biomolecules 2022, 12, 1366. https://doi.org/10.3390/biom12101366
Luedemann C, Reinersmann J-L, Klinger C, Degener S, Dreger NM, Roth S, Kaufmann M, Savelsbergh A. Prostate Cancer-Associated miRNAs in Saliva: First Steps to an Easily Accessible and Reliable Screening Tool. Biomolecules. 2022; 12(10):1366. https://doi.org/10.3390/biom12101366
Chicago/Turabian StyleLuedemann, Christoph, Jan-Ludwig Reinersmann, Claudia Klinger, Stephan Degener, Nici Markus Dreger, Stephan Roth, Michael Kaufmann, and Andreas Savelsbergh. 2022. "Prostate Cancer-Associated miRNAs in Saliva: First Steps to an Easily Accessible and Reliable Screening Tool" Biomolecules 12, no. 10: 1366. https://doi.org/10.3390/biom12101366
APA StyleLuedemann, C., Reinersmann, J.-L., Klinger, C., Degener, S., Dreger, N. M., Roth, S., Kaufmann, M., & Savelsbergh, A. (2022). Prostate Cancer-Associated miRNAs in Saliva: First Steps to an Easily Accessible and Reliable Screening Tool. Biomolecules, 12(10), 1366. https://doi.org/10.3390/biom12101366






