Design and Prototyping of Genetically Encoded Arsenic Biosensors Based on Transcriptional Regulator AfArsR
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA Construction and Mutagenesis
2.2. Protein Expression and Purification
2.3. Fluorescence Measurement
2.4. Computational Structure Prediction of Genetically Encoded As3+ Biosensors
2.5. Statistical Analysis
3. Results and Discussion
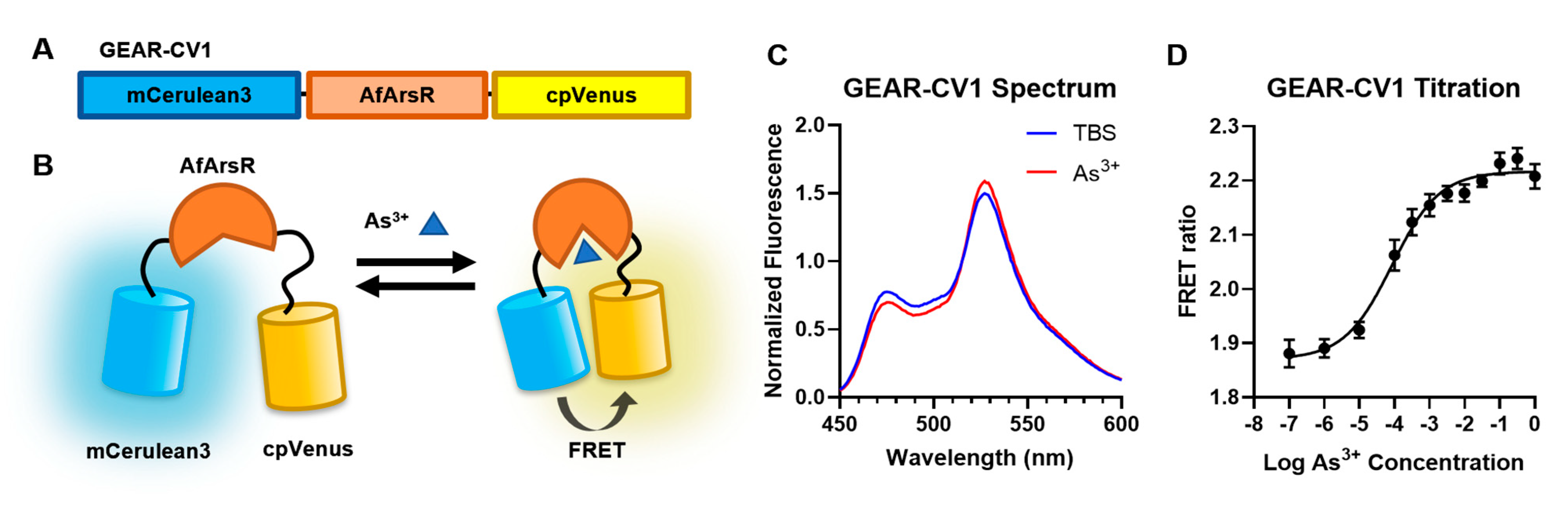
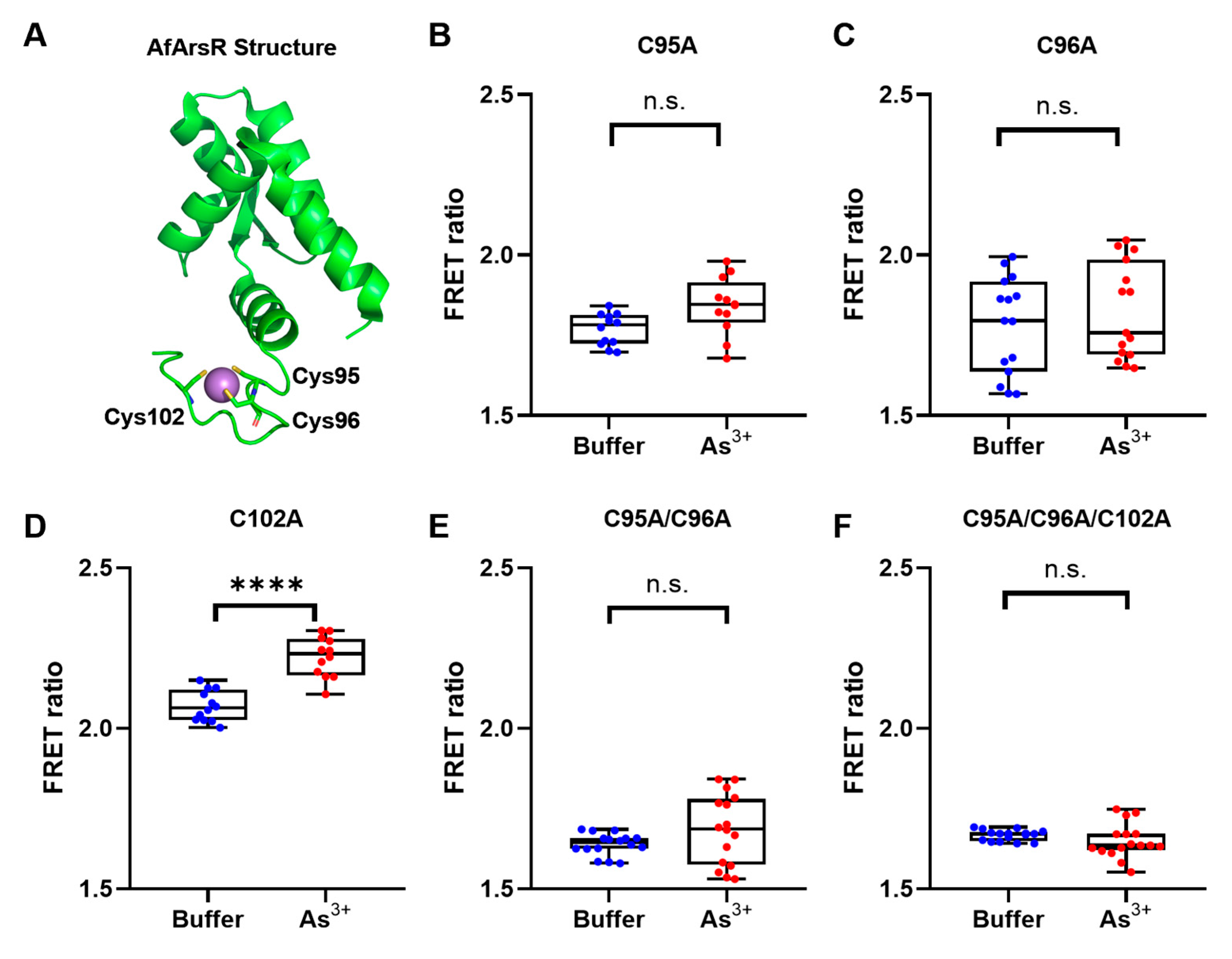
3.1. Development of Genetically Encoded As3+ Biosensors Based on FRET
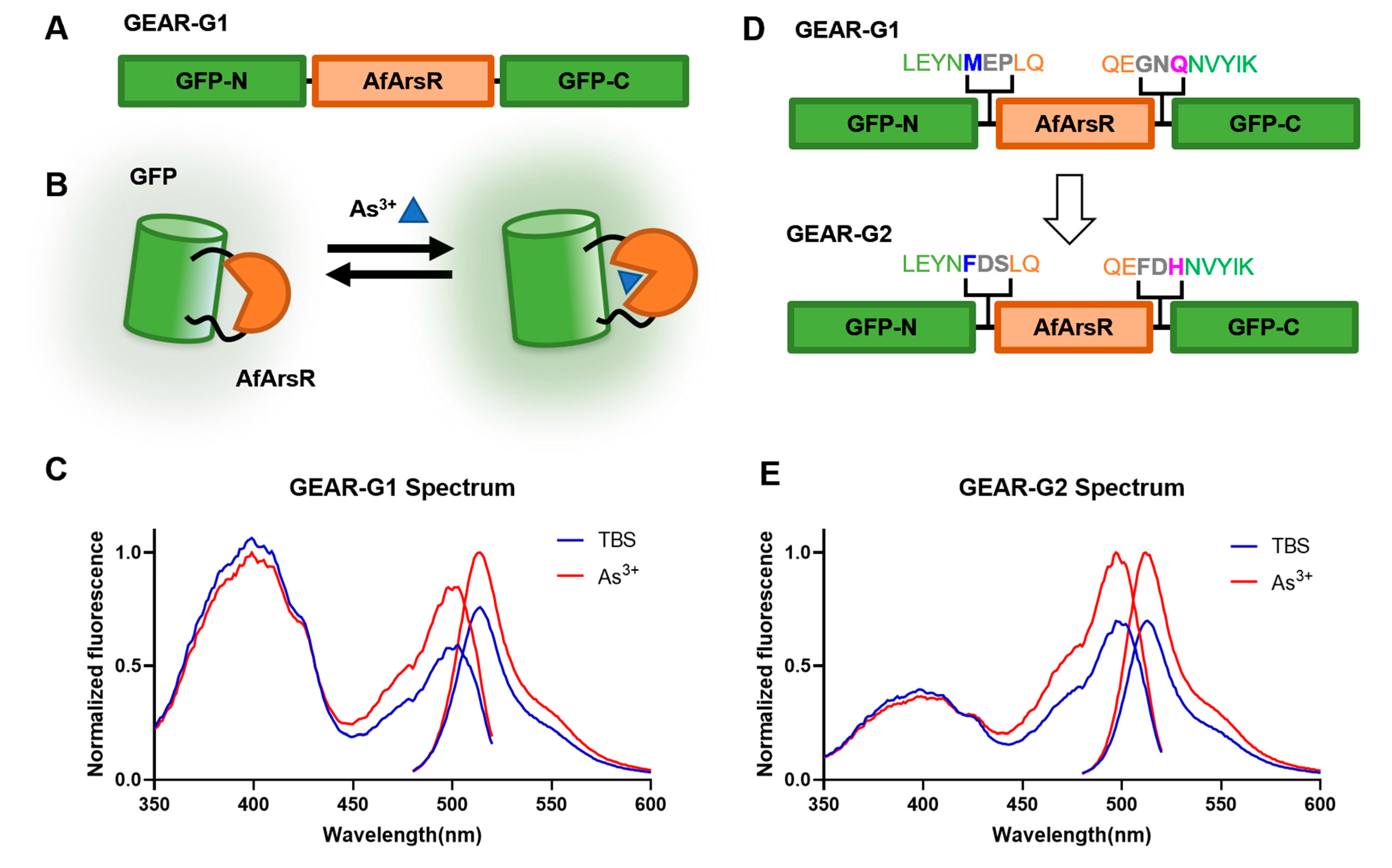
3.2. Development of Genetically Encoded As3+ Biosensors Based on a Single FP
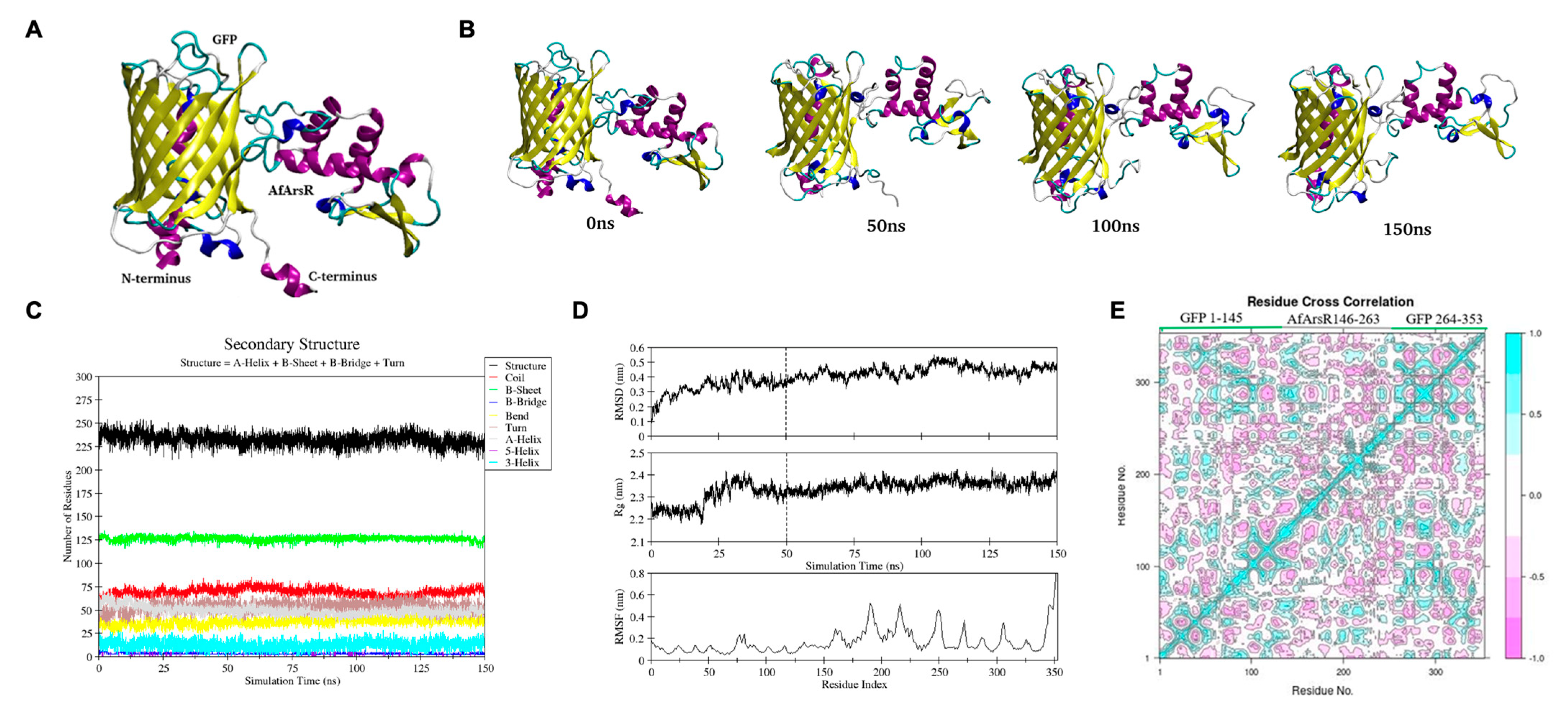
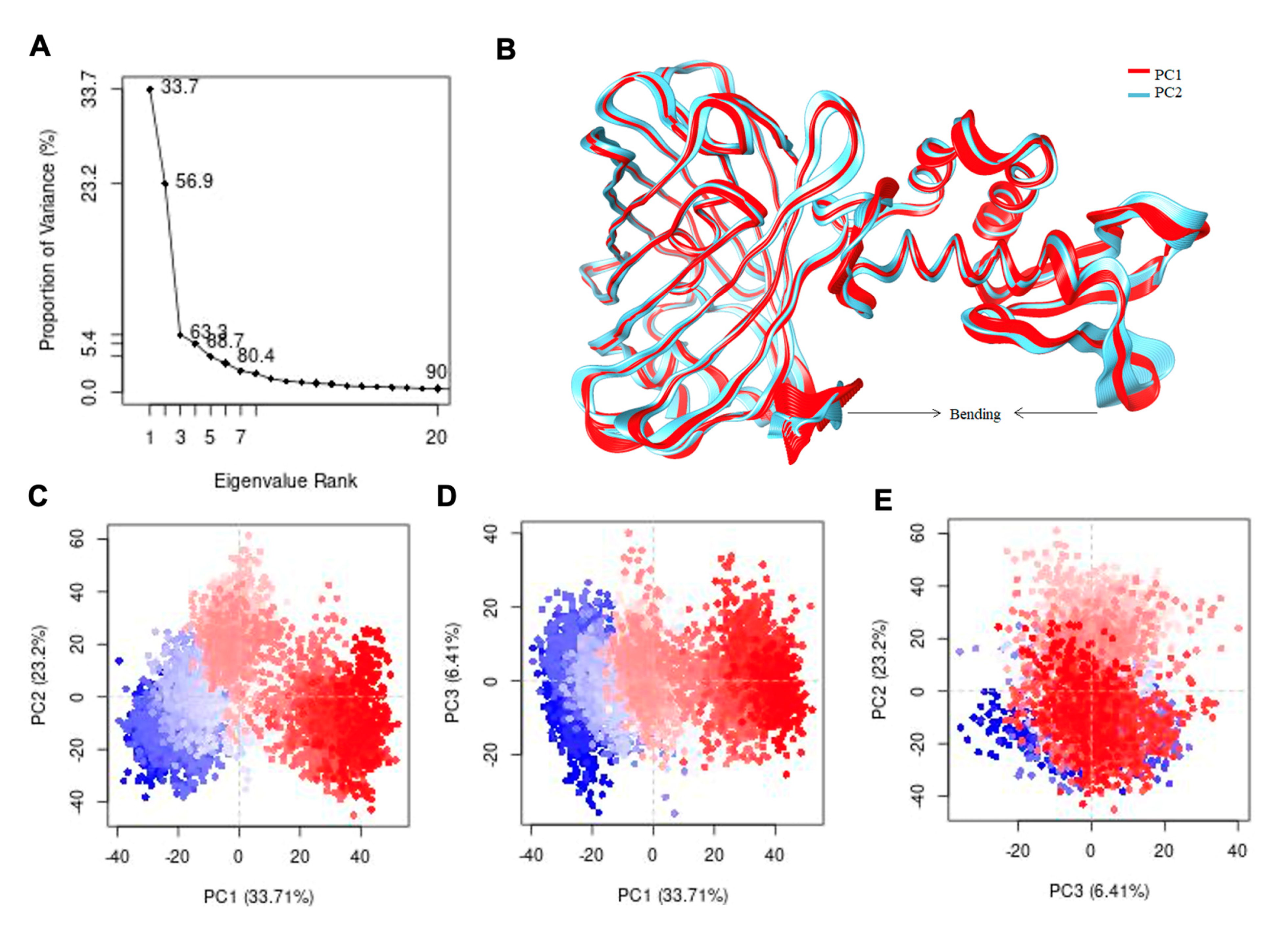
3.3. Computational Structure Prediction of Genetically Encoded As3+ Biosensors
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chung, J.-Y.; Yu, S.-D.; Hong, Y.-S. Environmental Source of Arsenic Exposure. J. Prev. Med. Public Health 2014, 47, 253–257. [Google Scholar] [CrossRef]
- Shahid, M.; Niazi, N.K.; Dumat, C.; Naidu, R.; Khalid, S.; Rahman, M.M.; Bibi, I. A Meta-Analysis of the Distribution, Sources and Health Risks of Arsenic-Contaminated Groundwater in Pakistan. Environ. Pollut. 2018, 242, 307–319. [Google Scholar] [CrossRef]
- Kuo, C.-C.; Moon, K.A.; Wang, S.-L.; Silbergeld, E.; Navas-Acien, A. The Association of Arsenic Metabolism with Cancer, Cardiovascular Disease, and Diabetes: A Systematic Review of the Epidemiological Evidence. Environ. Health Perspect. 2017, 125, 087001. [Google Scholar] [CrossRef]
- Rahman, M.A.; Rahman, A.; Khan, M.Z.K.; Renzaho, A.M.N. Human Health Risks and Socio-Economic Perspectives of Arsenic Exposure in Bangladesh: A Scoping Review. Ecotoxicol. Environ. Saf. 2018, 150, 335–343. [Google Scholar] [CrossRef]
- Di Giovanni, P.; Di Martino, G.; Scampoli, P.; Cedrone, F.; Meo, F.; Lucisano, G.; Romano, F.; Staniscia, T. Arsenic Exposure and Risk of Urothelial Cancer: Systematic Review and Meta-Analysis. Int. J. Environ. Res. Public Health 2020, 17, 3105. [Google Scholar] [CrossRef]
- Yogarajah, N.; Tsai, S.S.H. Detection of Trace Arsenic in Drinking Water: Challenges and Opportunities for Microfluidics. Environ. Sci.: Water Res. Technol. 2015, 1, 426–447. [Google Scholar] [CrossRef]
- Greenwald, E.C.; Mehta, S.; Zhang, J. Genetically Encoded Fluorescent Biosensors Illuminate the Spatiotemporal Regulation of Signaling Networks. Chem. Rev. 2018, 118, 11707–11794. [Google Scholar] [CrossRef]
- Campbell, R.E. Fluorescent-Protein-Based Biosensors: Modulation of Energy Transfer as a Design Principle. Anal. Chem. 2009, 81, 5972–5979. [Google Scholar] [CrossRef] [PubMed]
- Wiens, M.D.; Shen, Y.; Li, X.; Salem, M.A.; Smisdom, N.; Zhang, W.; Brown, A.; Campbell, R.E. A Tandem Green-Red Heterodimeric Fluorescent Protein with High FRET Efficiency. Chembiochem 2016, 17, 2361–2367. [Google Scholar] [CrossRef] [PubMed]
- Frommer, W.B.; Davidson, M.W.; Campbell, R.E. Genetically Encoded Biosensors Based on Engineered Fluorescent Proteins. Chem. Soc. Rev. 2009, 38, 2833–2841. [Google Scholar] [CrossRef] [PubMed]
- Murphy, J.N.; Saltikov, C.W. The ArsR Repressor Mediates Arsenite-Dependent Regulation of Arsenate Respiration and Detoxification Operons of Shewanella Sp. Strain ANA-3. J. Bacteriol. 2009, 191, 6722–6731. [Google Scholar] [CrossRef]
- Chen, J.; Rosen, B.P. Biosensors for Inorganic and Organic Arsenicals. Biosensors 2014, 4, 494–512. [Google Scholar] [CrossRef]
- Saltikov, C.W.; Olson, B.H. Homology of Escherichia Coli R773 arsA, arsB, and arsC Genes in Arsenic-Resistant Bacteria Isolated from Raw Sewage and Arsenic-Enriched Creek Waters. Appl. Environ. Microbiol. 2002, 68, 280–288. [Google Scholar] [CrossRef]
- Soleja, N.; Manzoor, O.; Khan, P.; Mohsin, M. Engineering Genetically Encoded FRET-Based Nanosensors for Real Time Display of Arsenic (As3+) Dynamics in Living Cells. Sci. Rep. 2019, 9, 11240. [Google Scholar] [CrossRef]
- Qin, J.; Fu, H.-L.; Ye, J.; Bencze, K.Z.; Stemmler, T.L.; Rawlings, D.E.; Rosen, B.P. Convergent Evolution of a New Arsenic Binding Site in the ArsR/SmtB Family of Metalloregulators. J. Biol. Chem. 2007, 282, 34346–34355. [Google Scholar] [CrossRef]
- Prabaharan, C.; Kandavelu, P.; Packianathan, C.; Rosen, B.P.; Thiyagarajan, S. Structures of Two ArsR As (III)-Responsive Transcriptional Repressors: Implications for the Mechanism of Derepression. J. Struct. Biol. 2019, 207, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Santha, S.; Pandaranayaka, E.P.J.; Rosen, B.P.; Thiyagarajan, S. Purification, Crystallization and Preliminary X-Ray Diffraction Studies of the Arsenic Repressor ArsR from Corynebacterium Glutamicum. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2011, 67, 1616–1618. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Ovchinnikov, S.; Kim, D.E.; DiMaio, F.; Baker, D. Protein Homology Model Refinement by Large-Scale Energy Optimization. Proc. Natl. Acad. Sci. USA 2018, 115, 3054–3059. [Google Scholar] [CrossRef] [PubMed]
- Lemkul, J.A. From Proteins to Perturbed Hamiltonians: A Suite of Tutorials for the GROMACS-2018 Molecular Simulation Package [Article v1.0]. Living J. Comput. Mol. Sci. 2019, 1, 5068. [Google Scholar] [CrossRef]
- Braun, E.; Moosavi, S.M.; Smit, B. Anomalous Effects of Velocity Rescaling Algorithms: The Flying Ice Cube Effect Revisited. J. Chem. Theory Comput. 2018, 14, 5262–5272. [Google Scholar] [CrossRef]
- Okumura, H.; Itoh, S.G.; Okamoto, Y. Explicit Symplectic Integrators of Molecular Dynamics Algorithms for Rigid-Body Molecules in the Canonical, Isobaric-Isothermal, and Related Ensembles. J. Chem. Phys. 2007, 126, 084103. [Google Scholar] [CrossRef]
- Kuznetsov, A.; Jarv, J. Mapping the ACE2 Binding Site on the SARS-CoV-2 Spike Protein S1: Molecular Recognition Pattern. Proc. Eston. Acad. Sci. 2020, 69, 228. [Google Scholar]
- Markwardt, M.L.; Kremers, G.-J.; Kraft, C.A.; Ray, K.; Cranfill, P.J.C.; Wilson, K.A.; Day, R.N.; Wachter, R.M.; Davidson, M.W.; Rizzo, M.A. An Improved Cerulean Fluorescent Protein with Enhanced Brightness and Reduced Reversible Photoswitching. PLoS ONE 2011, 6, e17896. [Google Scholar] [CrossRef] [PubMed]
- Nagai, T.; Yamada, S.; Tominaga, T.; Ichikawa, M.; Miyawaki, A. Expanded Dynamic Range of Fluorescent Indicators for Ca2+ by Circularly Permuted Yellow Fluorescent Proteins. Proc. Natl. Acad. Sci. USA 2004, 101, 10554–10559. [Google Scholar] [CrossRef] [PubMed]
- Thestrup, T.; Litzlbauer, J.; Bartholomaus, I.; Mues, M.; Russo, L.; Dana, H.; Kovalchuk, Y.; Liang, Y.; Kalamakis, G.; Laukat, Y.; et al. Optimized Ratiometric Calcium Sensors for Functional in Vivo Imaging of Neurons and T Lymphocytes. Nat. Methods 2014, 11, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Wu, S.-Y.; Rancic, V.; Aggarwal, A.; Qian, Y.; Miyashita, S.-I.; Ballanyi, K.; Campbell, R.E.; Dong, M. Genetically Encoded Fluorescent Indicators for Imaging Intracellular Potassium Ion Concentration. Commun. Biol. 2019, 2, 18. [Google Scholar] [CrossRef]
- Ai, H.W.; Olenych, S.G.; Wong, P.; Davidson, M.W.; Campbell, R.E. Hue-Shifted Monomeric Variants of Clavularia Cyan Fluorescent Protein: Identification of the Molecular Determinants of Color and Applications in Fluorescence Imaging. BMC Biol. 2008, 6, 13. [Google Scholar] [CrossRef]
- Griesbeck, O.; Baird, G.S.; Campbell, R.E.; Zacharias, D.A.; Tsien, R.Y. Reducing the Environmental Sensitivity of Yellow Fluorescent Protein. Mechanism and Applications. J. Biol. Chem. 2001, 276, 29188–29194. [Google Scholar] [CrossRef]
- Zhao, Y.; Shen, Y.; Wen, Y.; Campbell, R.E. High-Performance Intensiometric Direct- and Inverse-Response Genetically Encoded Biosensors for Citrate. ACS Cent. Sci. 2020, 6, 1441–1450. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Abdelfattah, A.S.; Zhou, H.; Ruangkittisakul, A.; Qian, Y.; Ballanyi, K.; Campbell, R.E. Genetically Encoded Glutamate Indicators with Altered Color and Topology. ACS Chem. Biol. 2018, 13, 1832–1837. [Google Scholar] [CrossRef]
- Zarowny, L.; Aggarwal, A.; Rutten, V.M.S.; Kolb, I.; GENIE Project; Patel, R.; Huang, H.-Y.; Chang, Y.-F.; Phan, T.; Kanyo, R.; et al. Bright and High-Performance Genetically Encoded Ca2+ Indicator Based on mNeonGreen Fluorescent Protein. ACS Sens 2020, 5, 1959–1968. [Google Scholar] [CrossRef]
- Nasu, Y.; Shen, Y.; Kramer, L.; Campbell, R.E. Structure- and Mechanism-Guided Design of Single Fluorescent Protein-Based Biosensors. Nat. Chem. Biol. 2021, 17, 509–518. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 Web Portal for Protein Modeling, Prediction and Analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Y. I-TASSER Server: New Development for Protein Structure and Function Predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef]
- Song, Y.; DiMaio, F.; Wang, R.Y.-R.; Kim, D.; Miles, C.; Brunette, T.; Thompson, J.; Baker, D. High-Resolution Comparative Modeling with RosettaCM. Structure 2013, 21, 1735–1742. [Google Scholar] [CrossRef]
- Klose, D.P.; Wallace, B.A.; Janes, R.W. 2Struc: The Secondary Structure Server. Bioinformatics 2010, 26, 2624–2625. [Google Scholar] [CrossRef]
- Spellmon, N.; Sun, X.; Sirinupong, N.; Edwards, B.; Li, C.; Yang, Z. Molecular Dynamics Simulation Reveals Correlated Inter-Lobe Motion in Protein Lysine Methyltransferase SMYD2. PLoS ONE 2015, 10, e0145758. [Google Scholar] [CrossRef]
- Lin, M.Z.; Schnitzer, M.J. Genetically Encoded Indicators of Neuronal Activity. Nat. Neurosci. 2016, 19, 1142–1153. [Google Scholar] [CrossRef]
- Shen, Y.; Nasu, Y.; Shkolnikov, I.; Kim, A.; Campbell, R.E. Engineering Genetically Encoded Fluorescent Indicators for Imaging of Neuronal Activity: Progress and Prospects. Neurosci. Res. 2020, 152, 3–14. [Google Scholar] [CrossRef]
- Shen, Y.; Rosendale, M.; Campbell, R.E.; Perrais, D. pHuji, a pH-Sensitive Red Fluorescent Protein for Imaging of Exo− and Endocytosis. J. Cell Biol. 2014, 207, 419–432. [Google Scholar] [CrossRef]
- Rizza, A.; Walia, A.; Lanquar, V.; Frommer, W.B.; Jones, A.M. In Vivo Gibberellin Gradients Visualized in Rapidly Elongating Tissues. Nat. Plants 2017, 3, 803–813. [Google Scholar] [CrossRef]
- Lobas, M.A.; Tao, R.; Nagai, J.; Kronschläger, M.T.; Borden, P.M.; Marvin, J.S.; Looger, L.L.; Khakh, B.S. A Genetically Encoded Single-Wavelength Sensor for Imaging Cytosolic and Cell Surface ATP. Nat. Commun. 2019, 10, 711. [Google Scholar] [CrossRef]
- Zhang, J.-F.; Liu, B.; Hong, I.; Mo, A.; Roth, R.H.; Tenner, B.; Lin, W.; Zhang, J.Z.; Molina, R.S.; Drobizhev, M.; et al. An Ultrasensitive Biosensor for High-Resolution Kinase Activity Imaging in Awake Mice. Nat. Chem. Biol. 2021, 17, 39–46. [Google Scholar] [CrossRef]
- Busenlehner, L.S.; Pennella, M.A.; Giedroc, D.P. The SmtB/ArsR Family of Metalloregulatory Transcriptional Repressors: Structural Insights into Prokaryotic Metal Resistance. FEMS Microbiol. Rev. 2003, 27, 131–143. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, S.S.; Shen, Y.; Fatmi, M.Q.; Campbell, R.E.; Bokhari, H. Design and Prototyping of Genetically Encoded Arsenic Biosensors Based on Transcriptional Regulator AfArsR. Biomolecules 2021, 11, 1276. https://doi.org/10.3390/biom11091276
Khan SS, Shen Y, Fatmi MQ, Campbell RE, Bokhari H. Design and Prototyping of Genetically Encoded Arsenic Biosensors Based on Transcriptional Regulator AfArsR. Biomolecules. 2021; 11(9):1276. https://doi.org/10.3390/biom11091276
Chicago/Turabian StyleKhan, Salma Saeed, Yi Shen, Muhammad Qaiser Fatmi, Robert E. Campbell, and Habib Bokhari. 2021. "Design and Prototyping of Genetically Encoded Arsenic Biosensors Based on Transcriptional Regulator AfArsR" Biomolecules 11, no. 9: 1276. https://doi.org/10.3390/biom11091276
APA StyleKhan, S. S., Shen, Y., Fatmi, M. Q., Campbell, R. E., & Bokhari, H. (2021). Design and Prototyping of Genetically Encoded Arsenic Biosensors Based on Transcriptional Regulator AfArsR. Biomolecules, 11(9), 1276. https://doi.org/10.3390/biom11091276





