Identification of Reference Genes for Circadian Studies on Brain Microvessels and Choroid Plexus Samples Isolated from Rats
Abstract
:1. Introduction
2. Materials and Methods
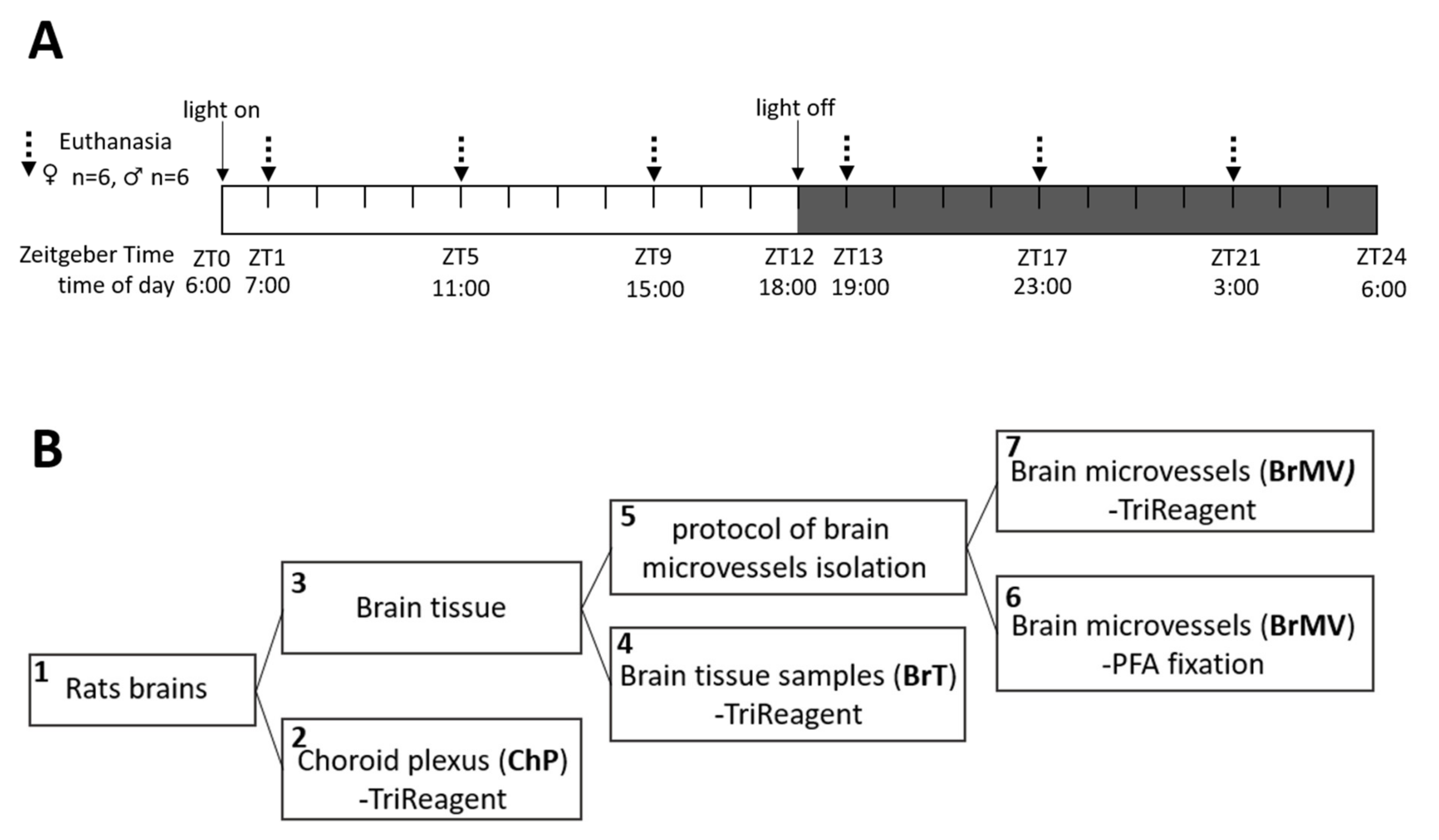
2.1. Animals and Tissue Collection
2.2. RNA Extraction, cDNA Preparation, and qPCR Analysis
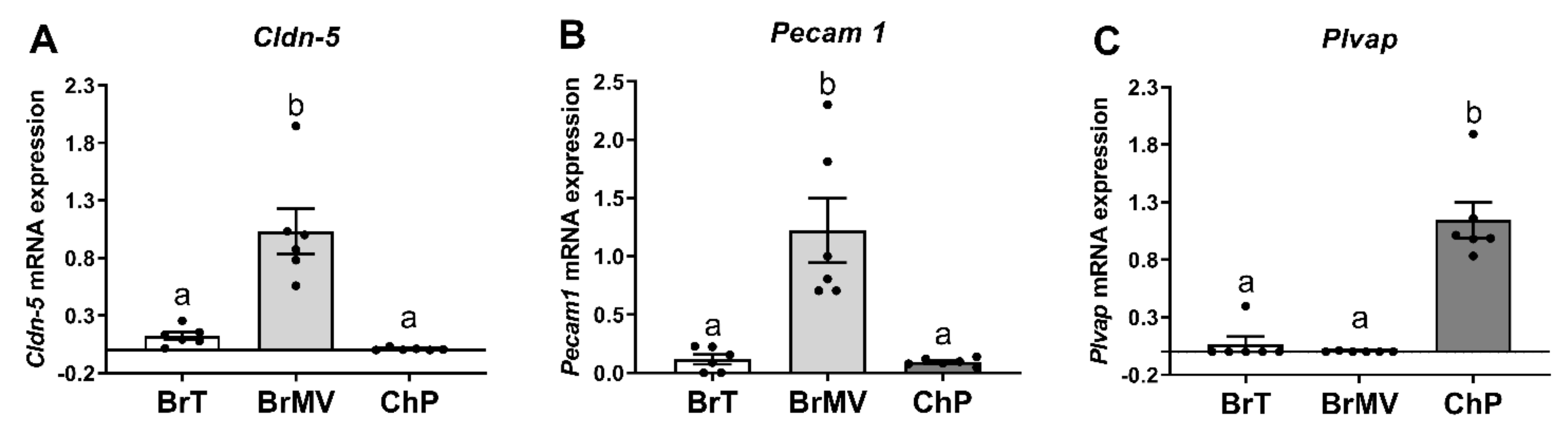
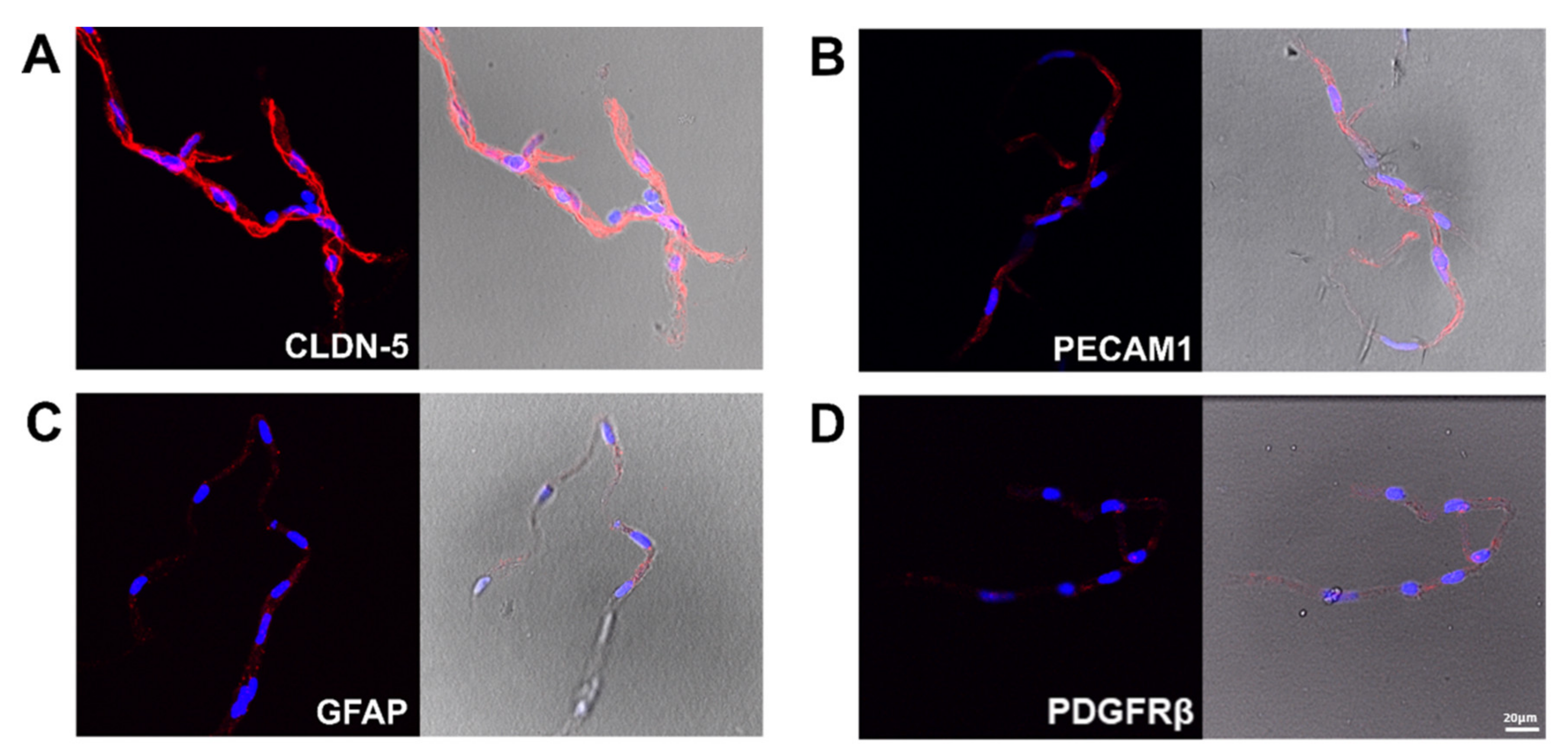
2.3. Verification of the Tissue Isolation Process
2.4. Identification and qPCR Verification of Optimal Reference Genes
| Gene | (Forward/Reverse) Sequence 5′→3′ | Amplicon Size (bp) | References/Sources | ||
|---|---|---|---|---|---|
| Reference genes | Actb | F: TGTCACCAACTGGGACGATA | R: GGGGTGTTGAAGGTCTCAAA | 165 | [26] |
| Apoe | F: CCTGAACCGCTTCTGGGATT | R: GCTCTTCCTGGACCTGGTCA | 65 | [27] | |
| Hmbs | F: TCCTGGCTTTACCATTGGAG | R: TGAATTCCAGGTGAGGGAAC | 176 | [26] | |
| Hprt1 | F: CCTGTTGATGTGGCCAGTAAAGA | R: ATCAAAAGGGACGCAGCAAC | 137 | [28] | |
| Pgk1 | F: ATGCAAAGACTGGCCAAGCTAC | R: AGCCACAGCCTCAGCATATTTC | 104 | [29] | |
| Ppia | F: TATCTGCACTGCCAAGACTGAGTG | R: CTTCTTGCTGGTCTTGCCATTCC | 126 | [29] | |
| Rplp2 | F: ATTGAGGATGTCATCGCTCAGG | R: TCTTTCTTCTCCTCTGCTGCAG | 137 | [28] | |
| Rps18 | F: ACGGACCAGAGCGAAAGCAT | R: TGTCAATCCTGTCCGTGTCC | 310 | [26] | |
| Tbp | F: TAATCCCAAGCGGTTTGCTG | R: TTCTTCACTCTTGGCTCCTGTG | 111 | [30] | |
| Ywhaz | F: TTCGCAGCCAGAAAGCAAAG | R: TTGTCATCACCAGCAGCAAC | 87 | [28] | |
| Marker genes | Plvap | F: ATAATCGGTTCATCGCCGCT | R: GCTTGAAGAGCAAGGCTTCG | 96 | NM_020086.1 |
| Cldn-5 | F: CTACAGGCTCTTGTGAGGACTTGAC | R: AGTAGGAACTGTTAGCGGCAGTTTG | 121 | [31] | |
| Pecam1 | F: GCCTCACCAAGAGAACGGAA | R: AATTGGATGGCTTGGCCTGA | 191 | NM_031591.1 | |
| Bmal1 | F: TGGACTGCAACCGCAAGAG | R: CCTTCCATGAGGGTCATCTTTG | 137 | [28] | |
2.5. Digital PCR
2.6. Statistical Analysis
3. Results
3.1. Characterization of the Isolated BrMV Fractions and ChP
3.2. Selection of Reference Genes and Efficiency of Primers
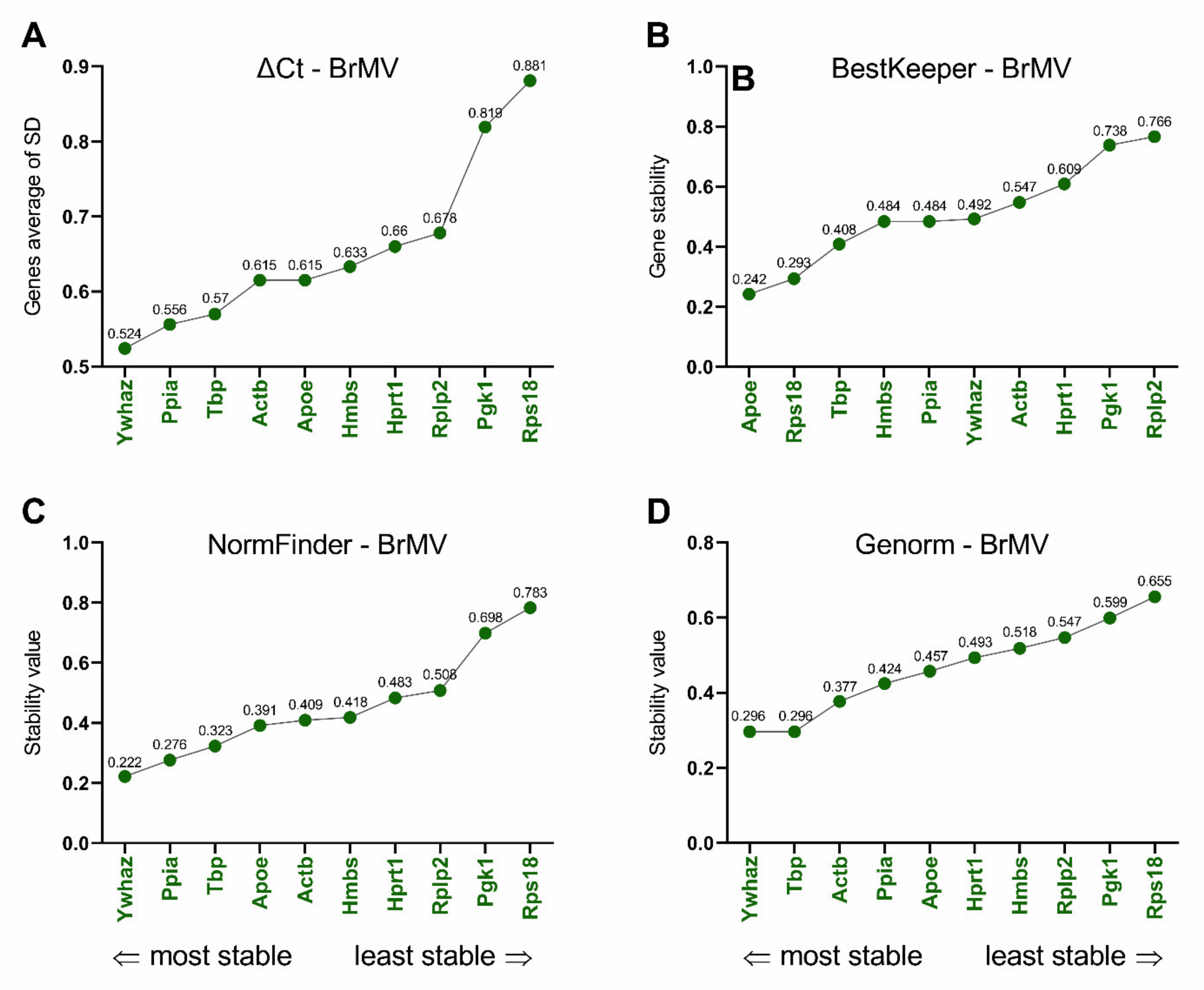
3.3. Selection and Verification of the Most Stable Reference Genes for Brain Microvessels
3.4. Selection and Verification of the Most Stable Reference Genes for Choroid Plexus
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abbott, N.J.; Patabendige, A.A.; Dolman, D.E.; Yusof, S.R.; Begley, D.J. Structure and function of the blood-brain barrier. Neurobiol. Dis. 2010, 37, 13–25. [Google Scholar] [CrossRef]
- Tietz, S.; Engelhardt, B. Brain barriers: Crosstalk between complex tight junctions and adherens junctions. J. Cell Biol. 2015, 209, 493–506. [Google Scholar] [CrossRef] [Green Version]
- Johanson, C.E.; Johanson, N.L. Choroid Plexus Blood-CSF Barrier: Major Player in Brain Disease Modeling and Neuromedicine. J. Neurol. Neuromed. 2018, 3, 39–58. [Google Scholar] [CrossRef]
- Dong, X. Current Strategies for Brain Drug Delivery. Theranostics 2018, 8, 1481–1493. [Google Scholar] [CrossRef]
- Santa-Maria, A.R.; Heymans, M.; Walter, F.R.; Culot, M.; Gosselet, F.; Deli, M.A.; Neuhaus, W. Transport Studies Using Blood-Brain Barrier In Vitro Models: A Critical Review and Guidelines. Handb. Exp. Pharm. 2020. [Google Scholar] [CrossRef]
- Pan, W.; Kastin, A.J. The Blood-Brain Barrier: Regulatory Roles in Wakefulness and Sleep. Neuroscientist 2017, 23, 124–136. [Google Scholar] [CrossRef] [PubMed]
- Myung, J.; Schmal, C.; Hong, S.; Tsukizawa, Y.; Rose, P.; Zhang, Y.; Holtzman, M.J.; De Schutter, E.; Herzel, H.; Bordyugov, G.; et al. The choroid plexus is an important circadian clock component. Nat. Commun. 2018, 9, 1062. [Google Scholar] [CrossRef]
- Guilding, C.; Piggins, H.D. Challenging the omnipotence of the suprachiasmatic timekeeper: Are circadian oscillators present throughout the mammalian brain? Eur. J. Neurosci. 2007, 25, 3195–3216. [Google Scholar] [CrossRef] [PubMed]
- Kondratova, A.A.; Kondratov, R.V. The circadian clock and pathology of the ageing brain. Nat. Rev. Neurosci. 2012, 13, 325–335. [Google Scholar] [CrossRef]
- Quintela, T.; Sousa, C.; Patriarca, F.M.; Gonçalves, I.; Santos, C.R. Gender associated circadian oscillations of the clock genes in rat choroid plexus. Brain Struct. Funct. 2015, 220, 1251–1262. [Google Scholar] [CrossRef]
- Quintela, T.; Furtado, A.; Duarte, A.C.; Gonçalves, I.; Myung, J.; Santos, C.R.A. The Role of Circadian Rhythm in Choroid Plexus Functions. Prog. Neurobiol. 2021, 31, 102129. [Google Scholar] [CrossRef] [PubMed]
- Carver, K.A.; Lourim, D.; Tryba, A.K.; Harder, D.R. Rhythmic expression of cytochrome P450 epoxygenases CYP4x1 and CYP2c11 in the rat brain and vasculature. Am. J. Physiol. Cell Physiol. 2014, 307, C989–C998. [Google Scholar] [CrossRef] [Green Version]
- Nakazato, R.; Kawabe, K.; Yamada, D.; Ikeno, S.; Mieda, M.; Shimba, S.; Hinoi, E.; Yoneda, Y.; Takarada, T. Disruption of Bmal1 Impairs Blood-Brain Barrier Integrity via Pericyte Dysfunction. J. Neurosci. 2017, 37, 10052–10062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.L.; Yue, Z.; Arnold, D.M.; Artiushin, G.; Sehgal, A. A Circadian Clock in the Blood-Brain Barrier Regulates Xenobiotic Efflux. Cell 2018, 173, 130–139.e10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deepak, S.; Kottapalli, K.; Rakwal, R.; Oros, G.; Rangappa, K.; Iwahashi, H.; Masuo, Y.; Agrawal, G. Real-Time PCR: Revolutionizing Detection and Expression Analysis of Genes. Curr. Genom. 2007, 8, 234–251. [Google Scholar] [CrossRef]
- Marcondes, F.K.; Bianchi, F.J.; Tanno, A.P. Determination of the estrous cycle phases of rats: Some helpful considerations. Braz. J. Biol. 2002, 62, 609–614. [Google Scholar] [CrossRef] [Green Version]
- Bowyer, J.F.; Thomas, M.; Patterson, T.A.; George, N.I.; Runnells, J.A.; Levi, M.S. A visual description of the dissection of the cerebral surface vasculature and associated meninges and the choroid plexus from rat brain. J. Vis. Exp. 2012, 69, e4285. [Google Scholar] [CrossRef]
- Yousif, S.; Marie-Claire, C.; Roux, F.; Scherrmann, J.M.; Declèves, X. Expression of drug transporters at the blood-brain barrier using an optimized isolated rat brain microvessel strategy. Brain Res. 2007, 1134, 1–11. [Google Scholar] [CrossRef]
- Chomczynski, P.; Sacchi, N. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: Twenty-something years on. Nat. Protoc. 2006, 1, 581–585. [Google Scholar] [CrossRef]
- Zhao, S.; Fernald, R.D. Comprehensive algorithm for quantitative real-time polymerase chain reaction. J. Comput. Biol. 2005, 12, 1047–1064. [Google Scholar] [CrossRef]
- Dai, M.; Lin, Y.; El-Amouri, S.S.; Kohls, M.; Pan, D. Comprehensive evaluation of blood-brain barrier-forming micro-vasculatures: Reference and marker genes with cellular composition. PLoS ONE 2018, 13, e0197379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, RESEARCH0034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersen, C.L.; Jensen, J.L.; Orntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [Green Version]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonefeld, B.E.; Elfving, B.; Wegener, G. Reference genes for normalization: A study of rat brain tissue. Synapse 2008, 62, 302–309. [Google Scholar] [CrossRef]
- Vanmierlo, T.; Rutten, K.; Dederen, J.; Bloks, V.W.; van Vark-van der Zee, L.C.; Kuipers, F.; Kiliaan, A.; Blokland, A.; Sijbrands, E.J.; Steinbusch, H.; et al. Liver X receptor activation restores memory in aged AD mice without reducing amyloid. Neurobiol. Aging 2011, 32, 1262–1272. [Google Scholar] [CrossRef]
- Takizawa, N.; Tanaka, S.; Oe, S.; Koike, T.; Matsuda, T.; Yamada, H. Hypothalamo-hypophysial system in rats with autotransplantation of the adrenal cortex. Mol. Med. Rep. 2017, 15, 3215–3221. [Google Scholar] [CrossRef]
- Langnaese, K.; John, R.; Schweizer, H.; Ebmeyer, U.; Keilhoff, G. Selection of reference genes for quantitative real-time PCR in a rat asphyxial cardiac arrest model. Bmc Mol. Biol. 2008, 9, 53. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Zhao, H.; Ni, J.; Pan, J.; Hua, H.; Wang, Y. Identification of suitable reference genes for gene expression studies in rat skeletal muscle following sciatic nerve crush injury. Mol. Med. Rep. 2019, 19, 4377–4387. [Google Scholar] [CrossRef]
- Burkhart, A.; Thomsen, L.B.; Thomsen, M.S.; Lichota, J.; Fazakas, C.; Krizbai, I.; Moos, T. Transfection of brain capillary endothelial cells in primary culture with defined blood-brain barrier properties. Fluids Barriers CNS 2015, 12, 19. [Google Scholar] [CrossRef] [Green Version]
- Christiansen, S.L.; Bouzinova, E.V.; Fahrenkrug, J.; Wiborg, O. Altered Expression Pattern of Clock Genes in a Rat Model of Depression. Int. J. Neuropsychopharmacol. 2016, 19, pyw061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nestorov, J.; Matic, G.; Elakovic, I.; Tanic, N. Gene expression studies: How to obtain accurate and reliable data by quantitative real-time RT PCR. J. Med. Biochem. 2013, 32, 325–338. [Google Scholar] [CrossRef] [Green Version]
- Bustin, S.A.; Benes, V.; Garson, J.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.; et al. The need for transparency and good practices in the qPCR literature. Nat. Methods 2013, 10, 1063–1067. [Google Scholar] [CrossRef] [PubMed]
- Szczepkowska, A.; Skowroński, M.T.; Kowalewska, M.; Milewski, S.; Skipor, J. Effect of Melatonin from Slow-Release Implants on Aquaporins (AQP1 and AQP4) in the Ovine Choroid Plexus. Czech. J. Anim. Sci. 2018, 63, 32–42. [Google Scholar] [CrossRef] [Green Version]
- Szczepkowska, A.; Kowalewska, M.; Skipor, J. Melatonin from slow-release implants upregulates claudin-2 in the ovine choroid plexus. J. Physiol. Pharm. 2019, 70, 249–254. [Google Scholar]
- Kowalewska, M.; Herman, A.P.; Szczepkowska, A.; Skipor, J. The effect of melatonin from slow-release implants on basic and TLR-4-mediated gene expression of inflammatory cytokines and their receptors in the choroid plexus in ewes. Res. Vet. Sci. 2017, 113, 50–55. [Google Scholar] [CrossRef]
- Kowalewska, M.; Szczepkowska, A.; Herman, A.P.; Pellicer-Rubio, M.T.; Jałyński, M.; Skipor, J. Melatonin from slow-release implants did not influence the gene expression of the lipopolysaccharide receptor complex in the choroid plexus of seasonally anoestrous adult ewes subjected or not to a systemic inflammatory stimulus. Small Rumin Res. 2017b, 147, 1–7. [Google Scholar] [CrossRef]
- Lisowski, P.; Pierzchala, M.; Joanna Gooecik, J.; Pareek, C.S.; Zwierzchowski, L. Evaluation of reference genes for studies of gene expression in the bovine liver, kidney, pituitary, and thyroid. J. Appl. Genet. 2008, 49, 367–372. [Google Scholar] [CrossRef]
- Vorachek, W.R.; Hugejiletu; Bobe, G.; Hall, J.A. Reference gene selection for quantitative PCR studies in sheep neutrophils. Int. J. Mol. Sci. 2013, 14, 11484–11495. [Google Scholar] [CrossRef] [Green Version]
- Danesh Mesgaran, S.; Sharbati, J.; Einspanier, R.; Gabler, C. mRNA expression pattern of selected candidate genes differs in bovine oviductal epithelial cells in vitro compared with the in vivo state and during cell culture passages. Reprod. Biol. Endocrinol. 2016, 14, 44. [Google Scholar] [CrossRef] [Green Version]
- Chapman, J.R.; Waldenstrom, J. With Reference to Reference Genes: A Systematic Review of Endogenous Controls in Gene Expression Studies. PLoS ONE 2015, 10, e0141853. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.J.; Zheng, D.; Balasubramanian, S.; Carriero, N.; Khurana, E.; Robilotto, R.; Gerstein, M.B. Comprehensive analysis of the pseudogenes of glycolytic enzymes in vertebrates: The anomalously high number of GAPDH pseudogenes highlights a recent burst of retrotrans-positional activity. Bmc Genom. 2009, 10, 480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kozera, B.; Rapacz, M. Reference genes in real-time PCR. J. Appl Genet. 2013, 54, 391–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.L.; Lahens, N.F.; Yue, Z.; Arnold, D.M.; Pakstis, P.P.; Schwarz, J.E.; Sehgal, A. A circadian clock regulates efflux by the blood-brain barrier in mice and human cells. Nat. Commun. 2021, 12, 617. [Google Scholar] [CrossRef]
- Müller, A.M.; Hermanns, M.I.; Skrzynski, C.; Nesslinger, M.; Müller, K.M.; Kirkpatrick, C.J. Expression of the endothelial markers PECAM-1, vWf, and CD34 in vivo and in vitro. Exp. Mol. Pathol. 2002, 72, 221–229. [Google Scholar] [CrossRef]
- Greene, C.; Hanley, N.; Campbell, M. Claudin-5: Gatekeeper of neurological function. Fluids Barriers CNS 2019, 16, 3. [Google Scholar] [CrossRef] [Green Version]
- Marques, F.; Sousa, J.C.; Coppola, G.; Gao, F.; Puga, R.; Brentani, H.; Geschwind, D.H.; Sousa, N.; Correia-Neves, M.; Palha, J.A. Transcriptome signature of the adult mouse choroid plexus. Fluids Barriers CNS 2011, 8, 10. [Google Scholar] [CrossRef] [Green Version]
- Pardridge, W.M. The Isolated Brain Microvessel: A Versatile Experimental Model of the Blood-Brain Barrier. Front. Physiol. 2020, 11, 398. [Google Scholar] [CrossRef]
- Zhang, Z.; Ma, Z.; Zou, W.; Guo, H.; Liu, M.; Ma, Y.; Zhang, L. The Appropriate Marker for Astrocytes: Comparing the Distribution and Expression of Three Astrocytic Markers in Different Mouse Cerebral Regions. Biomed. Res. Int. 2019, 2019, 9605265. [Google Scholar] [CrossRef]
- Winkler, E.A.; Bell, R.D.; Zlokovic, B.V. Pericyte-specific expression of PDGF beta receptor in mouse models with normal and deficient PDGF beta receptor signaling. Mol. Neurodegener 2010, 5, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quan, P.L.; Sauzade, M.; Brouzes, E. dPCR: A technology review. Sensors 2018, 18, 1271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakao, R.; Okauchi, H.; Hashimoto, C.; Wada, N.; Oishi, K. Determination of reference genes that are independent of feeding rhythms for circadian studies of mouse metabolic tissues. Mol. Genet. Metab. 2017, 121, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Hellemans, J.; Mortier, G.; De Paepe, A.; Speleman, F.; Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007, 8, R19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huggett, J.F.; Cowen, S.; Foy, C.A. Considerations for digital PCR as an accurate molecular diagnostic tool. Clin. Chem. 2015, 61, 79–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pomari, E.; Piubelli, C.; Perandin, F.; Bisoffi, Z. Digital PCR: A new technology for diagnosis of parasitic infections. Clin. Microbiol. Infect. 2019, 25, 1510–1516. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Xu, X.; Li, D.; Chen, K.; Zhan, Q.; Ge, M.; Zhou, X.; Liang, X.; Guan, M. Digital PCR-Based Detection of EGFR Mutations in Paired Plasma and CSF Samples of Lung Adenocarcinoma Patients with Central Nervous System Metastases. Target. Oncol. 2019, 14, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Schroder, A.L.; Pelch, K.E.; Nagel, S.C. Estrogen modulates expression of putative housekeeping genes in the mouse uterus. Endocrine 2009, 35, 211–219. [Google Scholar] [CrossRef]
- Nilsson, M.E.; Vandenput, L.; Tivesten, Å.; Norlén, A.K.; Lagerquist, M.K.; Windahl, S.H.; Börjesson, A.E.; Farman, H.H.; Poutanen, M.; Benrick, A.; et al. Measurement of a Comprehensive Sex Steroid Profile in Rodent Serum by High-Sensitive Gas Chromatography-Tandem Mass Spectrometry. Endocrinology 2015, 156, 2492–2502. [Google Scholar] [CrossRef] [Green Version]
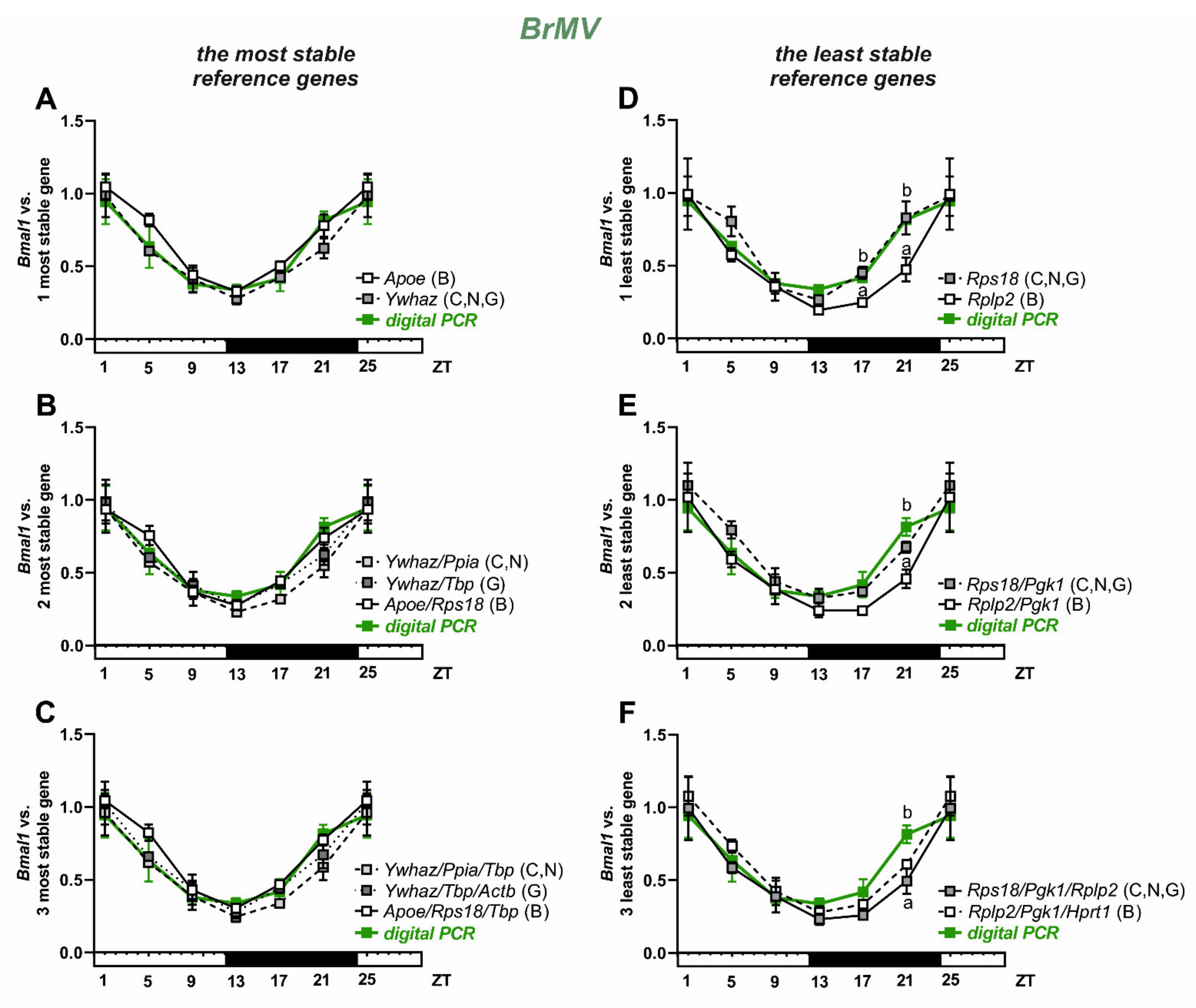
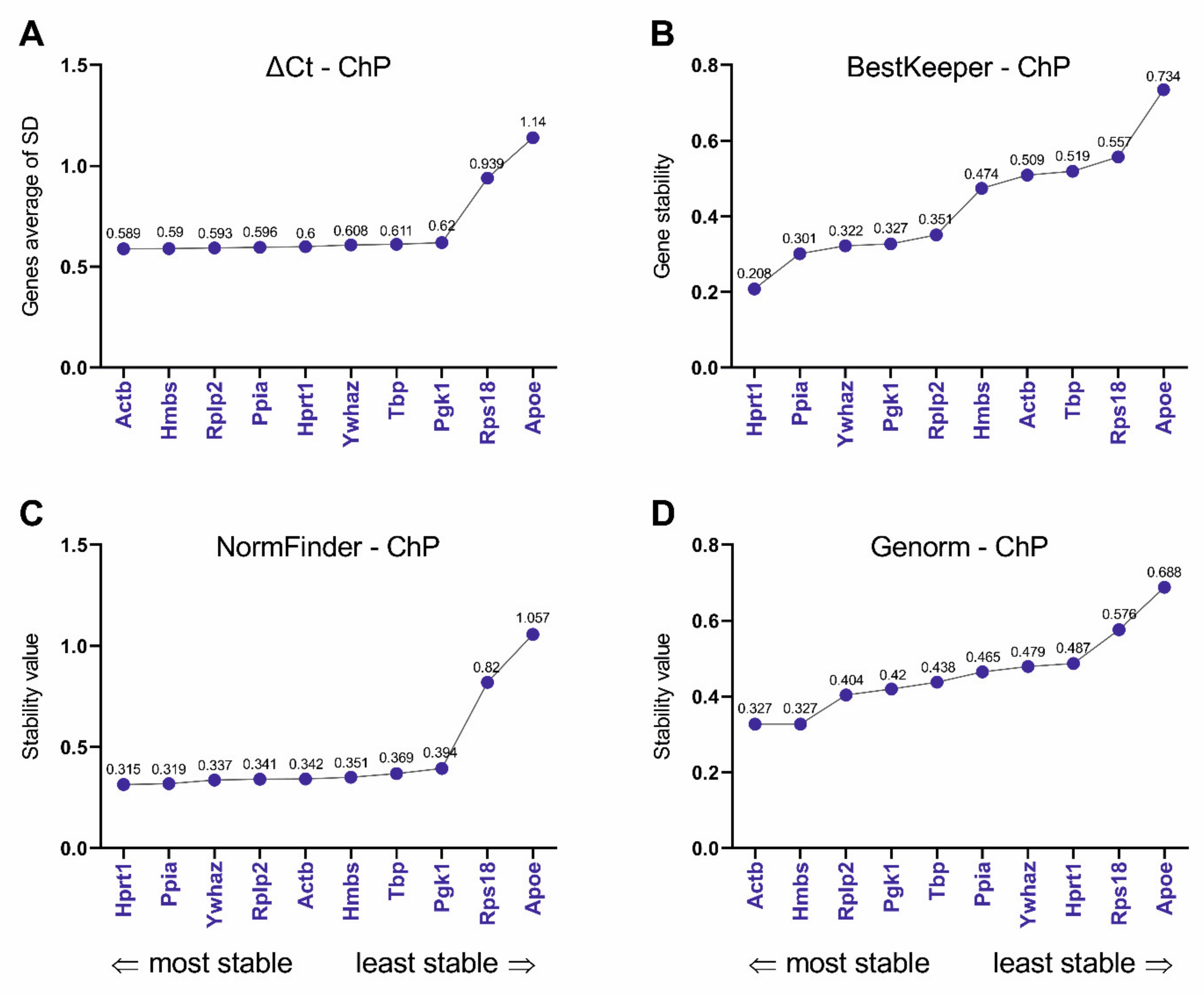
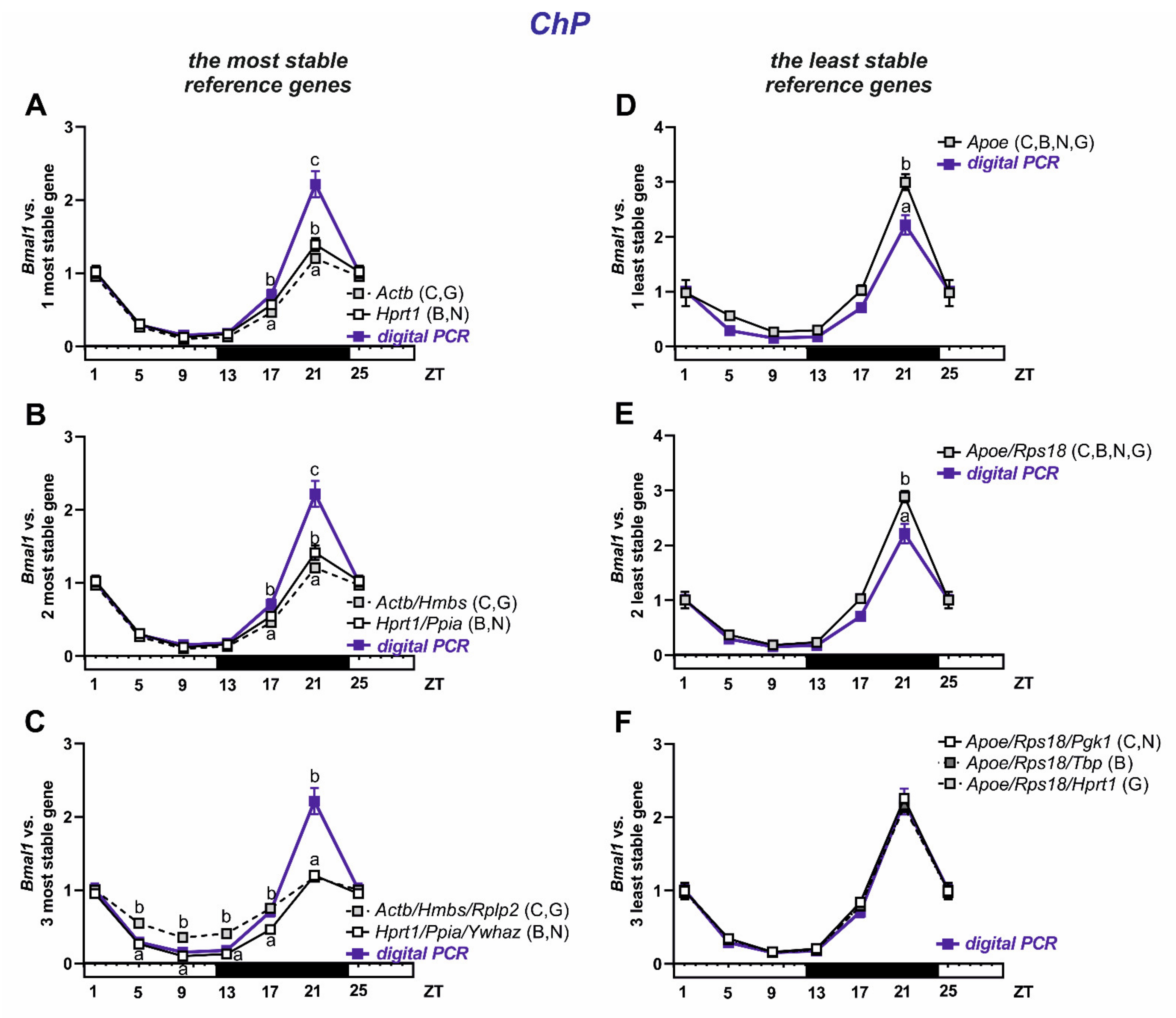
| Gene Symbol | Full Name | Function | Primers Efficiency | BrMV Cq Mean ± SEM | BrMV Cq Range | ChP Cq Mean ± SEM | ChP Cq Range |
|---|---|---|---|---|---|---|---|
| Actb | Beta actin | cytoskeletal structure protein | 85% | 19.56 ± 0.13 | 3.25 | 18.93 ± 0.08 | 2.12 |
| Apoe | Apolipoprotein E | transport of lipids, fat soluble vitamins, and cholesterol | 85% | 19.58 ± 0.08 | 1.95 | 22.77 ± 0.2 | 5.06 |
| Hmbs | Hydroxymethylbilane synthase | the third enzyme in the heme biosynthetic pathway | 85% | 26.5 ± 0.13 | 3.16 | 23.92 ± 0.09 | 2.36 |
| Hprt1 | Hypoxanthine-guanine phosphoribosyltransferase | recycle purines in cells | 86% | 23.43 ± 0.14 | 3.74 | 20.66 ± 0.07 | 2.20 |
| Pgk1 | Phosphoglycerate kinase 1 | enzyme involved in glycolysis process | 86% | 22.42 ± 0.13 | 3.02 | 19.63 ± 0.1 | 3.08 |
| Ppia | Peptidylprolyl isomerase A | catalyse the cis–trans isomerisation of peptide bonds N-terminal to proline residues in polypeptide chains | 84% | 18.82 ± 0.12 | 3.33 | 16.37 ± 0.07 | 2.15 |
| Rplp2 | Ribosomal protein lateral stalk subunit P2 | ribosomal phosphoprotein—component of the 60S subunit | 76% | 22.27 ± 0.14 | 3.91 | 21.71 ± 0.08 | 2.27 |
| Rps18 | Ribosomal protein S18 | ribosomal subunit | 83% | 17.36 ± 0.07 | 1.77 | 19.22 ± 0.08 | 1.92 |
| Tbp | TATA-box binding protein | transcription factor | 87% | 25.74 ± 0.15 | 2.78 | 24.08 ± 0.08 | 2.37 |
| Ywhaz | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta | involved in signal transduction pathways and role in tumor progression | 90% | 20.74 ± 0.11 | 2.56 | 20.41 ± 0.08 | 1.86 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szczepkowska, A.; Harazin, A.; Barna, L.; Deli, M.A.; Skipor, J. Identification of Reference Genes for Circadian Studies on Brain Microvessels and Choroid Plexus Samples Isolated from Rats. Biomolecules 2021, 11, 1227. https://doi.org/10.3390/biom11081227
Szczepkowska A, Harazin A, Barna L, Deli MA, Skipor J. Identification of Reference Genes for Circadian Studies on Brain Microvessels and Choroid Plexus Samples Isolated from Rats. Biomolecules. 2021; 11(8):1227. https://doi.org/10.3390/biom11081227
Chicago/Turabian StyleSzczepkowska, Aleksandra, András Harazin, Lilla Barna, Mária A. Deli, and Janina Skipor. 2021. "Identification of Reference Genes for Circadian Studies on Brain Microvessels and Choroid Plexus Samples Isolated from Rats" Biomolecules 11, no. 8: 1227. https://doi.org/10.3390/biom11081227
APA StyleSzczepkowska, A., Harazin, A., Barna, L., Deli, M. A., & Skipor, J. (2021). Identification of Reference Genes for Circadian Studies on Brain Microvessels and Choroid Plexus Samples Isolated from Rats. Biomolecules, 11(8), 1227. https://doi.org/10.3390/biom11081227







