Dissecting Multiple Pathways in the Relaxation Dynamics of Helix <==> Coil Transitions with Optimum Dimensionality Reduction
Abstract
1. Introduction
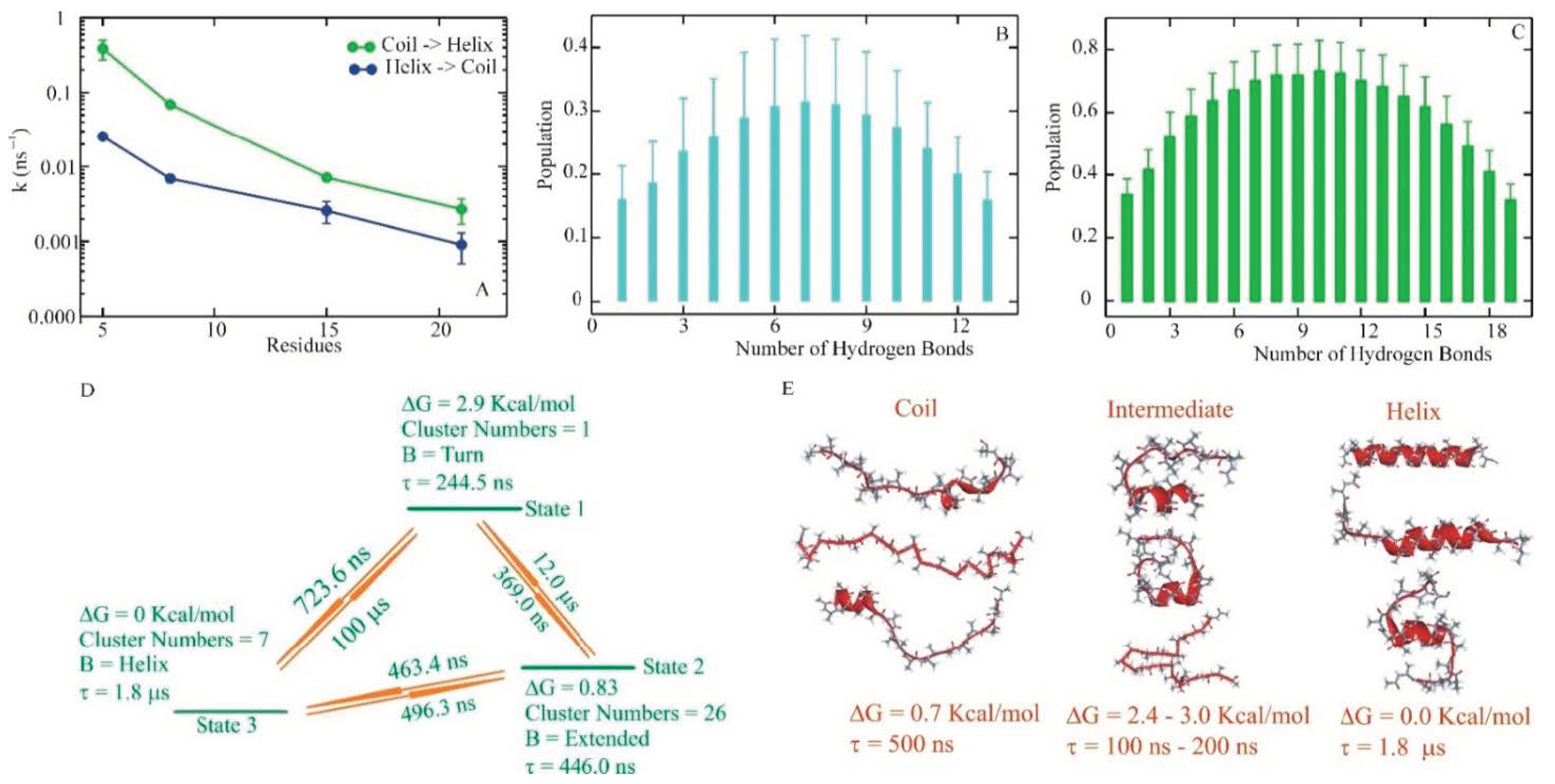
2. Helix–Coil Kinetics
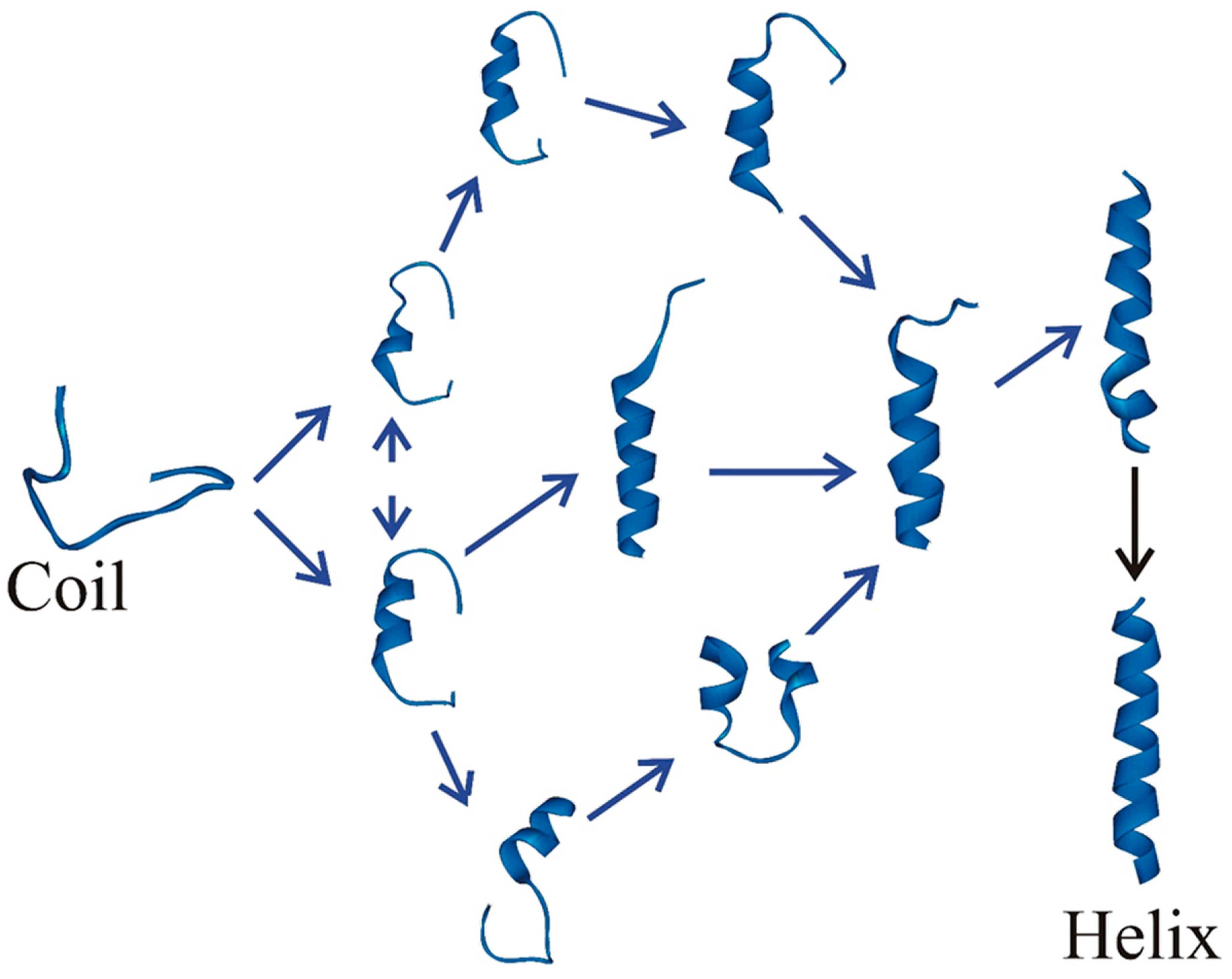
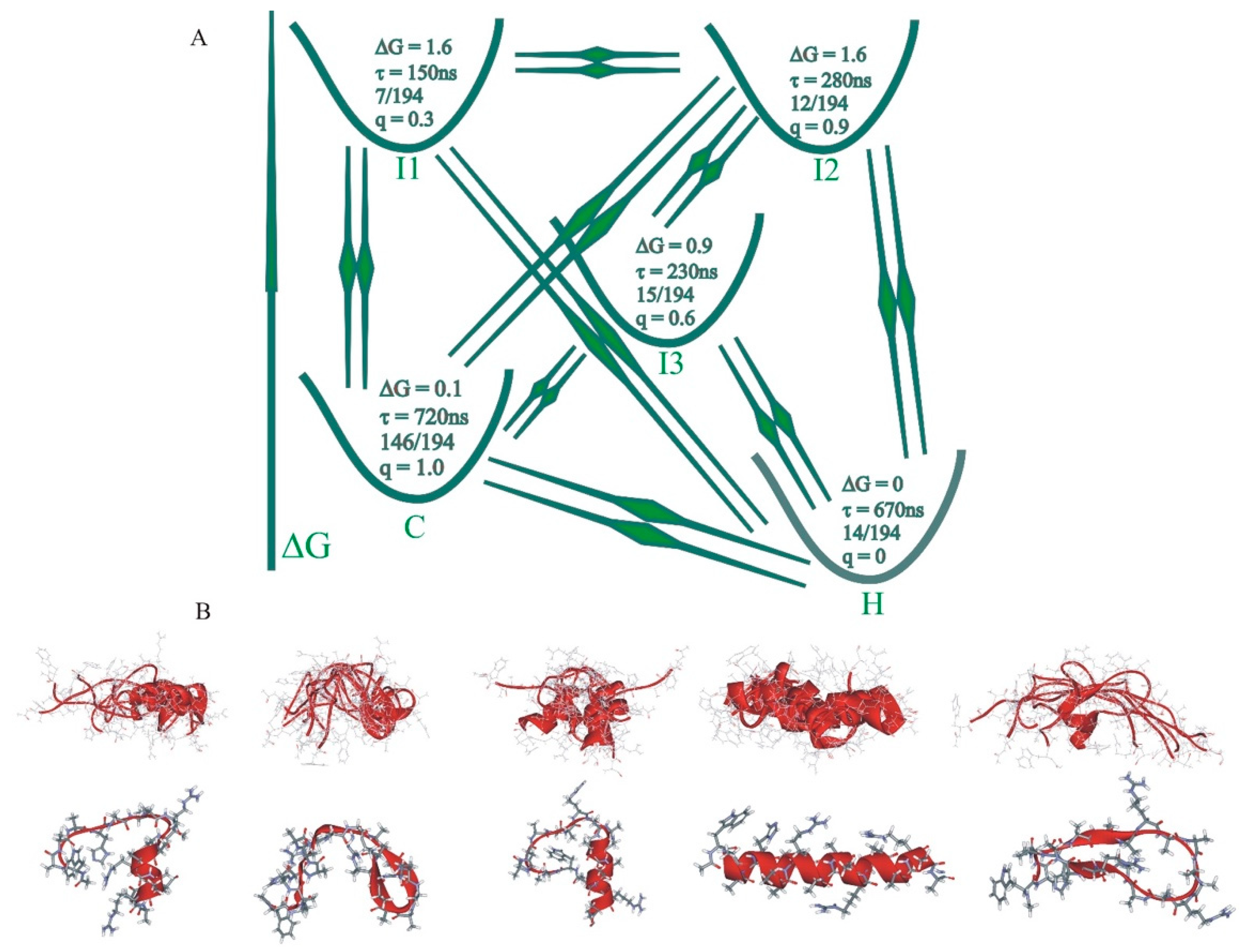
3. Multiple Pathways
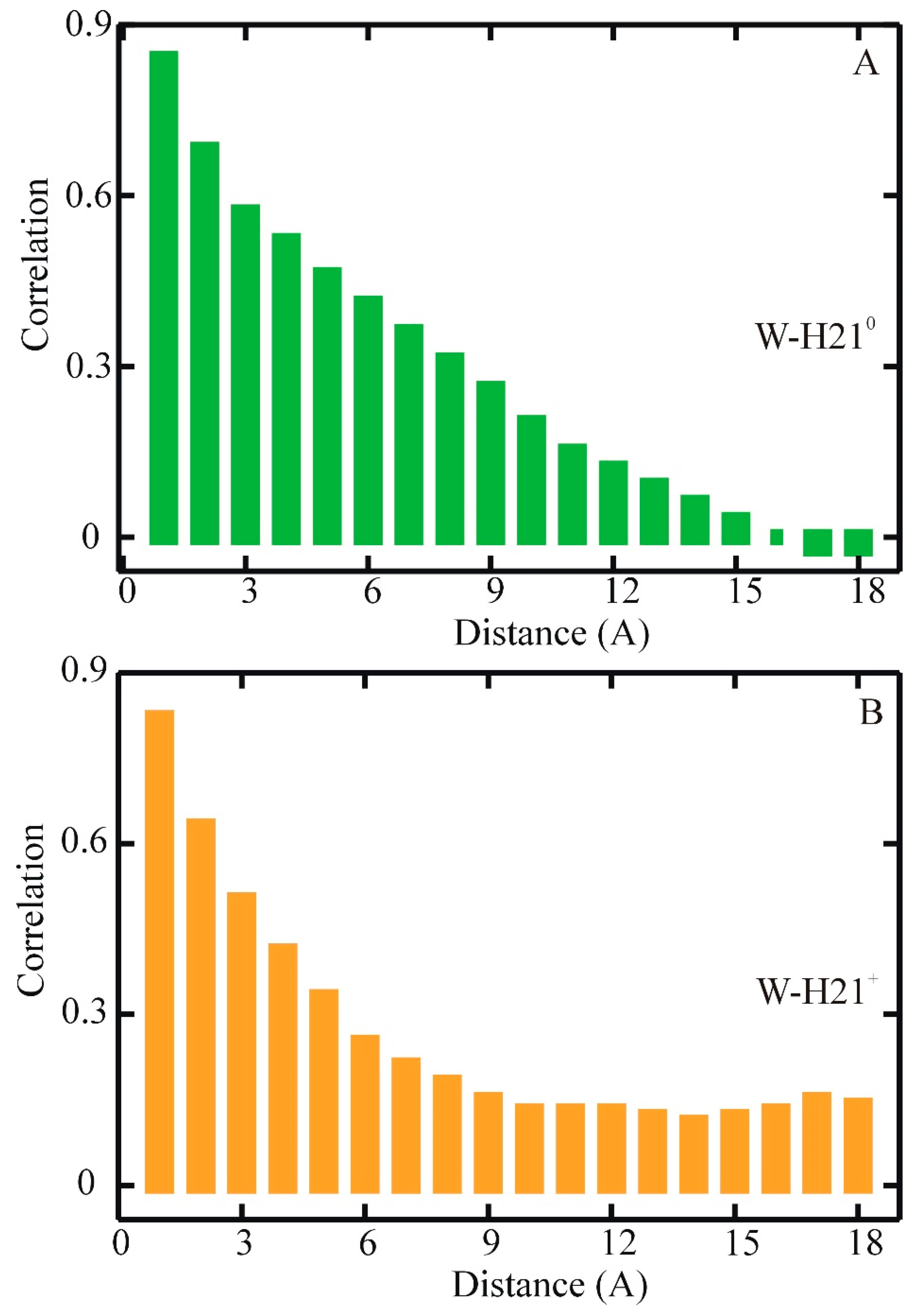
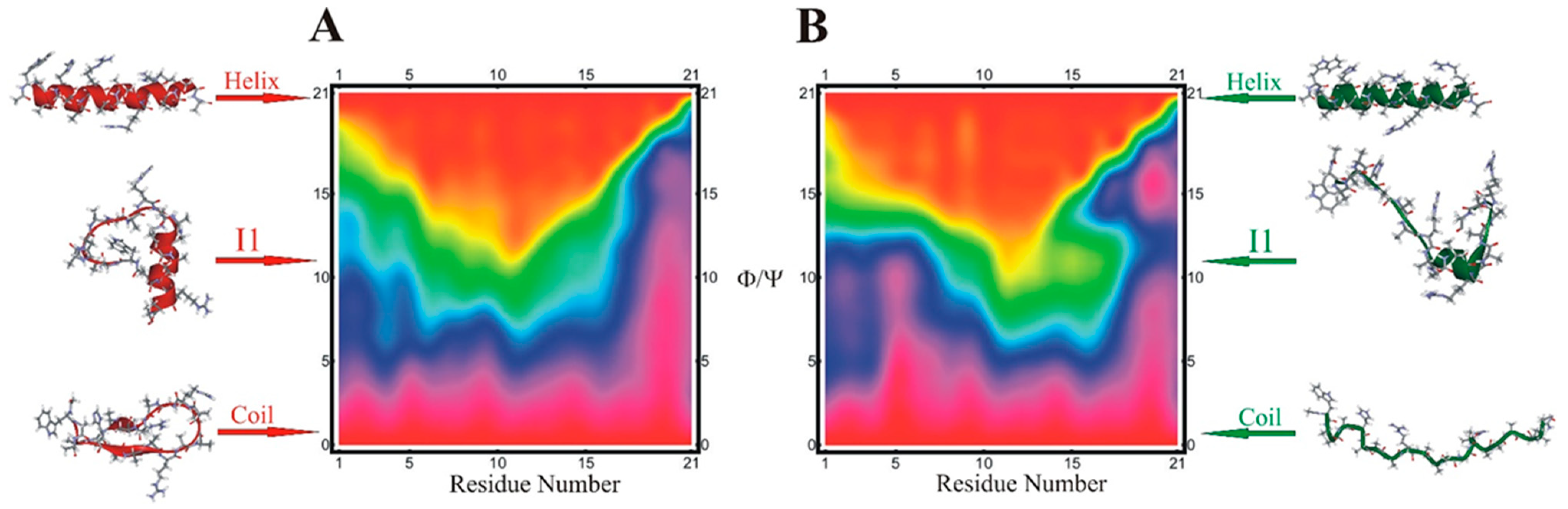
4. Optimum Dimensionality Reduction
5. Summary
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bryngelson, J.D.; Wolynes, P.G. Spin glasses and the statistical mechanics of protein folding. Proc. Natl. Acad. Sci. USA 1987, 84, 7524–7528. [Google Scholar] [CrossRef]
- Bryngelson, J.D.; Wolynes, P.G. Intermediates and barrier crossing in a random energy model (with applications to protein folding). J. Phys. Chem. 1989, 93, 6902–6915. [Google Scholar] [CrossRef]
- Alm, E.; Baker, D. Matching theory and experiment in protein folding. Curr. Opin. Struct. Biol. 1999, 9, 189–196. [Google Scholar] [CrossRef]
- Alm, E.; Baker, D. Prediction of protein-folding mechanisms from free-energy landscapes derived from native structures. Proc. Natl. Acad. Sci. USA 1999, 96, 11305–11310. [Google Scholar] [CrossRef] [PubMed]
- Allew, R.M.; Sabelko, J.; Gruebele, M. Direct observation of fast protein folding: The initial collapse of apomyoglobin. Proc. Natl. Acad. Sci. USA 1996, 93, 5759–5764. [Google Scholar] [CrossRef]
- Ballew, R.M.; Sabelko, J.; Gruebele, M. Observation of distinct nanosecond and microsecond protein folding events. Nat. Struct. Biol. 1996, 3, 923–926. [Google Scholar] [CrossRef] [PubMed]
- Bieri, O.; Wirz, J.; Hellrung, B.; Schutkowski, M.; Drewello, M.; Kiefhaber, T. The speed limit for protein folding measured by triplet-triplet energy transfer. Proc. Natl. Acad. Sci. USA 1999, 96, 9597–9601. [Google Scholar] [CrossRef]
- Blanco, F.J.; Rivas, G.; Serrano, L. A short linear peptide that folds into a β-hairpin in aqueous solution. Nat. Struct. Biol. 1994, 1, 584–590. [Google Scholar] [CrossRef]
- Brooks, C.L. Simulations of protein folding and unfolding. Curr. Opin. Struct. Biol. 1998, 8, 222–226. [Google Scholar] [CrossRef]
- Bryngelson, J.D.; Onuchic, J.N.; Socci, N.D.; Wolynes, P.G. Funnels, pathways, and the energy landscape of protein folding: A synthesis. Proteins: Struct. Funct. Bioinform. 1995, 21, 167–195. [Google Scholar] [CrossRef]
- Buckler, D.R.; Haas, E.; Scheraga, H.A. Analysis of the Structure of Ribonuclease A in Native and Partially Denatured States by Time-Resolved Nonradiative Dynamic Excitation Energy Transfer between Site-Specific Extrinsic Probes. Biochemistry 1995, 34, 15965–15978. [Google Scholar] [CrossRef] [PubMed]
- Burton, R.E.; Huang, G.S.; Daugherty, M.A.; Calderone, T.L.; Oas, T.G. The energy landscape of a fast-folding protein mapped by Ala-->Gly substitutions. Nat. Struct. Biol. 1997, 4, 305–310. [Google Scholar] [CrossRef]
- Burton, R.E.; Huang, G.S.; Daugherty, M.A.; Fullbright, P.W.; Oas, T.G. Microsecond protein folding through a compact transition state. J. Mol. Biol. 1996, 263, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Callender, R.H.; Dyer, R.; Gilmanshin, R.; Woodruff, W.H. FAST EVENTS IN PROTEIN FOLDING: The Time Evolution of Primary Processes. Annu. Rev. Phys. Chem. 1998, 49, 173–202. [Google Scholar] [CrossRef]
- Camacho, J.; Thirumalai, D. Theoretical predictions of folding pathways by using the proximity rule, with applications to bovine pancreatic trypsin inhibitor. Proc. Natl. Acad. Sci. USA 1995, 92, 1277–1781. [Google Scholar] [CrossRef] [PubMed]
- Chakrabartty, A.; Baldwin, R.L. Stability of α-helices. Adv. Prot. Chem. 1995, 46, 141–176. [Google Scholar]
- Chan, C.K.; Hofrichter, J.; Eaton, W.A.; Winkler, J.R.; Gray, H.B. Optical Triggers of Protein Folding. Science 1996, 274, 628–629. [Google Scholar] [CrossRef]
- Chan, C.-K.; Hu, Y.; Takahashi, S.; Rousseau, D.L.; Eaton, W.A.; Hofrichter, J. Submillisecond protein folding kinetics studied by ultrarapid mixing. Proc. Natl. Acad. Sci. USA 1997, 94, 1779–1784. [Google Scholar] [CrossRef]
- Chan, H.S.; Dill, K.A. Protein folding in the landscape perspective: Chevron plots and non-arrhenius kinetics. Proteins 1998, 30, 2–33. [Google Scholar] [CrossRef]
- Chen, E.; Wittung-Stafshede, P.; Kliger, D.S. Far-UV time-resolved circular dichroism detection of electron-transfer-triggered cytochrome c folding. J. Am. Chem. Soc. 1999, 121, 3811–3817. [Google Scholar] [CrossRef]
- Clarke, D.T.; Doig, A.J.; Stapley, B.J.; Jones, G.R. The α-helix folds on the millisecond time scale. Proc. Natl. Acad. Sci. USA 1999, 96, 7232–7237. [Google Scholar] [CrossRef]
- Daggett, V.; Levitt, M. Molecular-dynamics simulation of helix denaturation. J. Mol. Biol. 1992, 223, 1121–1138. [Google Scholar] [CrossRef]
- Daura, X.; Jaun, B.; Seebach, D.; van Gunsteren, W.F.; Mark, A.E. Reversible peptide folding in solution by molecular dynamics simulation. J. Mol. Biol. 1998, 280, 925–932. [Google Scholar] [CrossRef] [PubMed]
- Daura, X.; van Gunsteren, W.F.; Mark, A.E. Folding-unfolding thermodynamics of a beta-heptapeptide from equilibrium simulations. Proteins 1999, 34, 269–280. [Google Scholar] [CrossRef]
- Dill, K.A.; Shortle, D. Denatured states of proteins. Annu. Rev. Biochem. 1991, 60, 795–825. [Google Scholar] [CrossRef] [PubMed]
- Dill, K.A.; Stigter, D. Modeling protein stability as heteropolymer collapse. Adv. Prot. Chem. 1995, 46, 59–104. [Google Scholar]
- Dinner, A.R.; Lazaridis, T.; Karplus, M. Understanding β-hairpin formation. Proc. Natl. Acad. Sci. USA 1999, 96, 9068–9073. [Google Scholar] [CrossRef]
- Dobson, C.M. Protein misfolding, evolution and disease. Trends Biochem. Sci. 1999, 24, 329–332. [Google Scholar] [CrossRef]
- Dobson, C.M.; Sali, A.; Karplus, M. Protein folding: A perspective from theory and experiment. Angew. Chem. Int. Edit. 1998, 37, 868–893. [Google Scholar] [CrossRef]
- Duan, Y.; Kollman, P.A. Pathways to a protein folding intermediate observed in 1-microsecond simulation in aqueous solution. Science 1998, 282, 740–744. [Google Scholar] [CrossRef]
- Dyer, R.B.; Gai, F.; Woodruff, W.H.; Gilmanshin, R.; Callender, R.H. Infrared studies of fast events in protein folding. Acc. Chem. Res. 1998, 31, 709–716. [Google Scholar] [CrossRef]
- Eaton, W.A. Commentary: Searching for “downhill scenarios” in protein folding. Proc. Natl. Acad. Sci. USA 1999, 96, 5897–5899. [Google Scholar] [CrossRef] [PubMed]
- Elove, G.A.; Bhuyan, A.K.; Roder, H. Kinetic mechanism of cytochrome c folding: Involvement of the heme and its ligands. Biochemistry 1994, 33, 6925–6935. [Google Scholar] [CrossRef] [PubMed]
- Fersht, A.R. Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding; Freeman: San Francisco, CA, USA, 1998. [Google Scholar]
- Fersht, A.R.; Matouschek, A.; Serrano, L. The folding of an enzyme. I. Theory of protein engineering analysis of stability and pathway of protein folding. J. Mol. Biol. 1992, 224, 771–782. [Google Scholar] [CrossRef]
- Mayor, U.; Guydosh, N.R.; Johnson, C.M.; Grossmann, J.G.; Sato, S.; Jas, G.S.; Freund, S.M.V.; Alonso, D.O.V.; Daggett, V.; Fersht, A.R. The complete folding pathway of a protein from nanoseconds to microseconds. Nature 2003, 421, 863–867. [Google Scholar] [CrossRef]
- Baek, M.; DiMaio, F.; Anishchenko, I.; Dauparas, J.; Ovchinnikov, S.; Lee, G.R.; Wang, J.; Cong, Q.; Kinch, L.N.; Schaeffer, R.D.; et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science 2021, 373, 871–876. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Itzhaki, L.S.; Otzen, D.E.; Fersht, A.R. The structure of the transition state for folding of chymotrypsin inhibitor-2 analyzed by protein engineering methods evidence for a nucleation condensation mechanism for protein folding. J. Mol. Biol. 1995, 254, 260–288. [Google Scholar] [CrossRef]
- Jackson, S.E. How do small singledomain proteins fold? Fold. Des. 1998, 3, R81–R91. [Google Scholar] [CrossRef]
- Jackson, S.E.; Fersht, A.R. Folding of chymotrypsin inhibitor 2. 1. Evidence for a two-state transition. Biochemistry 1991, 30, 10428–10435. [Google Scholar] [CrossRef]
- Williams, S.; Causgrove, T.P.; Gilmanshin, R.; Fang, K.S.; Callender, R.H.; Woodruff, A.W.H.; Dyer, R. Fast Events in Protein Folding: Helix Melting and Formation in a Small Peptide. Biochemistry 1996, 35, 691–697. [Google Scholar] [CrossRef]
- Thompson, P.A.; Eaton, W.A.; Hofrichter, J. Laser temperature jump study of the helix <==> coil kinetics of an alanine peptide interpreted with a “kinetic zipper” model. Biochemistry 1997, 36, 9200–9210. [Google Scholar] [CrossRef] [PubMed]
- Thompson, P.A.; Munoz, V.; Jas, G.S.; Henry, E.R.; Eaton, W.A.; Hofrichter, J. The helix-coil kinetics of a heteropeptide. J. Phys. Chem. B 2000, 104, 378–389. [Google Scholar] [CrossRef]
- Mohammed, O.F.; Jas, G.S.; Lin, M.M.; Zewail, A.H. Primary Peptide Folding Dynamics Observed with Ultrafast Temperature Jump. Angew. Chem. Int. Edit. 2009, 48, 5628–5632. [Google Scholar] [CrossRef]
- Lin, M.M.; Mohammed, O.F.; Jas, G.S.; Zewail, A.H. Speed limit of protein folding evidenced in secondary structure dynamics. Proc. Natl. Acad. Sci. USA 2011, 108, 16622–16627. [Google Scholar] [CrossRef]
- Jas, G.S.; Kuczera, K. Computer simulations of helix folding in homo- and heteropeptides. Mol. Simulat. 2012, 38, 682–694. [Google Scholar] [CrossRef]
- Kuczera, K.; Jas, G.S.; Elber, R. Kinetics of Helix Unfolding: Molecular Dynamics Simulations with Milestoning. J. Phys. Chem. A 2009, 113, 7461–7473. [Google Scholar] [CrossRef] [PubMed]
- Kreuzer, S.M.; Elber, R.; Moon, T.J. Early Events in Helix Unfolding under External Forces: A Milestoning Analysis. J. Phys. Chem. B 2012, 116, 8662–8691. [Google Scholar] [CrossRef]
- Jas, G.S.; Middaugh, C.R.; Kuczera, K. Non-Exponential Kinetics and a Complete Folding Pathway of an α-Helical Heteropeptide: Direct Observation and Comprehensive Molecular Dynamics. J. Phys. Chem. B 2014, 118, 639–647. [Google Scholar] [CrossRef]
- Onuchic, J.; Luthey-Schulten, A.; Wolynes, P.G. Theory of protein folding: The energy landscape perspective. Annu. Rev.Phys. Chem. 1997, 48, 545–600. [Google Scholar] [CrossRef] [PubMed]
- Onuchic, J.; Socci, N.D.; Luthey-Schulten, Z.; Wolynes, P.G. Protein folding funnels: The nature of the transition state ensemble. Fold. Des. 1996, 1, 441–450. [Google Scholar] [CrossRef]
- Pande, V.S.; Grosberg, A.Y.; Tanaka, T.; Rokhsar, D.S. Pathways for protein folding: Is a new view needed? Curr. Opin. Struct. Biol. 1998, 8, 68–79. [Google Scholar] [CrossRef]
- Pande, V.S.; Rokhsar, D.S. Is the molten globule a third phase of proteins? Proc. Natl. Acad. Sci. USA 1998, 95, 1490–1494. [Google Scholar] [CrossRef]
- Pande, V.S.; Rokhsar, D.S. Molecular dynamics simulations of unfolding and refolding of a β-hairpin fragment of protein G. Proc. Natl. Acad. Sci. USA 1999, 96, 9062–9067. [Google Scholar] [CrossRef]
- Portman, J.J.; Takada, S.; Wolynes, P.G. Variational theory for site resolved protein folding free energy surfaces. Phys. Rev. Lett. 1998, 81, 5237–5240. [Google Scholar] [CrossRef]
- Muñoz, V.; Thompson, P.; Hofrichter, J.; Eaton, W.A. Folding dynamics and mechanism of β-hairpin formation. Nature 1997, 390, 196–199. [Google Scholar] [CrossRef]
- Jas, G.S.; Eaton, W.A.; Hofrichter, J. Effect of viscosity on the kinetics of α-helix and β-hairpin formation. J. Phys. Chem. B 2001, 105, 261–272. [Google Scholar] [CrossRef]
- Hegefeld, W.A.; Chen, S.E.; DeLeon, K.Y.; Kuczera, K.; Jas, G.S. Helix Formation in a Pentapeptide Experiment and Force-field Dependent Dynamics. J. Phys. Chem. A 2010, 114, 12391–12402. [Google Scholar] [CrossRef]
- Doig, A.J. The alpha-helix as the simplest protein model: Helix-Coil Theory, Stability and Design. In Protein Folding, Misfolding and Aggregation: Classical Themes and Novel Approaches; Muñoz, V., Ed.; Royal Society of Chemistry: London, UK, 2008. [Google Scholar]
- Huo, S.; Straub, J.E. Direct Computation of Long Time Processes in Peptides and Proteins: Reaction Path Study of the Coil-to-Helix Transition in Polyalanine. Proteins 1999, 36, 249–261. [Google Scholar] [CrossRef]
- Allen, M.P.; Tildesley, D.J. Computer Simulation of Liquids; Oxford University Press: Oxford, UK, 1987. [Google Scholar]
- Allen, R.J.; Frenkel, D.; ten Wolde, P.R. Forward flux sampling-type schemes for simulating rare events: Efficiency analysis. J. Chem. Phys. 2006, 124, 194111. [Google Scholar] [CrossRef]
- Aristoff, D.; Bello-Rivas, J.M.; Elber, R. A mathematical framework for exact milestoning. Multiscale Model. Simul. 2016, 14, 301–322. [Google Scholar] [CrossRef] [PubMed]
- Bahar, I.; Lezon, T.R.; Yang, L.W.; Eyal, E. Global dynamics of proteins: Bridging between structure and function. Annu. Rev. Biophys. 2010, 39, 23–42. [Google Scholar] [CrossRef]
- Bello-Rivas, J.M.; Elber, R. Exact milestoning. J. Chem. Phys. 2015, 142, 94102. [Google Scholar] [CrossRef]
- Bello-Rivas, J.M.; Elber, R. Simulations of thermodynamics and kinetics on rough energy landscapes with milestoning. J. Comput. Chem. 2016, 37, 602–613. [Google Scholar] [CrossRef]
- Bolhuis, P.G.; Chandler, D.; Dellago, C.; Geissler, P.L. Transition path sampling: Throwing ropes over rough mountain passes, in the dark. Annu. Rev. Phys. Chem. 2002, 53, 291–318. [Google Scholar] [CrossRef] [PubMed]
- Bowman, G.R.; Pande, V.S. An Introduction to Markov State Models and Their Applications to Long Timescale Molecular Simulations; Springer: Berlin, Germany, 2014. [Google Scholar]
- Cardenas, A.E.; Elber, R. Computational study of peptide permeation through membrane: Searching for hidden slow variables. Mol. Phys. 2013, 111, 3565–3578. [Google Scholar] [CrossRef]
- Cardenas, A.E.; Jas, G.S.; DeLeon, K.Y.; Hegefeld, W.A.; Kuczera, K.; Elber, R. Unassisted transport of N-acetyl-L-tryptophanamide through membrane: Experiment and simulation of kinetics. J. Phys. Chem. B 2012, 116, 2739–2750. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cardenas, A.E.; Elber, R. Modeling kinetics and equilibrium of membranes with fields: Milestoning analysis and implication to permeation. J. Chem. Phys. 2014, 141, 54101. [Google Scholar] [CrossRef]
- Dror, R.O.; Dirks, R.M.; Grossman, J.P.; Xu, H.; Shaw, D.E. A Computational Mi-croscope for Molecular Biology. Annu. Rev. Biophys. 2012, 41, 429–452. [Google Scholar] [CrossRef]
- Bowman, G.R. An Overview and Practical Guide to Building Markov State Models. In An Introduction to Markov State Models and Their Application to Long Timescale Molecular Simulation; Bowman, G.R., Pande, V.S., Noe, F., Eds.; Springer: Heidelberg, Germany, 2014; pp. 7–22. [Google Scholar]
- Kube, S.; Weber, M. A coarse graining method for the identification of transition rates between molecular conformations. J. Chem. Phys. 2007, 126, 24103. [Google Scholar] [CrossRef] [PubMed]
- Hummer, G.; Szabo, A. Optimal Dimensionality Reduction of Multistate Kinetic and Markov-State Models. J. Phys. Chem. B 2015, 119, 9029–9037. [Google Scholar] [CrossRef]
- Jas, G.S.; Vallejo-Calzada, R.; Johnson, C.K.; Kuczera, K. Dynamic elements and kinetics: Most favorable conformations of peptides in solution with measurements and simulations. J. Chem. Phys. 2019, 151, 225102. [Google Scholar] [CrossRef] [PubMed]
- Jas, G.S.; Kuczera, K. Helix-Coil Transition Courses through Multiple Pathways and Intermediates: Fast Kinetic Measurements and Dimensionality Reduction. J. Phys. Chem. B 2018, 122, 10806–10816. [Google Scholar] [CrossRef] [PubMed]
- Senne, M.; Trendelkamp-Schroer, B.; Mey, A.S.J.S.; Schutte, C.; Noe, F. EMMA: A Software Package for Markov Model Building and Analysis. J. Chem. Theory Comput. 2012, 8, 2223–2238. [Google Scholar] [CrossRef]
- Trendelkamp-Schroer, B.; Wu, H.; Paul, F.; Noe, F. Estimation’ and Uncertainty of Reversible Markov Models. J. Chem. Phys. 2015, 143, 174101. [Google Scholar] [CrossRef] [PubMed]
- Jas, G.S.; Childs, E.W.; Kuczera, K. Kinetic pathway analysis of an α-helix in two protonation states: Direct observation and optimal dimensionality reduction. J. Chem. Phys. 2019, 150, 74902. [Google Scholar] [CrossRef]
- Hess, B.; Kutzner, C.; van der Spoel, D.; Lindahl, E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced and Scalable Molecular Simulation. J. Chem. Theory Comput. 2008, 4, 435–447. [Google Scholar] [CrossRef]
- Brooks, B.R.; Brooks, C.L.; MacKerell, A.D.; Nilsson, L.; Petrella, R.J.; Roux, B.; Won, Y.; Archontis, G.; Bartels, C.; Boresch, S.; et al. CHARMM: The Biomolecular Simulation Program. J. Comput. Chem. 2009, 30, 1545–1614. [Google Scholar] [CrossRef]
- Krokhotin, A.; Liwo, A.; Maisuradze, G.; Niemi, J.; Scheraga, H.A. Kinks, loops and protein folding, with protein A as an example. J. Chem. Phys. 2014, 140, 25101. [Google Scholar] [CrossRef]
- Maisuradze, G.G.; Liwo, A.; Senet, P.; Scheraga, H.A. Local vs. global motions in protein folding. J. Chem. Theory Comput. 2013, 9, 2907–2921. [Google Scholar] [CrossRef][Green Version]
- Kuczera, K.; Szoszkiewicz, R.; He, J.; Jas, G.S. Length Dependent Folding Kinetics of Alanine-Based Helical Peptides from Optimal Dimensionality Reduction. Life 2021, 11, 385. [Google Scholar] [CrossRef] [PubMed]
- Tribello, G.A.; Gasparotto, P. Using Dimensionality Reduction to Analyze Protein Trajectories. Front. Mol. Biosci. 2019, 6, 1–11. [Google Scholar] [CrossRef]
- de Souza, V.C.; Goliatt, L.; Capriles, P.V.Z. Insight About Nonlinear Dimensionality Reduction Methods Applied to Protein Molecular Dynamics. In Bioinformatics and Biomedical Engineering; Rojas, I., Venzuela, O., Rojas, F., Ortuno, F., Eds.; Springer: Heidelberg, Germany, 2019; pp. 219–230. [Google Scholar]
- Stamati, H.; Clementi, C.; Kavrak, L.E. Application of nonlinear dimensionality reduction to characterize the conformational landscape of small peptides. Proteins 2010, 78, 223–235. [Google Scholar] [CrossRef] [PubMed]
- Jas, G.S.; Hegefeld, W.A.; Majek, P.; Kuczera, K.; Elber, R. Experiments and Comprehensive Simulations of the Formation of a Helical Turn. J. Phys. Chem. B 2012, 116, 6598–6610. [Google Scholar] [CrossRef] [PubMed][Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jas, G.S.; Childs, E.W.; Middaugh, C.R.; Kuczera, K. Dissecting Multiple Pathways in the Relaxation Dynamics of Helix <==> Coil Transitions with Optimum Dimensionality Reduction. Biomolecules 2021, 11, 1351. https://doi.org/10.3390/biom11091351
Jas GS, Childs EW, Middaugh CR, Kuczera K. Dissecting Multiple Pathways in the Relaxation Dynamics of Helix <==> Coil Transitions with Optimum Dimensionality Reduction. Biomolecules. 2021; 11(9):1351. https://doi.org/10.3390/biom11091351
Chicago/Turabian StyleJas, Gouri S., Ed W. Childs, C. Russell Middaugh, and Krzysztof Kuczera. 2021. "Dissecting Multiple Pathways in the Relaxation Dynamics of Helix <==> Coil Transitions with Optimum Dimensionality Reduction" Biomolecules 11, no. 9: 1351. https://doi.org/10.3390/biom11091351
APA StyleJas, G. S., Childs, E. W., Middaugh, C. R., & Kuczera, K. (2021). Dissecting Multiple Pathways in the Relaxation Dynamics of Helix <==> Coil Transitions with Optimum Dimensionality Reduction. Biomolecules, 11(9), 1351. https://doi.org/10.3390/biom11091351







