Brain-Derived Exosomal Proteins as Effective Biomarkers for Alzheimer’s Disease: A Systematic Review and Meta-Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Search Strategy
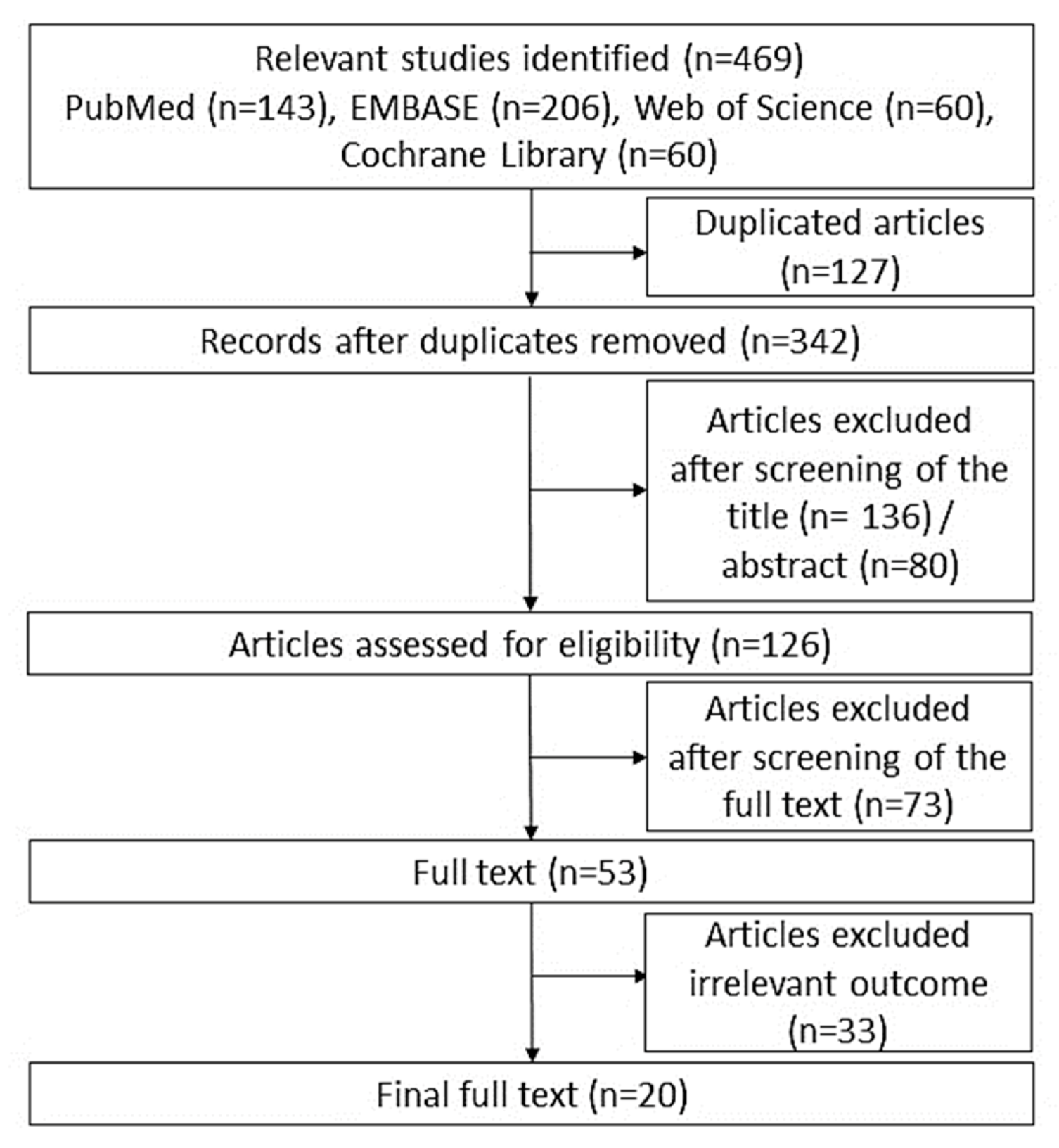
2.2. Study Selection
2.3. Data Extraction
2.4. Statistical Analysis
3. Results
3.1. Characteristics of the Eligible Studies
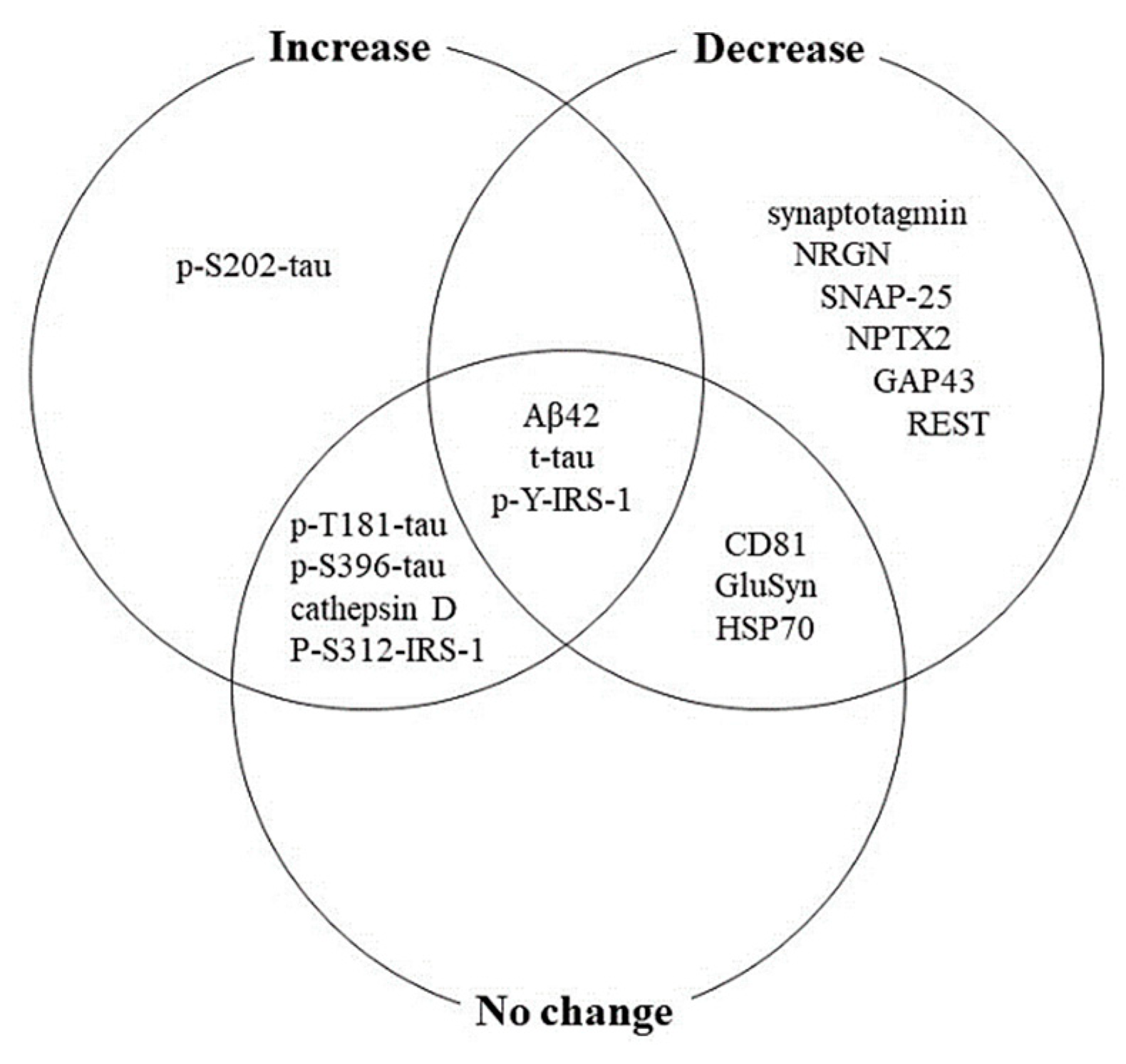
3.2. BDE Protein Changes in AD
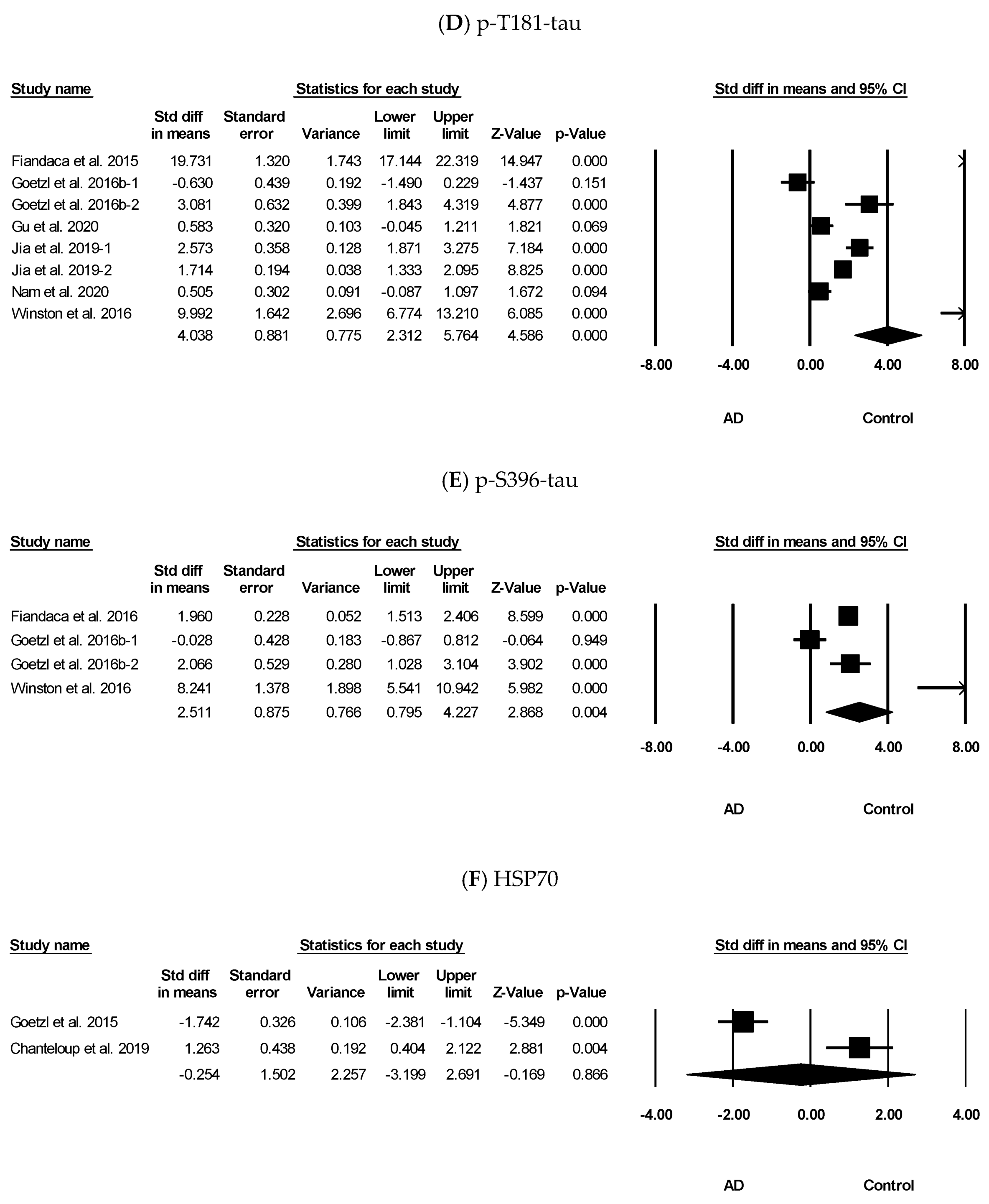
3.3. Meta-Analysis Results of Aβ42, t-tau, p-Y-IRS-1, p-T181-tau, p-S396-tau, and HSP70
4. Discussion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zetterberg, H.; Bendlin, B.B. Biomarkers for Alzheimer’s disease—Preparing for a new era of disease-modifying therapies. Mol. Psychiatry 2021, 26, 296–308. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Montagne, A.; Zhao, Z. Alzheimer’s pathogenic mechanisms and underlying sex difference. Cell. Mol. Life Sci. 2021, 78, 4907–4920. [Google Scholar] [CrossRef] [PubMed]
- Khan, T.K.; Alkon, D.L. Alzheimer’s Disease Cerebrospinal Fluid and Neuroimaging Biomarkers: Diagnostic Accuracy and Relationship to Drug Efficacy. J. Alzheimer’s Dis. 2015, 46, 817–836. [Google Scholar] [CrossRef]
- Mattson, M.P. Pathways towards and away from Alzheimer’s disease. Nature 2004, 430, 631–639. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Gu, B.J.; Masters, C.L.; Wang, Y.J. A systemic view of alzheimer diseaseinsights from amyloid-beta metabolism beyond the brain. Nat. Rev. Neurol. 2017, 13, 703. [Google Scholar] [CrossRef] [Green Version]
- Zetterberg, H. Blood-based biomarkers for Alzheimer’s disease—An update. J. Neurosci. Methods 2019, 319, 2–6. [Google Scholar] [CrossRef] [Green Version]
- Khan, T.K.; Alkon, D.L. Peripheral biomarkers of Alzheimer’s disease. J. Alzheimer’s Dis. 2015, 44, 729–744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bălașa, A.F.; Chircov, C.; Grumezescu, A.M. Body Fluid Biomarkers for Alzheimer’s Disease—An Up-To-Date Overview. Biomedicines 2020, 8, 421. [Google Scholar] [CrossRef] [PubMed]
- Janigro, D.; Bailey, D.M.; Lehmann, S.; Badaut, J.; O’Flynn, R.; Hirtz, C.; Marchi, N. Peripheral Blood and Salivary Biomarkers of Blood–Brain Barrier Permeability and Neuronal Damage: Clinical and Applied Concepts. Front. Neurol. 2021, 11, 577312. [Google Scholar] [CrossRef]
- Seol, W.; Kim, H.; Son, I. Urinary Biomarkers for Neurodegenerative Diseases. Exp. Neurobiol. 2020, 29, 325–333. [Google Scholar] [CrossRef]
- McGrowder, D.; Miller, F.; Vaz, K.; Nwokocha, C.; Wilson-Clarke, C.; Anderson-Cross, M.; Brown, J.; Anderson-Jackson, L.; Williams, L.; Latore, L.; et al. Cerebrospinal Fluid Biomarkers of Alzheimer’s Disease: Current Evidence and Future Perspectives. Brain Sci. 2021, 11, 215. [Google Scholar] [CrossRef] [PubMed]
- Vichianin, Y.; Khummongkol, A.; Chiewvit, P.; Raksthaput, A.; Chaichanettee, S.; Aoonkaew, N.; Senanarong, V. Accuracy of support-vector machines for diagnosis of alzheimer’s disease, using volume of brain obtained by structural mri at siriraj hospital. Front. Neurol. 2021, 12, 640696. [Google Scholar] [CrossRef] [PubMed]
- Bao, W.; Xie, F.; Zuo, C.; Guan, Y.; Huang, Y.H. Pet neuroimaging of Alzheimer’s disease: Radiotracers and their utility in clinical research. Front. Aging Neurosci. 2021, 13, 624330. [Google Scholar] [CrossRef]
- Blennow, K. A Review of Fluid Biomarkers for Alzheimer’s Disease: Moving from CSF to Blood. Neurol. Ther. 2017, 6, 15–24. [Google Scholar] [CrossRef]
- Paraskevaidi, M.; Allsop, D.; Karim, S.; Martin, F.L.; Crean, S. Diagnostic Biomarkers for Alzheimer’s Disease Using Non-Invasive Specimens. J. Clin. Med. 2020, 9, 1673. [Google Scholar] [CrossRef] [PubMed]
- Counts, S.E.; Ikonomovic, M.D.; Mercado, N.; Vega, I.E.; Mufson, E.J. Biomarkers for the Early Detection and Progression of Alzheimer’s Disease. Neurotherapeutics 2017, 14, 35–53. [Google Scholar] [CrossRef] [Green Version]
- Song, Z.; Xu, Y.; Deng, W.; Zhang, L.; Zhu, H.; Yu, P.; Qu, Y.; Zhao, W.; Han, Y.; Qin, C. Brain Derived Exosomes Are a Double-Edged Sword in Alzheimer’s Disease. Front. Mol. Neurosci. 2020, 13, 79. [Google Scholar] [CrossRef]
- Lizarraga-Valderrama, L.R.; Sheridan, G.K. Extracellular vesicles and intercellular communication in the central nervous system. FEBS Lett. 2021, 595, 1391–1410. [Google Scholar] [CrossRef]
- Abels, E.R.; Breakefield, X.O. Introduction to Extracellular Vesicles: Biogenesis, RNA Cargo Selection, Content, Release, and Uptake. Cell. Mol. Neurobiol. 2016, 36, 301–312. [Google Scholar] [CrossRef]
- Saheera, S.; Potnuri, A.G.; Krishnamurthy, P. Nano-Vesicle (Mis)Communication in Senescence-Related Pathologies. Cells 2020, 9, 1974. [Google Scholar] [CrossRef]
- Gassama, Y.; Favereaux, A. Emerging Roles of Extracellular Vesicles in the Central Nervous System: Physiology, Pathology, and Therapeutic Perspectives. Front. Cell. Neurosci. 2021, 15, 15. [Google Scholar] [CrossRef] [PubMed]
- Tricarico, C.; Clancy, J.; D’Souza-Schorey, C. Biology and biogenesis of shed microvesicles. Small GTPases 2017, 8, 220–232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malloci, M.; Perdomo, L.; Veerasamy, M.; Andriantsitohaina, R.; Simard, G.; Martínez, M.C. Extracellular Vesicles: Mechanisms in Human Health and Disease. Antioxid. Redox Signal. 2019, 30, 813–856. [Google Scholar] [CrossRef]
- Hornung, S.; Dutta, S.; Bitan, G. CNS-Derived Blood Exosomes as a Promising Source of Biomarkers: Opportunities and Challenges. Front. Mol. Neurosci. 2020, 13, 38. [Google Scholar] [CrossRef] [Green Version]
- Zetterberg, H.; Burnham, S.C. Blood-based molecular biomarkers for Alzheimer’s disease. Mol. Brain 2019, 12, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Watson, L.S.; Hamlett, E.D.; Stone, T.D.; Sims-Robinson, C. Neuronally derived extracellular vesicles: An emerging tool for understanding Alzheimer’s disease. Mol. Neurodegener. 2019, 14, 22. [Google Scholar] [CrossRef]
- Sinha, M.S.; Ansell-Schultz, A.; Civitelli, L.; Hildesjö, C.; Larsson, M.; Lannfelt, L.; Ingelsson, M.; Hallbeck, M. Alzheimer’s disease pathology propagation by exosomes containing toxic amyloid-beta oligomers. Acta Neuropathol. 2018, 136, 41–56. [Google Scholar] [CrossRef] [Green Version]
- Fiandaca, M.S.; Kapogiannis, D.; Mapstone, M.; Boxer, A.; Eitan, E.; Schwartz, J.B.; Abner, E.L.; Petersen, R.C.; Federoff, H.J.; Miller, B.L.; et al. Identification of preclinical alzheimer’s disease by a profile of pathogenic proteins in neurally derived blood exosomes: A case-control study. Alzheimer’s Dement. J. Alzheimer’s Assoc. 2015, 11, 600–607.e601. [Google Scholar] [CrossRef] [Green Version]
- Goetzl, E.J.; Boxer, A.; Schwartz, J.B.; Abner, E.L.; Petersen, R.C.; Miller, B.L.; Kapogiannis, D. Altered lysosomal proteins in neural-derived plasma exosomes in preclinical Alzheimer disease. Neurology 2015, 85, 40–47. [Google Scholar] [CrossRef] [Green Version]
- Kapogiannis, D.; Boxer, A.; Schwartz, J.B.; Abner, E.L.; Biragyn, A.; Masharani, U.; Frassetto, L.; Petersen, R.C.; Miller, B.L.; Goetzl, E.J. Dysfunctionally phosphorylated type 1 insulin receptor substrate in neural-derived blood exosomes of preclinical alzheimer’s disease. FASEB J. 2015, 29, 589–596. [Google Scholar] [CrossRef] [Green Version]
- Abner, E.L.; Jicha, G.A.; Shaw, L.M.; Trojanowski, J.Q.; Goetzl, E.J. Plasma neuronal exosomal levels of Alzheimer’s disease biomarkers in normal aging. Ann. Clin. Transl. Neurol. 2016, 3, 399–403. [Google Scholar] [CrossRef]
- Goetzl, E.J.; Kapogiannis, D.; Schwartz, J.B.; Lobach, I.V.; Goetzl, L.; Abner, E.L.; Jicha, G.A.; Karydas, A.M.; Boxer, A.; Miller, B.L. Decreased synaptic proteins in neuronal exosomes of frontotemporal dementia and Alzheimer’s disease. FASEB J. 2016, 30, 4141–4148. [Google Scholar] [CrossRef] [Green Version]
- Goetzl, E.J.; Mustapic, M.; Kapogiannis, D.; Eitan, E.; Lobach, I.V.; Goetzl, L.; Schwartz, J.B.; Miller, B.L. Cargo proteins of plasma astrocyte-derived exosomes in Alzheimer’s disease. FASEB J. 2016, 30, 3853–3859. [Google Scholar] [CrossRef] [Green Version]
- Winston, C.N.; Goetzl, E.J.; Akers, J.C.; Carter, B.S.; Rockenstein, E.M.; Galasko, D.; Masliah, E.; Rissman, R.A. Prediction of conversion from mild cognitive impairment to dementia with neuronally derived blood exosome protein profile. Alzheimer’s Dement. Diagn. Assess. Dis. Monit. 2016, 3, 63–72. [Google Scholar] [CrossRef] [Green Version]
- Guix, F.X.; Corbett, G.T.; Cha, D.J.; Mustapic, M.; Liu, W.; Mengel, D.; Chen, Z.; Aikawa, E.; Young-Pearse, T.; Kapogiannis, D.; et al. Detection of Aggregation-Competent Tau in Neuron-Derived Extracellular Vesicles. Int. J. Mol. Sci. 2018, 19, 663. [Google Scholar] [CrossRef] [Green Version]
- Goetzl, E.J.; Abner, E.L.; Jicha, G.A.; Kapogiannis, D.; Schwartz, J.B. Declining levels of functionally specialized synaptic proteins in plasma neuronal exosomes with progression of alzheimer’s disease. FASEB J. 2018, 32, 888–893. [Google Scholar] [CrossRef] [Green Version]
- Jia, L.; Qiu, Q.; Zhang, H.; Chu, L.; Du, Y.; Zhang, J.; Zhou, C.; Liang, F.; Shi, S.; Wang, S.; et al. Concordance between the assessment of Aβ42, T-tau, and P-T181-tau in peripheral blood neuronal-derived exosomes and cerebrospinal fluid. Alzheimer’s Dement. 2019, 15, 1071–1080. [Google Scholar] [CrossRef]
- Agliardi, C.; Guerini, F.R.; Zanzottera, M.; Bianchi, A.; Nemni, R.; Clerici, M. SNAP-25 in Serum Is Carried by Exosomes of Neuronal Origin and Is a Potential Biomarker of Alzheimer’s Disease. Mol. Neurobiol. 2019, 56, 5792–5798. [Google Scholar] [CrossRef] [PubMed]
- Chanteloup, G.; Cordonnier, M.; Moreno-Ramos, T.; Pytel, V.; Matías-Guiu, J.; Gobbo, J.; Cabrera-Martín, M.N.; Gómez-Pinedo, U.; Garrido, C.; Matías-Guiu, J.A. Exosomal HSP70 for Monitoring of Frontotemporal Dementia and Alzheimer’s Disease: Clinical and FDG-PET Correlation. J. Alzheimer’s Dis. 2019, 71, 1263–1269. [Google Scholar] [CrossRef] [PubMed]
- Cicognola, C.; Brinkmalm, G.; Wahlgren, J.; Portelius, E.; Gobom, J.; Cullen, N.C.; Hansson, O.; Parnetti, L.; Constantinescu, R.; Wildsmith, K.; et al. Novel tau fragments in cerebrospinal fluid: Relation to tangle pathology and cognitive decline in Alzheimer’s disease. Acta Neuropathol. 2019, 137, 279–296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goetzl, E.J.; Nogueras-Ortiz, C.; Mustapic, M.; Mullins, R.; Abner, E.L.; Schwartz, J.B.; Kapogiannis, D. Deficient neurotrophic factors of CSPG4-type neural cell exosomes in Alzheimer disease. FASEB J. 2019, 33, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Kapogiannis, D.; Mustapic, M.; Shardell, M.D.; Berkowitz, S.; Diehl, T.C.; Spangler, R.D.; Tran, J.; Lazaropoulos, M.P.; Chawla, S.; Gulyani, S.; et al. Association of Extracellular Vesicle Biomarkers with Alzheimer Disease in the Baltimore Longitudinal Study of Aging. JAMA Neurol. 2019, 76, 1340–1351. [Google Scholar] [CrossRef] [PubMed]
- Gu, D.; Liu, F.; Meng, M.; Zhang, L.; Gordon, M.L.; Wang, Y.; Cai, L.; Zhang, N. Elevated matrix metalloproteinase-9 levels in neuronal extracellular vesicles in Alzheimer’s disease. Ann. Clin. Transl. Neurol. 2020, 7, 1681–1691. [Google Scholar] [CrossRef]
- Jia, L.; Zhu, M.; Kong, C.; Pang, Y.; Zhang, H.; Qiu, Q.; Wei, C.; Tang, Y.; Wang, Q.; Li, Y.; et al. Blood neuro-exosomal synaptic proteins predict Alzheimer’s disease at the asymptomatic stage. Alzheimer’s Dement. 2020, 17, 49–60. [Google Scholar] [CrossRef]
- Nam, E.; Lee, Y.-B.; Moon, C.; Chang, K.-A. Serum Tau Proteins as Potential Biomarkers for the Assessment of Alzheimer’s Disease Progression. Int. J. Mol. Sci. 2020, 21, 5007. [Google Scholar] [CrossRef] [PubMed]
- Perrotte, M.; Haddad, M.; Le Page, A.; Frost, E.H.; Fulöp, T.; Ramassamy, C. Profile of pathogenic proteins in total circulating extracellular vesicles in mild cognitive impairment and during the progression of Alzheimer’s disease. Neurobiol. Aging 2020, 86, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Picciolini, S.; Gualerzi, A.; Carlomagno, C.; Cabinio, M.; Sorrentino, S.; Baglio, F.; Bedoni, M. An SPRi-based biosensor pilot study: Analysis of multiple circulating extracellular vesicles and hippocampal volume in Alzheimer’s disease. J. Pharm. Biomed. Anal. 2021, 192, 113649. [Google Scholar] [CrossRef] [PubMed]
- Omar, S.H.; Preddy, J. Advantages and Pitfalls in Fluid Biomarkers for Diagnosis of Alzheimer’s Disease. J. Pers. Med. 2020, 10, 63. [Google Scholar] [CrossRef]
- Jin, Q.; Wu, P.; Zhou, X.; Qian, H.; Xu, W. Extracellular Vesicles: Novel Roles in Neurological Disorders. Stem Cells Int. 2021, 2021, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Eren, E.; Hunt, J.F.V.; Shardell, M.; Chawla, S.; Tran, J.; Gu, J.; Vogt, N.M.; Johnson, S.C.; Bendlin, B.B.; Kapogiannis, D. Extracellular vesicle biomarkers of alzheimer’s disease associated with sub-clinical cognitive decline in late middle age. Alzheimer’s Dement. J. Alzheimer’s Assoc. 2020, 16, 1293–1304. [Google Scholar] [CrossRef]
- Soto-Rojas, L.; Pacheco-Herrero, M.; Martínez-Gómez, P.; Campa-Córdoba, B.; Apátiga-Pérez, R.; Villegas-Rojas, M.; Harrington, C.; de la Cruz, F.; Garcés-Ramírez, L.; Luna-Muñoz, J. The Neurovascular Unit Dysfunction in Alzheimer’s Disease. Int. J. Mol. Sci. 2021, 22, 2022. [Google Scholar] [CrossRef] [PubMed]
- Sagare, A.P.; Bell, R.D.; Zlokovic, B.V. Neurovascular defects and faulty amyloid-beta vascular clearance in Alzheimer’s disease. J. Alzheimer’s Dis. 2013, 33 (Suppl. 1), S87–S100. [Google Scholar] [CrossRef] [Green Version]
- Yamazaki, Y.; Kanekiyo, T. Blood-Brain Barrier Dysfunction and the Pathogenesis of Alzheimer’s Disease. Int. J. Mol. Sci. 2017, 18, 1965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koelsch, G. BACE1 Function and Inhibition: Implications of Intervention in the Amyloid Pathway of Alzheimer’s Disease Pathology. Molecules 2017, 22, 1723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breijyeh, Z.; Karaman, R. Comprehensive Review on Alzheimer’s Disease: Causes and Treatment. Molecules 2020, 25, 5789. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, R.A. The molecular biology of senile plaques and neurofibrillary tangles in alzheimer’s disease. Folia Neuropathol. 2009, 47, 289–299. [Google Scholar]
- Chen, G.-F.; Xu, T.-H.; Yan, Y.; Zhou, Y.-R.; Jiang, Y.; Melcher, K.; Xu, H.E. Amyloid beta: Structure, biology and structure-based therapeutic development. Acta Pharmacol. Sin. 2017, 38, 1205–1235. [Google Scholar] [CrossRef]
- Tabaton, M.; Piccini, A. Role of water-soluble amyloid-beta in the pathogenesis of Alzheimer’s disease. Int. J. Exp. Pathol. 2005, 86, 139–145. [Google Scholar] [CrossRef]
- Eshraghi, M.; Adlimoghaddam, A.; Mahmoodzadeh, A.; Sharifzad, F.; Yasavoli-Sharahi, H.; Lorzadeh, S.; Albensi, B.; Ghavami, S. Alzheimer’s Disease Pathogenesis: Role of Autophagy and Mitophagy Focusing in Microglia. Int. J. Mol. Sci. 2021, 22, 3330. [Google Scholar] [CrossRef]
- Iqbal, K.; Liu, F.; Gong, C.-X.; Grundke-Iqbal, I. Tau in Alzheimer Disease and Related Tauopathies. Curr. Alzheimer Res. 2010, 7, 656–664. [Google Scholar] [CrossRef] [Green Version]
- Milà-Alomà, M.; Suárez-Calvet, M.; Molinuevo, J.L. Latest advances in cerebrospinal fluid and blood biomarkers of Alzheimer’s disease. Ther. Adv. Neurol. Disord. 2019, 12, 1756286419888819. [Google Scholar] [CrossRef] [Green Version]
- Toombs, J.; Zetterberg, H. In the blood: Biomarkers for amyloid pathology and neurodegeneration in Alzheimer’s disease. Brain Commun. 2020, 2. [Google Scholar] [CrossRef]
- Zetterberg, H.; Wilson, D.; Andreasson, U.; Minthon, L.; Blennow, K.; Randall, J.; Hansson, O. Plasma tau levels in Alzheimer’s disease. Alzheimers Res. Ther. 2013, 5, 9. [Google Scholar] [CrossRef] [PubMed]
- Xing, W.; Gao, W.; Lv, X.; Xu, X.; Zhang, Z.; Yan, J.; Mao, G.; Bu, Z. The diagnostic value of exosome-derived biomarkers in Alzheimer’s disease and mild cognitive impairment: A meta-analysis. Front. Aging Neurosci. 2021, 13, 637218. [Google Scholar] [CrossRef] [PubMed]
- Rastogi, S.; Sharma, V.; Bharti, P.S.; Rani, K.; Modi, G.P.; Nikolajeff, F.; Kumar, S. The Evolving Landscape of Exosomes in Neurodegenerative Diseases: Exosomes Characteristics and a Promising Role in Early Diagnosis. Int. J. Mol. Sci. 2021, 22, 440. [Google Scholar] [CrossRef]
- Crotti, A.; Sait, H.R.; McAvoy, K.M.; Estrada, K.; Ergun, A.; Szak, S.; Marsh, G.; Jandreski, L.; Peterson, M.; Reynolds, T.L.; et al. BIN1 favors the spreading of Tau via extracellular vesicles. Sci. Rep. 2019, 9, 1–20. [Google Scholar] [CrossRef]
- Mullins, R.J.; Mustapic, M.; Goetzl, E.J.; Kapogiannis, D. Exosomal biomarkers of brain insulin resistance associated with regional atrophy in Alzheimer’s disease. Human Brain Mapp. 2017, 38, 1933–1940. [Google Scholar] [CrossRef] [Green Version]
- Cersosimo, E.; DeFronzo, R.A. Insulin resistance and endothelial dysfunction: The road map to cardiovascular diseases. Diabetes Metab. Res. Rev. 2006, 22, 423–436. [Google Scholar] [CrossRef] [PubMed]
- Mullins, R.; Diehl, T.C.; Chia, C.W.; Kapogiannis, D. Insulin Resistance as a Link between Amyloid-Beta and Tau Pathologies in Alzheimer’s Disease. Front. Aging Neurosci. 2017, 9, 118. [Google Scholar] [CrossRef]
- Talbot, K.; Wang, H.-Y.; Kazi, H.; Han, L.-Y.; Bakshi, K.P.; Stucky, A.; Fuino, R.L.; Kawaguchi, K.R.; Samoyedny, A.J.; Wilson, R.S.; et al. Demonstrated brain insulin resistance in Alzheimer’s disease patients is associated with IGF-1 resistance, IRS-1 dysregulation, and cognitive decline. J. Clin. Investig. 2012, 122, 1316–1338. [Google Scholar] [CrossRef] [Green Version]
- Boudesco, C.; Cause, S.; Jego, G.; Garrido, C. Hsp70: A Cancer Target Inside and Outside the Cell. Methods Mol Biol. 2018, 1709, 371–396. [Google Scholar] [CrossRef]
- Evans, C.G.; Wisen, S.; Gestwicki, J.E. Heat shock proteins 70 and 90 inhibit early stages of amyloid beta-(1-42) aggregation in vitro. J. Biol. Chem. 2006, 281, 33182–33191. [Google Scholar] [CrossRef] [Green Version]
- Magrane, J.; Smith, R.C.; Walsh, K.; Querfurth, H.W. Heat shock protein 70 participates in the neuroprotective response to intracellularly expressed beta-amyloid in neurons. J. Neurosci. 2004, 24, 1700–1706. [Google Scholar] [CrossRef] [Green Version]
- Yuyama, K.; Igarashi, Y. Exosomes as carriers of Alzheimer’s amyloid-ss. Front. Neurosci. 2017, 11, 229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuyama, K.; Sun, H.; Sakai, S.; Mitsutake, S.; Okada, M.; Tahara, H.; Furukawa, J.; Fujitani, N.; Shinohara, Y.; Igarashi, Y. Decreased amyloid-beta pathologies by intracerebral loading of glycosphingolipid-enriched exosomes in alzheimer model mice. J. Biol. Chem. 2014, 289, 24488–24498. [Google Scholar] [CrossRef] [Green Version]
- Yuyama, K.; Sun, H.; Mitsutake, S.; Igarashi, Y. Sphingolipid-modulated exosome secretion promotes clearance of amyloid-beta by microglia. J. Biol. Chem. 2012, 287, 10977–10989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, K.; Klyubin, I.; Kim, Y.; Jung, J.H.; Mably, A.J.; O’Dowd, S.T.; Lynch, T.; Kanmert, D.; Lemere, C.A.; Finan, G.M.; et al. Exosomes neutralize synaptic-plasticity-disrupting activity of abeta assemblies in vivo. Mol. Brain 2013, 6, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.K.T.; Langfelder, P.; Horvath, S.; Palazzolo, M.J. Exosomes and Homeostatic Synaptic Plasticity Are Linked to Each other and to Huntington’s, Parkinson’s, and Other Neurodegenerative Diseases by Database-Enabled Analyses of Comprehensively Curated Datasets. Front. Neurosci. 2017, 11, 149. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Zhang, Y.; Du, X.-F.; Li, J.; Zi, H.-X.; Bu, J.-W.; Yan, Y.; Han, H.; Du, J.-L. Neurons secrete miR-132-containing exosomes to regulate brain vascular integrity. Cell Res. 2017, 27, 882–897. [Google Scholar] [CrossRef]
- Kalani, A.; Tyagi, A.; Tyagi, N. Exosomes: Mediators of Neurodegeneration, Neuroprotection and Therapeutics. Mol. Neurobiol. 2013, 49, 590–600. [Google Scholar] [CrossRef] [Green Version]
- Kalani, A.; Tyagi, N. Exosomes in neurological disease, neuroprotection, repair and therapeutics: Problems and perspectives. Neural Regen. Res. 2015, 10, 1565–1567. [Google Scholar] [CrossRef]
- Wang, S.; Cesca, F.; Loers, G.; Schweizer, M.; Buck, F.; Benfenati, F.; Schachner, M.; Kleene, R. Synapsin I Is an Oligomannose-Carrying Glycoprotein, Acts As an Oligomannose-Binding Lectin, and Promotes Neurite Outgrowth and Neuronal Survival When Released via Glia-Derived Exosomes. J. Neurosci. 2011, 31, 7275–7290. [Google Scholar] [CrossRef]
- Long, X.; Yao, X.; Jiang, Q.; Yang, Y.; He, X.; Tian, W.; Zhao, K.; Zhang, H. Astrocyte-derived exosomes enriched with miR-873a-5p inhibit neuroinflammation via microglia phenotype modulation after traumatic brain injury. J. Neuroinflamm. 2020, 17, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Niu, F.; Yao, H.; Liao, K.; Chen, X.; Kook, Y.; Ma, R.; Hu, G.; Buch, S. Exosomal miR-9 Released from HIV Tat Stimulated Astrocytes Mediates Microglial Migration. J. Neuroimmune Pharmacol. 2018, 13, 330–344. [Google Scholar] [CrossRef] [PubMed]
- Luarte, A.; Cisternas, P.; Caviedes, A.; Batiz, L.F.; Lafourcade, C.; Wyneken, U.; Henzi, R. Astrocytes at the Hub of the Stress Response: Potential Modulation of Neurogenesis by miRNAs in Astrocyte-Derived Exosomes. Stem Cells Int. 2017, 2017, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Bakhti, M.; Winter, C.; Simons, M. Inhibition of Myelin Membrane Sheath Formation by Oligodendrocyte-derived Exosome-like Vesicles. J. Biol. Chem. 2011, 286, 787–796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frühbeis, C.; Fröhlich, D.; Kuo, W.P.; Amphornrat, J.; Thilemann, S.; Saab, A.S.; Kirchhoff, F.; Möbius, W.; Goebbels, S.; Nave, K.-A.; et al. Neurotransmitter-Triggered Transfer of Exosomes Mediates Oligodendrocyte–Neuron Communication. PLoS Biol. 2013, 11, e1001604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, G.; Zhao, S.; Xia, X.; Li, C.; Li, C.; Ji, C.; Sheng, S.; Tang, Y.; Zhu, J.; Wang, Y.; et al. Glutaminase C Regulates Microglial Activation and Pro-inflammatory Exosome Release: Relevance to the Pathogenesis of Alzheimer’s Disease. Front. Cell. Neurosci. 2019, 13, 264. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Ge, X.; Yu, J.; Han, Z.; Yin, Z.; Li, Y.; Chen, F.; Wang, H.; Zhang, J.; Lei, P. Increased mir-124-3p in microglial exosomes following traumatic brain injury inhibits neuronal inflammation and contributes to neurite outgrowth via their transfer into neurons. FASEB J. 2018, 32, 512–528. [Google Scholar] [CrossRef] [Green Version]
- Reynolds, J.L.; Mahajan, S.D. Transmigration of Tetraspanin 2 (Tspan2) siRNA Via Microglia Derived Exosomes across the Blood Brain Barrier Modifies the Production of Immune Mediators by Microglia Cells. J. Neuroimmune Pharmacol. 2019, 15, 554–563. [Google Scholar] [CrossRef] [PubMed]
- Yin, Q.Q.; Ji, X.; Lv, R.; Pei, J.-J.; Du, Y.; Shen, C.; Hou, X. Targetting Exosomes as a New Biomarker and Therapeutic Approach for Alzheimer’s Disease. Clin. Interv. Aging 2020, 15, 195–205. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Study | Location | Specimen | Method | Patients | Sample (M/F) | Age (y) (M ± SD) | MMSE (M ± SD) | BDE Proteins | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | Case | Control | Case | Control | ||||||
| Fiandaca et al. 2015 [28] | USA | Plasma | ExoQuick/ELISA | AD | 30/27 | 30/27 | 79.5 ± 6.1 | 79.6 ± 6.0 | Aβ42, t-tau, p-T181-tau, p-S396-tau | ||
| Goetzl et al. 2015 [29] | USA | Plasma | ExoQuick/ELISA | AD (include MCI) | 13/13 | 13/13 | 75.4 ± 7.9 | 75.8 ± 7.9 | 22.5 ± 1.5 | cathepsin D, LAMP-1, HSP70, ubiquitinylated protein | |
| Kapogiannis et al. 2015 [30] | USA | Plasma | ExoQuick/ELISA | AD | 13/13 | 13/13 | 74.3 ± 7.5 | 74.3 ± 7.5 | t-IRS-1, p-S312-IRS-1, p-Y-IRS-1, p-S312-IRS-1/p-Y-IRS-1 | ||
| Dementia | 10 | 20.5 ± 2.2 | |||||||||
| Abner et al. 2016 [31] | USA | Plasma | ExoQuick/ELISA | AD | 5/5 | 10/10 | 77.6 | 77.6 | 29.4 ± 0.8 | Aβ42, p-T181-tau, NRGN, cathepsin D, REST | |
| Goetzl et al. 2016a [32] | USA | Plasma | ExoQuick/ELISA | AD | 6/6 | 6/6 | 74.4 ± 2.0 | 74.4 ± 2.0 | 26.3 ± 1.0 | 29.8 ± 0.1 | Synaptophysin, synaptotagmin, synaptopodin, NRGN, p-S9-synapsin 1, GAP43, synapsin 1, MOG, GAP43 |
| AD2 (after diagnosis of dementia) | 2/7 | 2/7 | 87.8 ± 2.5 | 82.2 ± 2.3 | 21.4 ± 1.6 | 28.3 ± 1.0 | |||||
| Goetzl et al. 2016b [33] | USA | Plasma | ExoQuick/ELISA | AD (include amnestic mild cognitive impairment and early dementia) | 12 | 10 | Aβ42, sAPPα, sAPPβ, BACE-1, γ-secretase, p-T181-tau, p-S396-tau, GDNF, GFAP, GluSyn, NF-Lch, NS-enolase, CD81, Septin-8 | ||||
| Winston et al. 2016 [34] | USA | Plasma | ExoQuick/ELISA | AD | 11/9 | 10 | 75.4 ± 6.8 | 17.7 ± 0.7 | Aβ42, p-T181-tau, p-S396-tau, NRGN, REST | ||
| Guix et al. 2018 [35] | USA | Plasma | ExoQuick/ELISA | AD (mild) | 3/7 | 3/7 | 75.6 ± 12.9 | 75.9 ± 8.7 | 75.6 ± 12.9 | 29.7 ± 0.5 | Aβ42, p-T181-tau, MR tau, FL tau |
| AD (moderate) | 4/6 | 75.6 ± 12.9 | 75.6 ± 12.9 | ||||||||
| Goetzl et al. 2018 [36] | USA | Plasma | ExoQuick/ELISA | AD | 12/16 | 12/16 | 73.1 ± 1.4 | 73.2 ± 1.5 | 25.6 ± 0.8 | 29.7 ± 0.1 | AMPA4 receptor, NPTX2, NLGN1, |
| AD2 (after diagnosis of dementia) | 10/8 | 10/8 | 78.2 ± 1.8 | 70.1 ± 1.7 | 20.2 ± 1.5 | 28.3 ± 1.0 | NRXN2 | ||||
| Jia et al. 2019 [37] | China | Plasma | ExoQuick/ELISA | AD | 39/42 | 35/37 | 65 ± 6 | 64 ± 5 | 19.6 ± 3.1 | 29.3 ± 1.2 | Aβ42, p-T181-tau |
| Agliardi et al. 2019 [38] | Italy | Serum | ExoQuick/ELISA | AD | 8/16 | 4/13 | 77.7 ± 1.4 | 76.5 ± 1.5 | 21.9 ± 0.9 | 28.7 ± 0.4 | SNAP-25 |
| Chanteloup et al. 2019 [39] | Spain | Plasma | ExoQuick/ELISA | AD | 21 | 13 | 77.1 ± 8.2 | 75.2 ± 6.7 | HSP70 | ||
| Cicognola et al. 2019 [40] | Sweden | Serum | ExoQuick/SIMOA | AD | 4 | 4 | 79.5 | 67 | >15 | N-224 tau, N-123 tau | |
| Goetzl et al. 2019 [41] | USA | Plasma | ExoQuick/ELISA | AD | 9/15 | 9/15 | 73.1 ± 1.6 | 73.1 ± 1.8 | 26.1 ± 0.9 | 29.3 ± 0.2 | AMPA4 receptor, FGF-2, FGF-13, HGF, IGF-1, GluSyn, CD81 |
| AD2 (after conversion to moderate dementia) | 7/8 | 7/8 | 84.5 ± 1.7 | 80.2 ± 1.8 | 24.3 ± 0.9 | 29.4 ± 0.6 | |||||
| Kapogiannis et al. 2019 [42] | USA | Plasma | ExoQuick/ELISA, SIMOA | AD (future) | 60/68 | 112/110 | 79.1 ± 7.0 | 76.2 ± 7.4 | 27.5 ± 1.8 | 28.4 ± 1.8 | t-tau, p-T181-tau, p-T231-tau, p-S312-IRS-1, p-Y-IRS-1, TSG101 |
| Serum | AD | 17/18 | 6/23 | 74.0 ± 8.7 | 72.1 ± 7.9 | 23.9 ± 3.0 | 29.8 ± 0.6 | ||||
| Gu et al. 2020 [43] | China | Plasma | ExoQuick/ELISA | AD | 8/23 | 5/10 | 68.6 ± 8.0 | 64.8 ± 6.0 | 15.9 ± 6.6 | 27.7 ± 1.7 | Aβ42, p-T181-tau, p-S396-tau, IL-6, MMP-9, CD81 |
| Jia et al. 2020 [44] | China | Plasma | ExoQuick/ELISA | AD | 59/62 | 74/86 | 66 ± 5 | 54 ± 6 | 20.7 ± 2.9 | 29.1 ± 1.1 | Synaptotagmin, NRGN, SNAP-25, GAP43 |
| Nam et al. 2020 [45] | Korea | Serum | ExoQuick/ELISA | AD | 3/17 | 17/9 | 76.6 ± 1.3 | 73.9 ± 0.9 | 16.6 ± 0.5 | 27.7 ± 0.2 | Aβ42, t-tau, p-T181-tau, p-S202-tau, p-tau/t-tau |
| Perrotte et al. 2020 [46] | Canada | Plasma | Exosome isolation kit/Luminex | AD (mild) | 1/11 | 3/9 | 75.6 ± 1.3 | 68.8 ± 1.5 | 24.0 ± 0.5 | 29.4 ± 0.3 | Aβ42, APP, Aβ42/p-T181-tau, t-tau, p-T181-tau, p- T181-tau/t-tau, t-tau/Aβ42 |
| AD (moderate) | 4/8 | 79.1 ± 1.1 | 19.9 ± 1.4 | ||||||||
| AD (severe) | 2/10 | 83.0 ± 1.6 | |||||||||
| Picciolini et al. 2021 [47] | Italy | Plasma | Chromatography using qEV columns/ELISA | AD | 4/6 | 5/5 | 73.9 ± 3.0 | 62.6 ± 2.0 | TSPO, GM1 | ||
| Category | Level | Exosomal Proteins |
|---|---|---|
| Aβ targeted | Increase | Aβ42, APP, sAPPβ, BACE-1 |
| Decrease | Aβ42, APP | |
| No change | Aβ42, APP, sAPPα, sAPPβ, BACE-1, Aβ42/p-T181-tau, γ-secretase | |
| Tau targeted | Increase | t-tau, p-T181-tau, p-T231-tau, p-S202-tau, p-S396-tau, p-tau/t-tau, p-T181-tau/t-tau,-tau/Aβ42 |
| Decrease | t-tau | |
| No change | t-tau, p-T181-tau, p-S396-tau, p-T181-tau/t-tau, t-tau/Aβ42, N-224 tau, N-123 tau, MR tau, FL tau | |
| Synaptic protein | Decrease | NRGN, synaptophysin, synaptotagmin, synaptopodin, SNAP-25, AMPA4 receptor, NPTX2, NLGN1, NRXN2, p-S9-synapsin 1, synapsin 1, MOG |
| Autolysosomal | Increase | cathepsin D, LAMP-1 |
| No change | cathepsin D, LAMP-1 | |
| Growth/trophic | Increase | GDNF |
| Decrease | GDNF, FGF-2, FGF-13, HGF, IGF-1 | |
| Brain insulin resistance | Increase | p-Y-IRS-1, p-S312-IRS-1, p-S312-IRS-1/p-Y-IRS-1 |
| Decrease | p-Y-IRS-1 | |
| No change | t-IRS-1, p-Y-IRS-1, p-S312-IRS-1, p-S312-IRS-1/p-Y-IRS-1 | |
| Inflammatory related | Increase | MMP-9, TSPO |
| No change | IL-6 | |
| Molecular chaperone | Increase | ubiquitinylated protein |
| Decrease | HSP70 | |
| No change | HSP70, ubiquitinylated protein | |
| Transcriptional repressor | Decrease | REST |
| Cell type marker | Increase | GFAP, NF-Lch, NS-enolase |
| Decrease | GFAP, GluSyn | |
| No change | GluSyn, NF-Lch, NS-enolase | |
| Exosome marker | Decrease | CD81 |
| No change | CD81, TSG101 | |
| Other | Increase | GM1 |
| Decrease | GAP43, Septin-8 | |
| No change | Septin-8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, K.Y.; Shin, K.Y.; Chang, K.-A. Brain-Derived Exosomal Proteins as Effective Biomarkers for Alzheimer’s Disease: A Systematic Review and Meta-Analysis. Biomolecules 2021, 11, 980. https://doi.org/10.3390/biom11070980
Kim KY, Shin KY, Chang K-A. Brain-Derived Exosomal Proteins as Effective Biomarkers for Alzheimer’s Disease: A Systematic Review and Meta-Analysis. Biomolecules. 2021; 11(7):980. https://doi.org/10.3390/biom11070980
Chicago/Turabian StyleKim, Ka Young, Ki Young Shin, and Keun-A Chang. 2021. "Brain-Derived Exosomal Proteins as Effective Biomarkers for Alzheimer’s Disease: A Systematic Review and Meta-Analysis" Biomolecules 11, no. 7: 980. https://doi.org/10.3390/biom11070980
APA StyleKim, K. Y., Shin, K. Y., & Chang, K.-A. (2021). Brain-Derived Exosomal Proteins as Effective Biomarkers for Alzheimer’s Disease: A Systematic Review and Meta-Analysis. Biomolecules, 11(7), 980. https://doi.org/10.3390/biom11070980







