1. Introduction
Insulin secretion is essentially controlled by the interaction of two incretin hormones, the glucose-dependent insulinotropic polypeptide (GIP) and the glucagon-like peptide-1 (GLP-1) with their receptors GIPR and GLP-1R, respectively. GLP-1 has attracted attention for its use in the treatment of type 2 diabetes. GLP-1 is highly homologous to several other peptide hormones involved in glycemic control and energy metabolism, such as glucagon, GLP-2, oxyntomodulin and GIP. The binding of all these peptides to their receptors, all members of the class B G-protein coupled receptor family, happens in a mainly α-helical conformation in a very similar manner [
1]. Another commonality of these peptides is a highly conserved hydrophobic moiety FhxWL (with h denoting any hydrophobic and x denoting any amino acid, respectively) (
Figure S1), which has been previously suggested to play a role in the aggregation of glucagon [
2].
A major drawback hindering the pharmaceutical use of GLP-1 is its rapid degradation in vivo by the enzyme dipeptidyl-peptidase IV. This has stimulated the search for peptidase resistant analogues. The first stable marketed GLP-1 mimetic was exendin-4, a peptide of 39 AA (amino acid) residues that was originally isolated from the venom of the lizard
Heloderma suspectum. Exendin-4 shares about 50% sequence identity with GLP-1. Since then, several advanced unimolecular dual or triple agonist peptides targeting more than one of the receptors indicated above have been developed (see [
3] for a review). In line with this, we recently characterized the potent dual GLP-1/glucagon agonist Dual-Cex ([
4], assigned as peptide 7 in [
5]), which was accomplished by engineering sequence sections of oxyntomodulin into exendin-4.
A series of studies published by the Andersen lab in the early 2000s [
6,
7,
8,
9,
10] provided basic structural information of exendin-4 concerning secondary and tertiary structure. Notably, peptide conformation is highly dependent on solvent conditions. In principle, the amino acids 9–27 of exendin-4 form an α-helix under various solvent conditions, while the nine N-terminal amino acids are essentially unstructured [
7]. The superposition of unbound exendin-4 in the presence of 30% trifluoroethanol (TFE) obtained from NMR [
10] and receptor-bound exendin-4(9–39) in crystals shows striking similarities [
11]. This suggests that the helical structure preferences of exendin-4 in solution are related to its receptor binding affinity. Exendin-4 further harbors a C-terminal segment, which can fold into the so-called tryptophan (Trp-) cage, a particular structural motif, that was clearly identified in aqueous solutions containing TFE [
10]. Under these conditions, the C-terminal segment of exendin-4 folds back on the central α-helix and forms what has been designated the smallest stable known protein tertiary structure. Here, the two hydrophobic bulky amino acids Phe22 and Trp25, which are part of the conserved hydrophobic moiety FhxWL, are closely associated with Gly26 and Pro31, Pro37 and Pro38, resulting in the essential enclosure of Trp25 [
9]. This fold is only partially populated in aqueous glycol solvent [
8] and not formed in the presence of dodecylphosphocholine micelles, where residues 31–39 are unstructured and highly dynamic. Independent of the formation of the actual Trp-cage conformation, a hydrophobic cluster between Trp25 and Pro31 has been suggested to provide a stable capping of the exendin-4 α-helix [
8]. To avoid misinterpretation, we will refer to the specific conformation of the C-terminal segment in 30% TFE as the ‘Trp-cage conformation’ and use the term ‘hydrophobic moiety’ when referring to other, less strictly defined conformations of this segment in the following.
Based on the stable Trp-cage conformation of exendin-4 in 30% TFE, a variety of Trp-cage miniproteins have been designed. Most of them were derived from the initial Trp-cage variety TC5b [
9], which was mainly realized by N-terminal truncation of the exendin-4 backbone and several point mutations. TC5b represents a non-fluxional, compact, and globular structure in aqueous environments [
9]. Trp-cage miniproteins have been the subject of numerous studies concerning solvent interactions, folding processes, stability, and aggregation tendency [
12,
13,
14,
15,
16], mainly due to their small size being a helpful property in computer simulations. To our knowledge, literature on the oligomeric state of Trp-cage miniproteins is limited to a single study using a cyclized Trp-cage variant [
17], facilitating analytical ultracentrifugation (AUC).
Likewise, reports on the oligomerization of exendin-4 are scarce and structural details of the resulting oligomers are not available to date. And yet, self-assembly of pharmaceutical peptides is not only of general basic interest, but is fundamental for biological activity, stability, absorption, and lifetime under formulation conditions. The existence of oligomeric states of exendin-4 in buffer or under formulation conditions and their possible characteristics have been suggested only indirectly from NMR- and CD-spectroscopic data. Also, the state of exendin-4 in aqueous solutions containing TFE has not been directly determined up to now. From NMR measurements of translational diffusion coefficients Wang et al. [
18] proposed the existence of dimers or trimers. In aqueous solution, Hudson suggested the oligomerization of exendin-4 via amphipathic helix–helix interactions, yielding helix-bundles [
7], which is still a matter of debate.
In contrast to these previous reports, we use combined static and dynamic light scattering (SLS/DLS) for direct determination of the molecular mass and hydrodynamic radius. This enables us to unequivocally and directly determine the association behavior of exendin-4 in different solvent and concentration conditions. We relate our experimental results to a molecular structural hypothesis, which was built on an alignment of exendin-4 with the crystal structure of a cyclized Trp-cage variant [
17] to describe molecular details of exendin-4 oligomers. We discuss the validity of this hypothesis based on spectroscopic and light scattering data. We put special focus on the role of the conserved hydrophobic moiety FhxWL in the association of exendin-4, utilizing a monomeric exendin-4 derived peptide Dual-Cex [
4,
5] and a set of derived mutants. We further exploit differences in the properties of exendin-4 and Dual-Cex to discuss the role of helix–helix interactions in self-assembly.
4. Discussion
A first important matter of this work is a detailed characterization of the association state of exendin-4 under appropriate solution conditions. Up to now, the association state of exendin-4 has not been well defined and controversial opinions about the appearance of oligomerization, the type of oligomers, and the mechanisms of formation have been published [
6,
7,
18]. Hudson and Andersen [
6,
7] concluded the existence of oligomeric species in strictly aqueous media from the influence of concentration on NMR and CD data. Wang et al. [
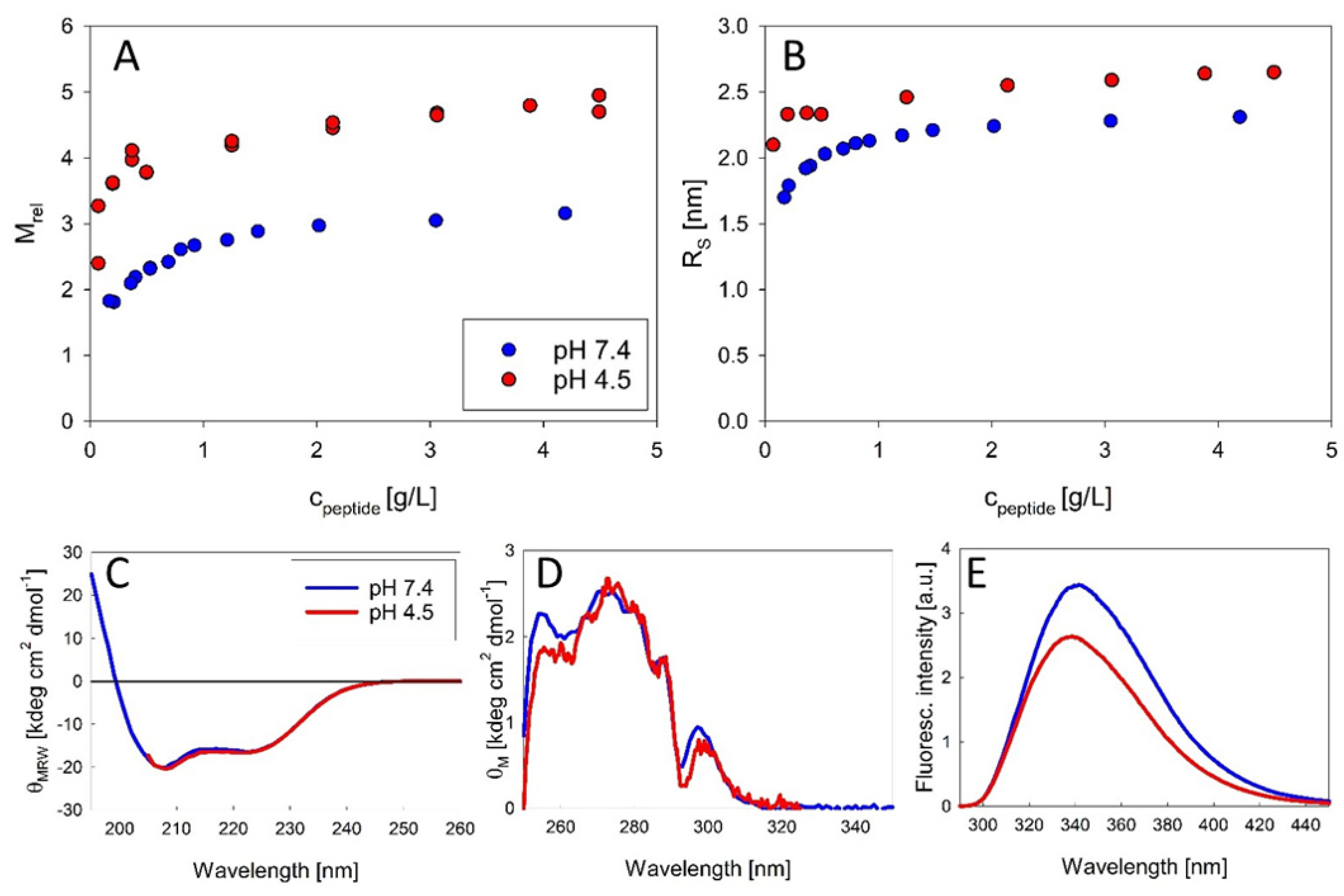
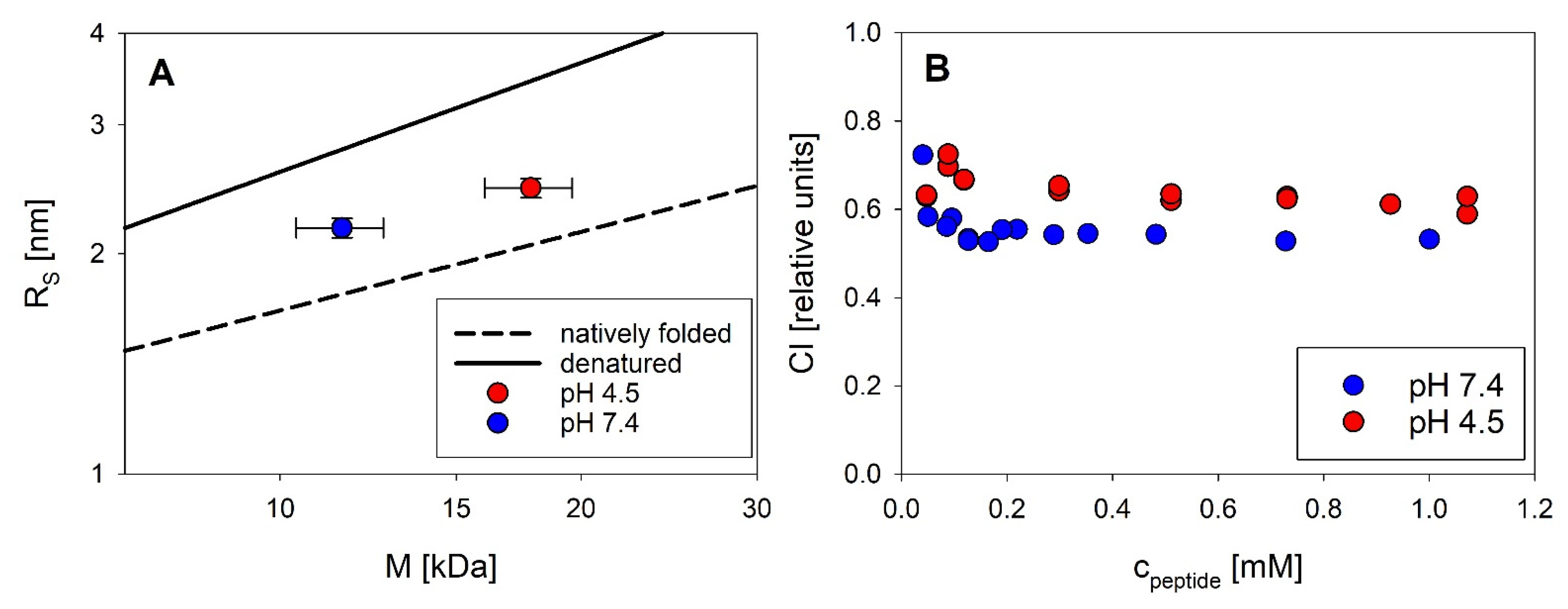
18] tried to estimate the size of putative oligomers from NMR diffusion-ordered spectroscopy (DOSY), suggesting the formation of trimers. From our SLS data we can directly calculate the mass and thus the number of associated peptide molecules without any assumptions about the arrangement of monomers. Our study therefore provides the first published association equilibria of exendin-4 in aqueous solutions. The oligomeric state of exendin-4 is clearly pH dependent. At pH 7.4, we find an apparently unimodal association into trimers, which is observed over a sufficiently wide concentration range (
Figure 2A,B). Therefore, our data substantially add to the knowledge on the association of exendin-4 available from previous studies. The postulation of a trimeric main species is supported by published NMR data, which suggest that a defined oligomeric species is populated over a wide range of concentrations [
7]. At pH 4.5, close to the isoelectric point of exendin-4 (pI = 4.86 [
26]), oligomer formation is biphasic with the formation of trimers being reinforced, as the trimeric state is already approached at lower peptide concentrations compared with pH 7.4. With increasing concentration, larger oligomers, likely a mixture of trimers and dimers of trimers are obvious. This differential oligomerization behavior is attributable to a different degree of electrostatic interactions. At both pH regimes, dissociation becomes evident at low concentrations, but complete dissociation into monomers could not be verified, since it likely happens below the detection limit of SLS. The conformation of monomeric subunits regarding the secondary and tertiary structure level is comparable at both pH regimes (
Figure 2C–E). The conserved hydrophobic moiety is essentially covered from solvent during trimer formation (
Figure 5).
These findings on the global solution structure of exendin-4 stimulated us to establish a structural hypothesis of the smallest oligomer of exendin-4, the trimer at pH 7.4, using molecular modelling based on the crystal structure of an exendin-4 derived Trp-cage miniprotein (
Figure 3). Importantly, this hypothesis is fully compatible with all so far presented experimental data and moreover supported by comparing calculated and measured R
S. In the following, we will facilitate experimental data of selected exendin-4 derived peptides in order to discuss the validity of molecular details of the association of exendin-4 as suggested by this hypothesis. We will restrict this discussion to the smallest oligomer at pH 7.4, but a dimer of trimers at low pH, relevant in the pharmaceutical formulation of exendin-4, may also in principle be rationalized following this hypothesis.
A key structural element of exendin-4 is a central amphipathic α-helix, which has been found in solution [
10] and also in the crystal structure of N-terminally truncated exendin-4(9–39) bound to the isolated extracellular domain of human GLP-1R [
11]. Our structural hypothesis suggests an interaction of monomeric subunits of exendin-4 in the trimer via helix–helix contacts of the C-terminal half of this helix. Similarly, Hudson et al. concluded an association of exendin-4 via interhelical interaction based on the extensive broadening of NMR resonances in the Gln13 to Leu26 stretch [
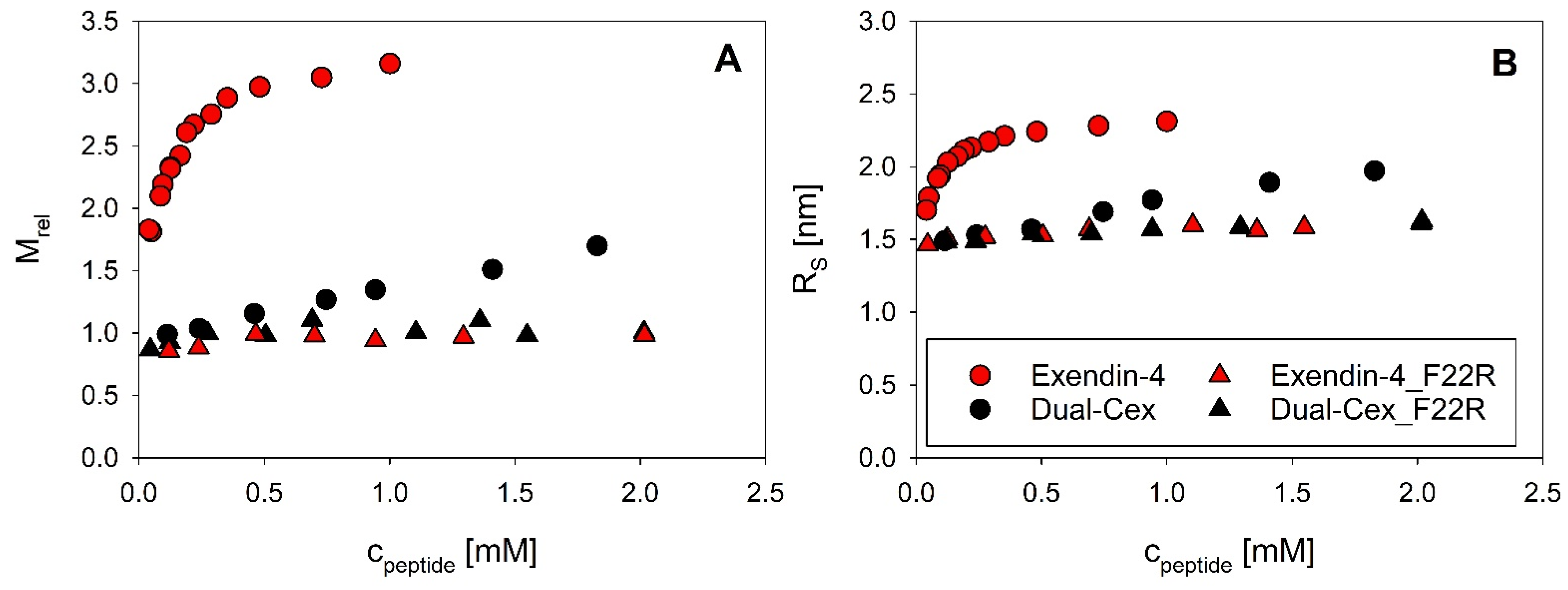
7]. Interestingly, precisely these amino acids are situated in the oligomer interface in our structural hypothesis and will be denoted as helical interface segments in the following. The high importance of helix–helix interactions of this helical interface segment for association of exendin-4 is directly evidenced by the largely monomeric nature of the exendin-4 derived Dual-Cex, which carries modifications within the respective segment. These modifications encompass Asp15 to Asp21 and apparently heavily impact charge and hydrophobic moment (
Figure 4D–F). It is thus conceivable, that the oligomerization of exendin-4 necessitates hydrophobic interactions between the amphipathic α-helical segments. Comparable hydrophobic interactions are not established among Dual-Cex monomers, which is markedly less α-helical in buffer and the α-helix, if established, is considerably less amphipathic. Taken together, this hinders self-assembly of Dual-Cex. Hudson initially suggested a helix-bundle or even coiled-coil state for exendin-4 oligomers. This idea was based on the lack of an isodichroic point in the CD spectra of exendin-4 during melting and a comparatively high melting temperature [
7]. The classical picture of a helix-bundle is associated with knobs-into-holes packing of hydrophobic residues of adjacent helices as originally introduced by Crick [
27]. This often results in almost parallel or antiparallel orientation of the interacting helices, which are frequently characterized by conserved patterns of amino acids [
27]. However, the actual angle of interacting helices constituting a helix-bundle can be relatively wide [
28]. As both diagnostics suggested for a helix bundle state by Hudson [
7] would also apply for a rather triangular orientation of the helical segments situated in the interface of a trimer architecture, our structural hypothesis is not in conflict with the data presented by Hudson [
7]. However, a typical coiled-coil structure of exendin-4 oligomers is not supported by our data. This is because the rather defined oligomers of exendin-4 in aqueous solution are fairly extended, which points towards a certain degree of structural flexibility (
Figure 10), with the pH 7.4 trimers being slightly more extended than the dimers of trimers observed at pH 4.5. In contrast to the quite extended nature of exendin-4 oligomers, coiled-coils should be rather compact structures. In addition, we don’t have indications for coiled-coil structures from the measured CD spectra. Typical coiled-coils have CD spectra differing from that of α-helical structures by a stronger negative ellipticity at 222 nm leading to θ
222/θ
208 ratios greater than 1 [
29]. Furthermore, coiled-coils are rather stable structures that unfold in a cooperative way [
30]. This was not observed for exendin-4 at concentrations corresponding to oligomers ([
6] and
Figure S4).
The C-terminal part of the exendin-4 helix is constituted by the hydrophobic moiety FhxWL comprising AA 22–26, which is highly conserved among related peptides. Thus, the general statement of helix–helix interactions in the helical interface segment (AA13–26) can be partially specified as interactions of this conserved hydrophobic moiety, which are evident in the exendin-4 trimer. This can be derived from progressive shielding of Trp25 going along with oligomerization (
Figure 5). Actual evidence for the importance of interactions of this conserved hydrophobic moiety for oligomerization is impressively evidenced by the F22R substituted isoforms of exendin-4 (exendin-4_F22R) and Dual-Cex (Dual-Cex_F22R), in which oligomerization is completely disabled. This accounts for both the trimerization of exendin-4 as well as the marginal association of Dual-Cex and might be enhanced by electrostatic repulsion between the introduced Arg residues. Therefore, it is clear that the conserved hydrophobic moiety is not only interacting in the trimer, but also presents an important driving force for oligomerization. This argues in favor of our structural hypothesis, which suggests strong hydrophobic attraction between the F22 residues of all monomeric subunits in the center of the trimer. It is important to note that the F22R mutation has only a minor impact on helix stability, which clearly allows us to link the loss of oligomerizability of exendin-4_F22R with the specific absence of Phe22 rather than an overall reduction in helicity. Interestingly, Dual-Cex, which is modified in AA15–21, harbors the conserved hydrophobic moiety without any modifications and is essentially monomeric, indicating that interactions of the conserved hydrophobic moieties alone are not sufficient for oligomerization. This proves the essential role of further helix–helix interactions in the oligomerization of exendin-4 and can be rationalized by our structural hypothesis, which suggests close contact and thus hydrophobic interaction of Trp25 with Val19. The latter is replaced by a less hydrophobic Ala in Dual-Cex, which is a possible explanation for the lack of oligomerization of Dual-Cex. This argues for the suggested triangular oligomer architecture of exendin-4 with Val19 functioning as an anchor for neighboring exendin-4 subunits in the trimer.
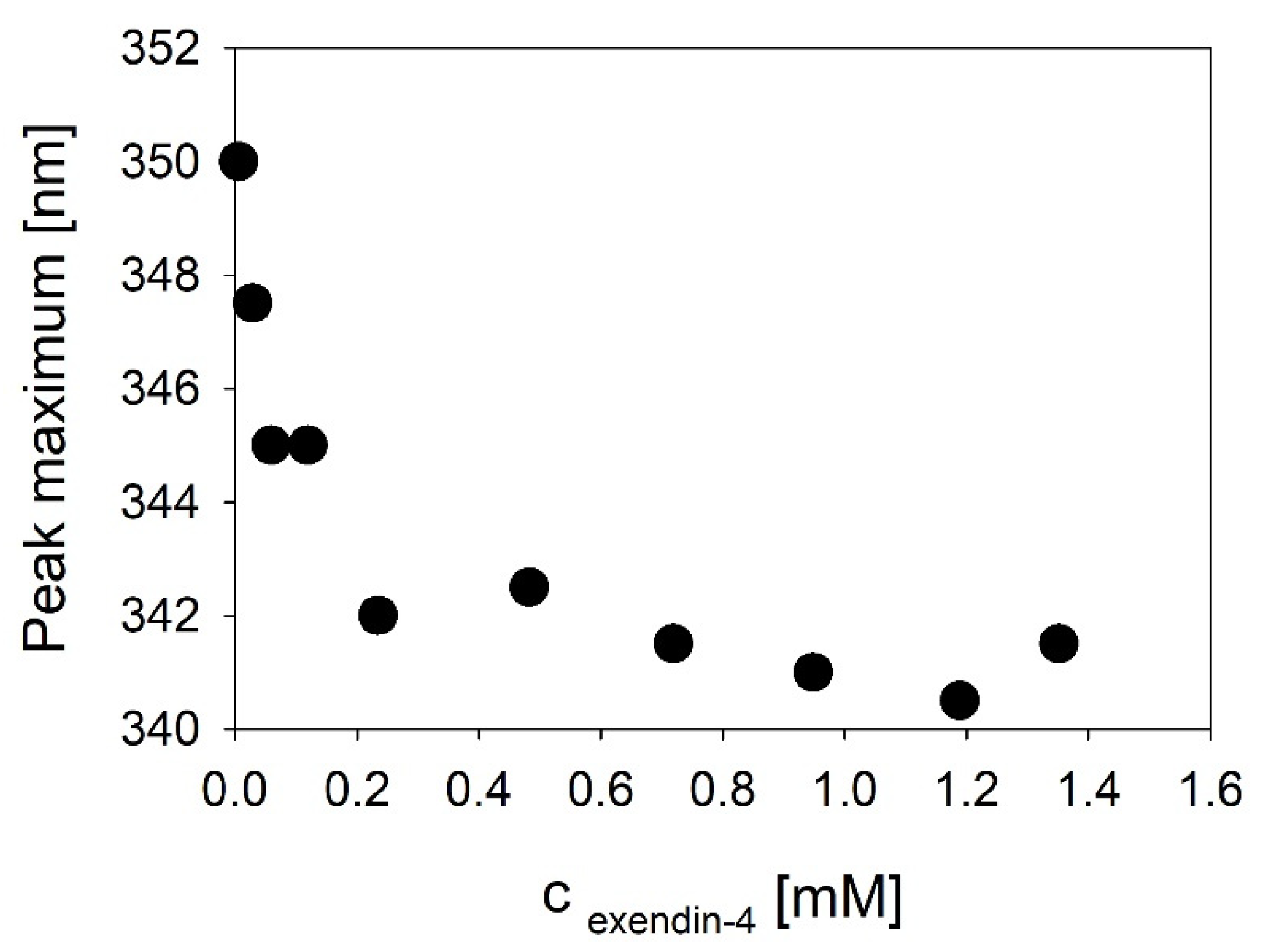
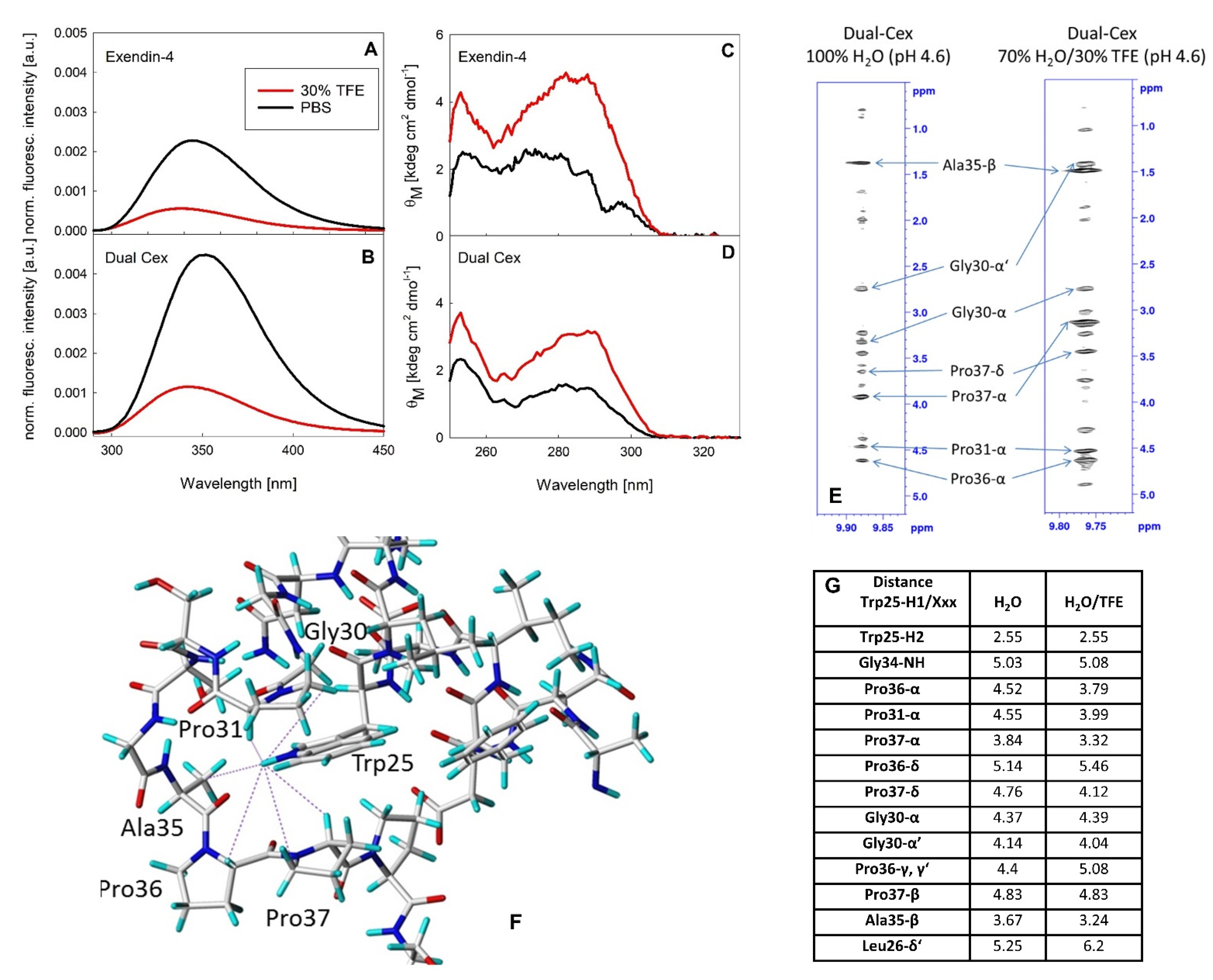
Our structural hypothesis facilitates an NMR derived monomer structure of exendin-4 in 30% TFE. The impact of TFE on the association state of exendin-4 has not been finally resolved to date. Wang et al. [
18] have tried to estimate the size of putative oligomers from NMR DOSY, but the results in the presence of TFE (dimers or trimers) are questionable. Our SLS data (
Figure S2A) unequivocally show that exendin-4 has reached a fully monomeric state already at a TFE concentration of 20%. The hydrodynamic dimensions of the monomers are rather large, typical of extended proteins (
Figure S5). The absolute ratio of helicity estimated from our CD spectra in 30% TFE is similar to the one of the NMR derived monomer structure, accounting for about 70%. It is obvious that in 30% TFE, exendin-4 has about 20% more α-helical structure than in aqueous buffer (
Figure 4B). It is reasonable to assume that this is due to a more elongated conformation of monomers mainly brought about by the central helical part which is stabilized in the presence of TFE. TFE has been described as accumulating near peptide surfaces and to preferentially interact with amino acid side chains [
31], therefore under these conditions, the helix can be regarded as essentially covered with TFE molecules. In this perfectly covered TFE-state, exendin-4 is not able to form oligomers. The superior helicity in 30% TFE and the fact that the specific Trp-cage conformation of the C-terminus of exendin-4 is formed under these conditions, but not in aqueous buffer, challenges the suitability of the NMR derived structure of the exendin-4 monomer in 30% TFE as a building block for our molecular modelling approach. Our structural hypothesis specifically relies on the stability of the C-terminal half of the α-helix of exendin-4 in aqueous buffer. A hydrophobic cluster between Trp25 and Pro31, which forms independently of the Trp-cage already in aqueous buffer, has been suggested to provide a stable C-terminal capping of the exendin-4 α-helix previously [
7]. Therefore, it is conceivable that TFE stabilizes the helix towards the N-terminal part of the peptide, which is not involved in inter-monomer interaction in our exendin-4 trimer model. The addition of 30% TFE to solutions of exendin-4 and Dual-Cex results in a substantial quenching of Trp fluorescence and an increase in molar ellipticity around 290 nm, indicating the shielding of Trp25 from solvent and a concomitantly reduced flexibility. Trp fluorescence quenching has been recognized as a diagnostic marker for formation of the Trp-cage [
32,
33]. Therefore, our data clearly evidence the formation of the Trp-cage in the presence of TFE for Dual-Cex and exendin-4 (
Figure 9) but cannot resolve to what degree the Trp-cage exists in strictly aqueous media. A direct comparison of the structures of the exendin-4 monomer in water and 30% TFE in single-residue resolution has been previously hampered by the oligomeric nature of exendin-4 and the concomitantly poor-quality NMR spectra [
7]. Encouraged by the similar signal changes in the fluorescence and near-UV CD spectra of exendin-4 and Dual-Cex, we took advantage of the monomeric nature of Dual-Cex and recorded NOESY spectra in aqueous buffer and 30% TFE (
Figure 9E–G). For exendin-4, formation of the Trp-cage in 30% TFE leads to large highfield-shifts for Gly30-Hα2, Pro37-Hα, and Pro38-CδH
2 [
7]. In the case of Dual-Cex, the addition of 30% TFE induces an equivalent highfield-shift of these protons (
Table S1), thus confirming the same Trp-cage conformation for both peptides. However, in the absence of TFE clearly smaller highfield-shifts and some differences in the interproton distances are obtained (
Figure 9G and
Table S2). This indicates a slightly different geometry of the residues surrounding the side chain of Trp25 and thus a different conformation of the C-terminus and the conserved hydrophobic moiety. It is conceivable that in aqueous buffer the Trp-cage conformation is either transiently sampled, leading to a more open conformation, or represents an alternative defined fold characterized by a different local energy minimum. This interesting question should be addressed by time-resolved structure analytics in future studies. An important hint arguing against the Trp-cage conformation being stabilized in the exendin-4 oligomer comes from Hudson et al., who found the C-terminus from Ser33–Ser39 being rather flexible in the oligomer, similar to statistical coils. They thus excluded the interaction of Trp25 with Pro37, which is one of the key characteristics for the Trp-cage conformation. The authors concluded from these data, that hydrophobic surface burial is apparently better accomplished by oligomerization than by formation of the Trp-cage [
7]. Taken together, these data suggest that the NMR monomer structure of exendin-4 in 30% TFE is certainly not an ideal monomeric building block for modelling. However, it is undoubtedly well-suited concerning the dimension of the helical segment involved in inter-monomer interaction in our model. In addition, the geometry of the conserved hydrophobic moiety can be regarded as sufficiently similar in aqueous buffer and in 30% TFE, while the seven C-terminal amino acids certainly have a substantially higher degree of freedom in the exendin-4 oligomer than indicated in our structural model.
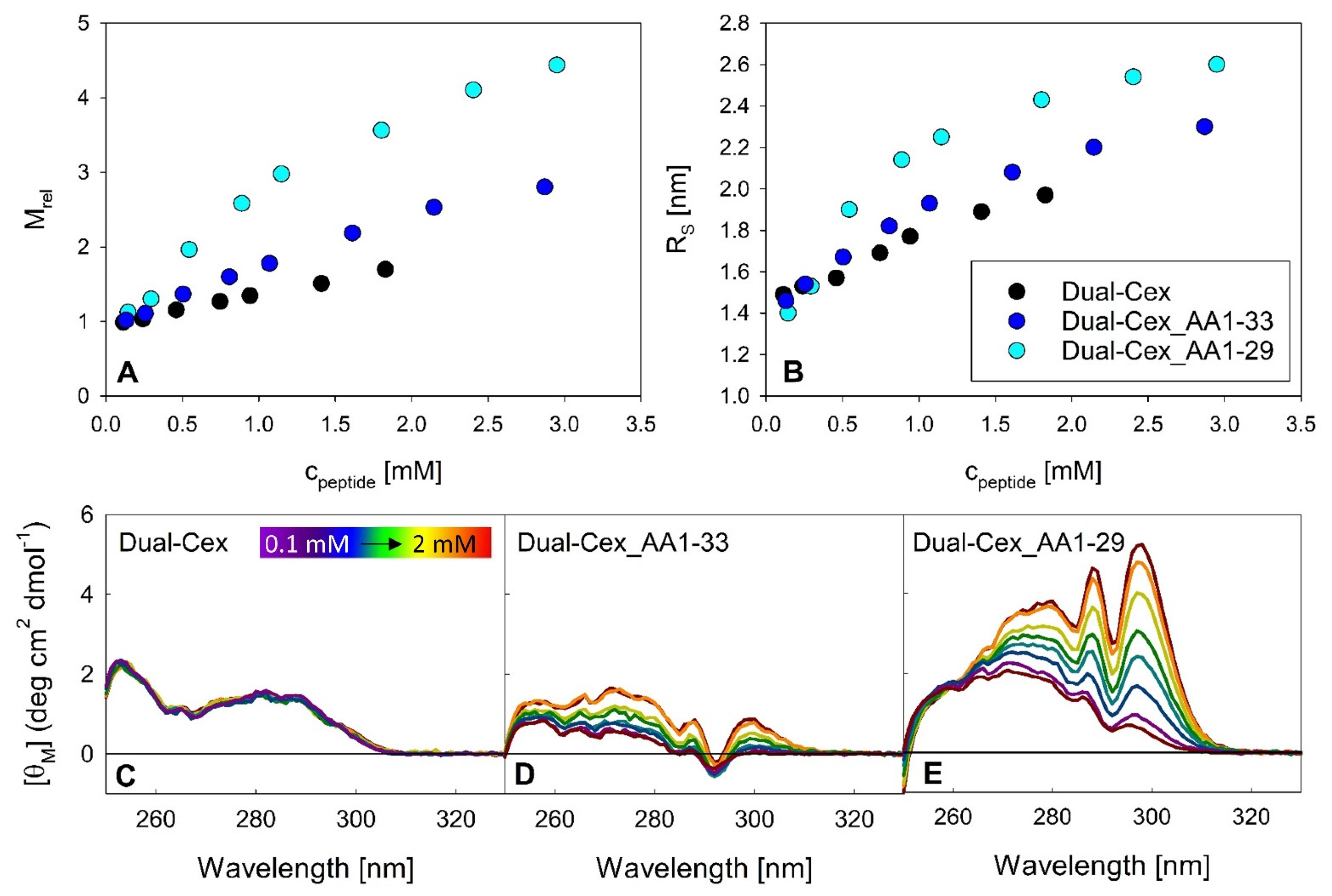
The latter might be of importance as these amino acids may play a role in shielding the conserved hydrophobic moiety. In this context, our C-terminally truncated Dual-Cex isoforms are highly instructive. Our data clearly evidence that further exposure of the conserved hydrophobic moiety by stepwise C-terminal truncation is sufficient for oligomerization of Dual-Cex (
Figure 7). Assuming that the conformation of the conserved hydrophobic moiety is identical in exendin-4 and Dual-Cex, this implies that it must be at least partially shielded already in aqueous buffer. Interestingly, this exposure of the conserved hydrophobic moiety leads to a different kind of oligomerization than that which we observe for exendin-4, as it is less defined, suggesting constant addition of monomeric subunits with increasing concentration, likely via the presentation of hydrophobic interfaces to the surrounding solvent. While not as detailed as in our study, a role of the conserved hydrophobic moiety in the oligomerization of related peptide has been evidenced in earlier studies. In this context, the addition of the nine C-terminal amino acids of exendin-4 to glucagon results in dramatically improved solubility and stability of the highly aggregation-prone native glucagon peptide [
2]. It is important to note that all exendin-4 derived assembly structures presented in our study are essentially stable, in contrast to glucagon. The GLP-2 analog teduglutide, which also exhibits the conserved hydrophobic moiety, self-assembles into defined pentamers, as shown by AUC [
34]. Following NMR structures determined in water-TFE solutions, this cluster is solvent-exposed in the teduglutide monomer [
35]. Self-assembly induces not only α-helical structure, but also a distinct near-UV CD signal around 300 nm, indicating a clustering of Trp residues, but not Phe residues in the oligomer [
34]. This is different from our C-terminally truncated Dual-Cex variants in two aspects, first the rather defined nature of the teduglutide oligomers and second the apparent differences in the near-UV CD signals, which indicate involvement of both, Phe and Trp residues in the oligomerization of the C-terminally truncated Dual-Cex variants. Importantly, the effect of truncation of residues 34–39 of Dual-Cex on self-association and concomitant conformational changes of the conserved hydrophobic moiety is relatively low, while the effect of truncation of residues 30–39 is much more drastic. This suggests that a previously described interaction of Pro31 with Trp25 in aqueous buffer [
7] is highly efficient in shielding Trp25, while contributions of farther C-terminally localized amino acids, likely Pro36–38, are less important. These considerations are in line with our structural hypothesis, which suggests that Trp25 of exendin-4 monomers do not interact with each other in the trimer, but rather with Pro31 and Pro36–38 of their own C-termini. Apparently, the actual impact of Pro36–38 on shielding of Trp25 might be overrated in our structural hypothesis due to the use of the TFE monomer structure with its stable Trp-cage conformation.
Additional information about the complicated interplay between the different forces driving association of Dual-Cex can be derived from different Dual-Cex isoforms conjugated to a palmitic acid. We have previously shown that palmitic acid fusion coerces Dual-Cex into an oligomeric state [
4]. It is necessary to note that these assemblies differ from those of exendin-4 and might rather be imagined as quasi-micellar structures. Palmitic acid conjugation also leads to self-assembly of all Dual-Cex isoforms used in our study, but the resulting assembly structures are remarkably different from each other in terms of size and stability towards dilution (
Figure 8). Apparently, an intact conserved hydrophobic moiety is mandatory for the formation of stable self-assemblies, as in contrast to Dual-Cex_C16, both palmitic acid conjugated Dual-Cex isoforms harboring the F22R mutation (Dual-Cex-C16_F22R and Dual-C16_F22R) show substantial dissociation upon dilution. Likely, overall hydrophobicity of the conserved moiety is no longer sufficient for stable self-assembly upon loss of Phe22, underlining the fundamental role of Phe22 interaction in the exendin-4 trimer, as suggested in our structural hypothesis. On the other hand, the exposure of the conserved hydrophobic moiety (Dual-C16) affects the molecular architecture of the fatty acid stabilized self-assemblies, resulting in dilution-stable oligomers with a larger number of monomeric subunits. The latter might be rationalized via the micelle-like nature of the palmitic acid enforced assemblies, allowing a higher number of C-terminally truncated monomeric subunits, a hypothesis which needs further research.