Regulation of S100A10 Gene Expression
Abstract
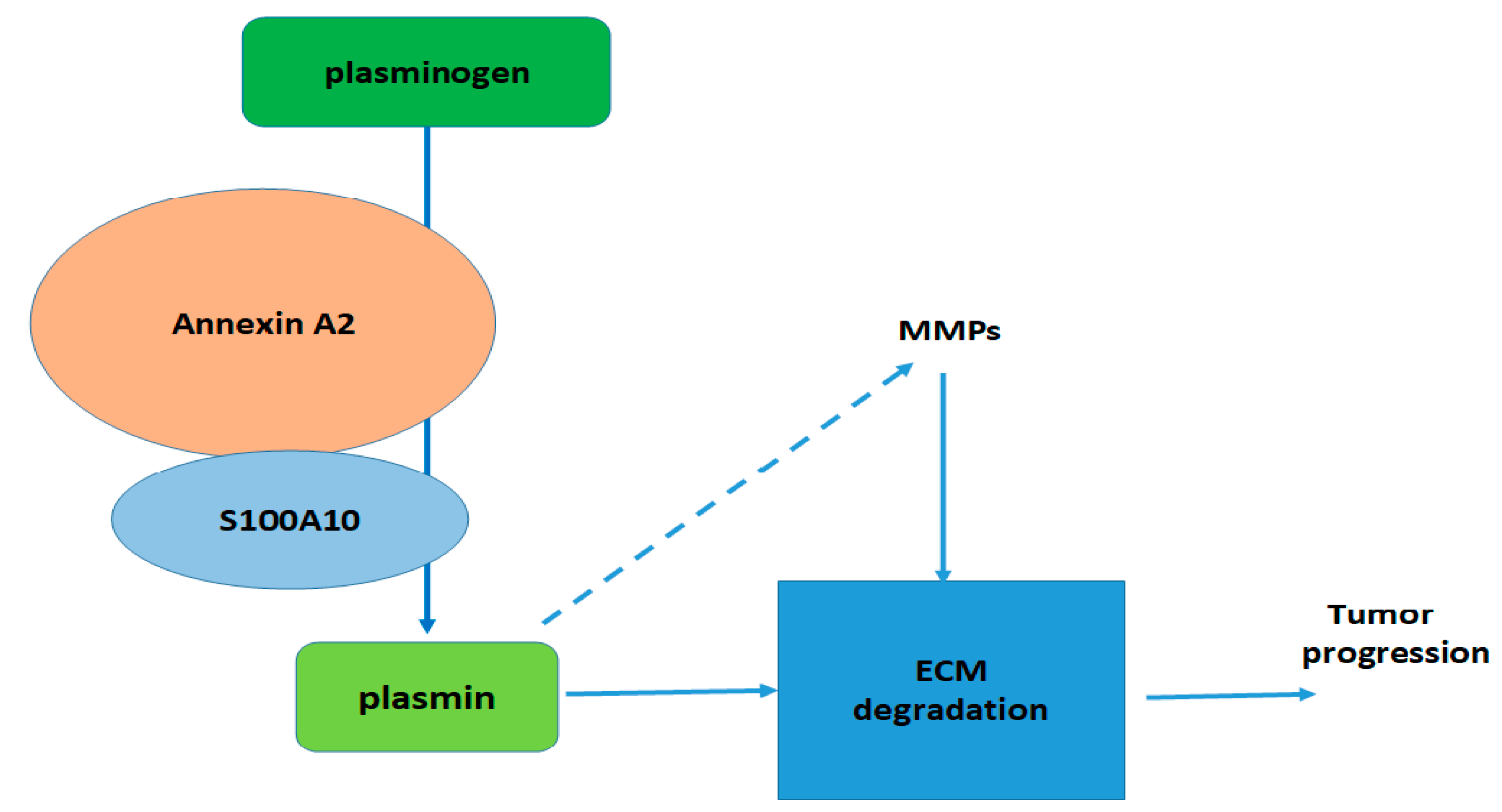
1. Introduction
2. Regulation of S100A10 Expression—Literature Data
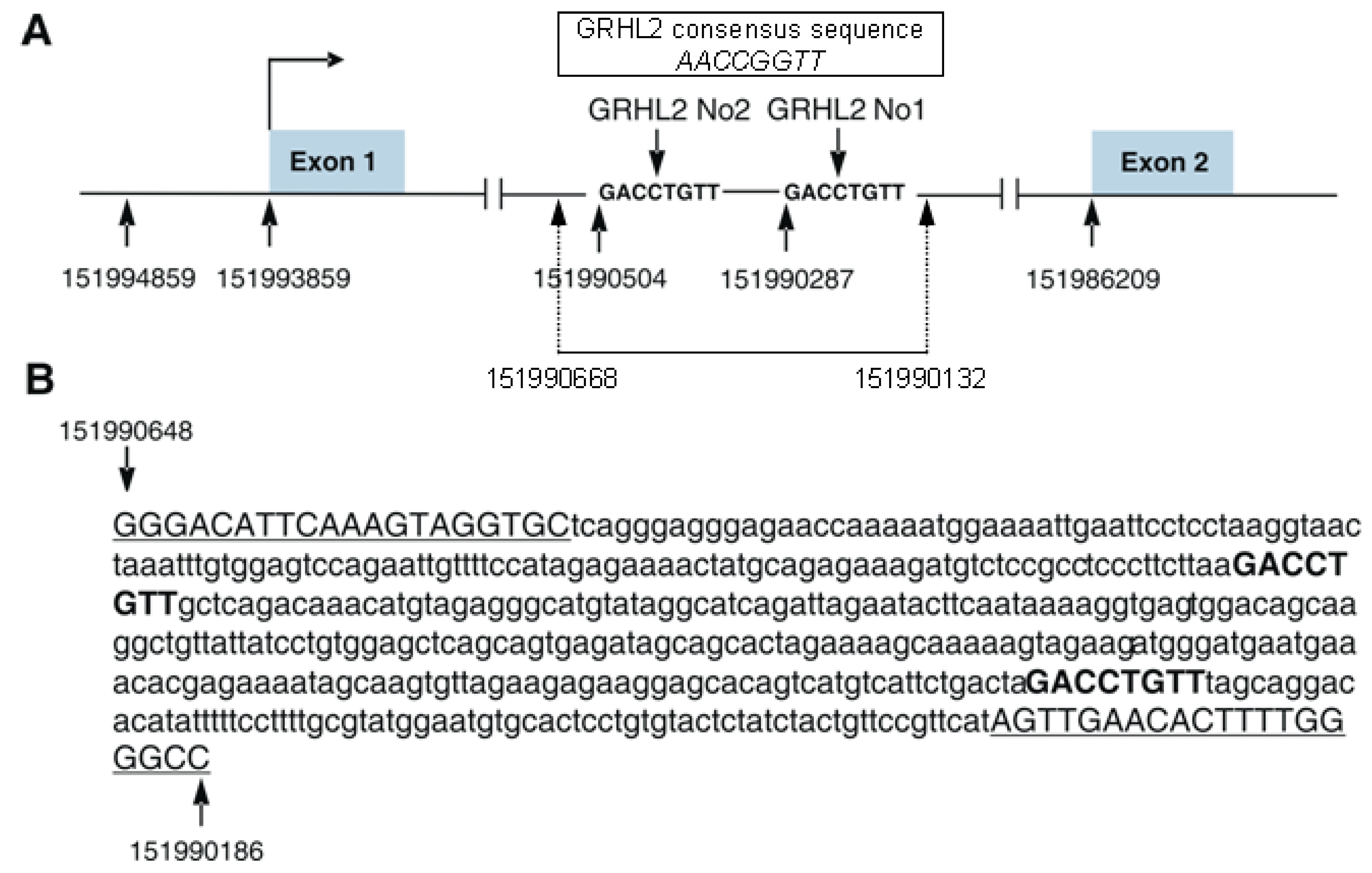
3. Regulation of S100A10 Expression by GRHL2 Transcription Factor—Original Data
3.1. Influence of GRHL2 on S100A10 Expression
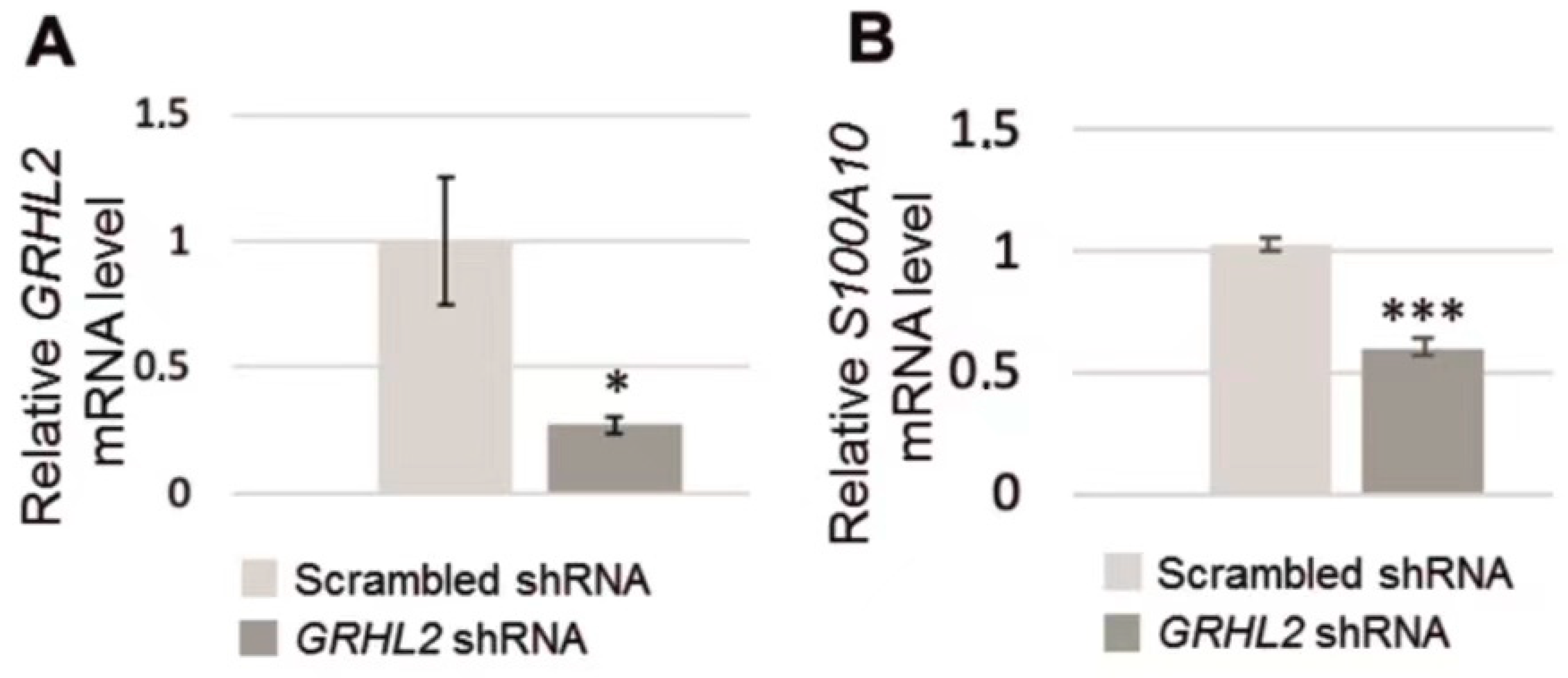
3.2. Analysis of S100A10 mRNA in RC-124 Cells with Silenced GRHL2 Expression
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BDNF | brain-derived neurotrophic factor |
| BSA | bovine serum albumin |
| ChIP | chromatin immunoprecipitation assay |
| DTT | dithiothreitol |
| EDTA | ethylenediaminetetraacetic acid |
| EGF | epidermal growth factor |
| FGF2 | fibroblast growth factor 2 |
| bFGF | basic fibroblast growth factor |
| GAS | interferon γ activation site |
| GD | gonadotrophin |
| GR | glucocorticoid receptor |
| GRE | glucocorticoid response element |
| GRHL2 | grainyhead-like 2 |
| hTERT | human telomerase reverse transcriptase |
| JNK | c-Jun N-terminal kinase |
| L-1β | interleukin 1β |
| IP3 | 1,4,5-trisphosphate inositol |
| γIRE | interferon γ response element |
| MAP | mitogen activated protein |
| TGF-β | transforming growth factor-β |
References
- Gonzalez, L.L.; Garrie, K.; Turner, M.D. Role of S100 proteins in health and disease. Biochim. Biophys. Acta 2020, 1867, 118677. [Google Scholar] [CrossRef]
- Schäfer, B.W.; Wicki, R.; Engelkamp, D.; Mattei, M.G.; Heizmann, C.W. Isolation of a YAC clone covering a cluster of nine S100 genes on human chromosome 1q21: Rationale for a new nomenclature of the S100 calcium-binding protein family. Genomics 1995, 25, 638–643. [Google Scholar] [CrossRef]
- Marenholz, I.; Heizmann, C.W.; Fritz, G. S100 proteins in mouse and man: From evolution to function and pathology (including an update of the nomenclature). Biochem. Biophys. Res. Commun. 2004, 322, 1111–1122. [Google Scholar] [CrossRef] [PubMed]
- Leśniak, W. Epigenetic regulation of S100 protein expression. Clin. Epigenet. 2011, 2, 77–83. [Google Scholar] [CrossRef][Green Version]
- Gross, S.R.; Sin, C.G.; Barraclough, R.; Rudland, P.S. Joining S100 proteins and migration: For better or for worse, in sickness and in health. Cell Mol. Life Sci. 2014, 71, 1551–1579. [Google Scholar] [CrossRef]
- Heizmann, C.W. Ca2+-Binding Proteins of the EF-Hand Superfamily: Diagnostic and Prognostic Biomarkers and Novel Therapeutic Targets. Methods Mol. Biol. 2019, 1929, 157–186. [Google Scholar]
- Rescher, U.; Gerke, V. S100A10/p11: Family, friends and functions. Pflug. Arch. 2008, 455, 575–582. [Google Scholar] [CrossRef]
- Bharadwaj, A.; Bydoun, M.; Hollow, R.; Waisman, D. Annexin A2 heterotetramer: Structure and function. Int. J. Mol. Sci. 2013, 14, 6259–6305. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Ma, Y.; Fei, F.; Zheng, M.; Li, Z.; Zhao, Q.; Du, J.; Liu, K.; Lu, R.; Zhang, S. Critical role and its underlying molecular events of the plasminogen receptor, S100A10 in malignant tumor and non-tumor diseases. J. Cancer 2020, 11, 826–836. [Google Scholar] [CrossRef]
- Sato, K.; Saiki, Y.; Arai, K.; Ishizawa, K.; Fukushige, S.; Aoki, K.; Abe, J.; Takahashi, S.; Sato, I.; Sakurada, A.; et al. S100A10 upregulation associates with poor prognosis in lung squamous cell carcinoma. Biochem. Biophys. Res. Commun. 2018, 505, 466–470. [Google Scholar] [CrossRef] [PubMed]
- Tantyo, N.A.; Karyadi, A.S.; Rasman, S.Z.; Salim, M.R.G.; Devina, A.; Sumarpo, A. The prognostic value of S100A10 expression in cancer. Oncol. Lett. 2019, 17, 1417–1424. [Google Scholar] [CrossRef]
- Saiki, Y.; Ho, A. Multiple functions of S100A10, an important cancer promoter. Pathol. Int. 2019, 69, 629–636. [Google Scholar] [CrossRef]
- Madureira, P.; O’Connell, P.A.; Surette, A.P.; Miller, V.A.; Waisman, D.M. The biochemistry and regulation of S100A10: A multifunctional plasminogen receptor involved in oncogenesis. J. Biomed. Biotechnol. 2012, 2012, 353687. [Google Scholar] [CrossRef]
- Seo, J.S.; Svenningsson, P. Modulation of Ion Channels and Receptors by p11 (S100A10). Trends Pharmacol. Sci. 2020, 41, 487–497. [Google Scholar] [CrossRef] [PubMed]
- Warner-Schmidt, J.L.; Chen, E.Y.; Zhang, X.; Marshall, J.J.; Morozov, A.; Svenningsson, P.; Greengard, P. A role for p11 in the antidepressant action of brain-derived neurotrophic factor. Biol. Psychiatry 2010, 68, 528–535. [Google Scholar] [CrossRef] [PubMed]
- Chottekalapanda, R.U.; Kalik, S.; Gresack, J.; Ayala, A.; Gao, M.; Wang, W.; Meller, S.; Aly, A.; Schaefer, A.; Greengard, P. AP-1 controls the p11-dependent antidepressant response. Mol. Psychiatry 2020, 25, 1364–1381. [Google Scholar] [CrossRef] [PubMed]
- Pawliczak, R.; Cowan, M.J.; Huang, X.; Nanavaty, U.B.; Alsaaty, S.; Logun, C.; Shelhamer, J.H. p11 expression in human bronchial epithelial cells is increased by nitric oxide in a cGMP-dependent pathway involving protein kinase G activation. J. Biol. Chem. 2001, 276, 44613–44621. [Google Scholar] [CrossRef]
- García-Morales, V.; Rodríguez-Bey, G.; Gómez-Pérez, L.; Domínguez-Vías, G.; González-Forero, D.; Portillo, F.; Campos-Caro, A.; Gento-Caro, Á.; Issaoui, N.; Soler, R.M.; et al. Sp1-regulated expression of p11 contributes to motor neuron degeneration by membrane insertion of TASK1. Nat. Commun. 2019, 10, 3784. [Google Scholar] [CrossRef]
- Huang, X.L.; Pawliczak, R.; Yao, X.L.; Cowan, M.J.; Gladwin, M.T.; Walter, M.J.; Holtzman, M.J.; Madara, P.; Logun, C.; Shelhamer, J.H. Interferon-gamma induces p11 gene and protein expression in human epithelial cells through interferon-gamma-activated sequences in the p11 promoter. J. Biol. Chem. 2003, 278, 9298–9308. [Google Scholar] [CrossRef] [PubMed]
- Chédeville, A.L.; Lourdusamy, A.; Monteiro, A.R.; Hill, R.; Madureira, P.A. Investigating Glioblastoma Response to Hypoxia. Biomedicines 2020, 8, 310. [Google Scholar] [CrossRef]
- Lu, H.; Xie, Y.; Tran, L.; Lan, J.; Yang, Y.; Murugan, N.L.; Wang, R.; Wang, Y.J.; Semenza, G.L. Chemotherapy-induced S100A10 recruits KDM6A to facilitate OCT4-mediated breast cancer stemness. J. Clin. Investig. 2020, 130, 4607–4623. [Google Scholar] [CrossRef]
- Zhang, L.; Li, H.; Su, T.P.; Barker, J.L.; Maric, D.; Fullerton, C.S.; Webster, M.J.; Hough, C.J.; Li, X.X. Traumatic Stress Brain Study Group, Ursano R. p11 is up-regulated in the forebrain of stressed rats by glucocorticoid acting via two specific glucocorticoid response elements in the p11 promoter. Neuroscience 2008, 153, 1126–1134. [Google Scholar] [CrossRef] [PubMed]
- Auden, A.; Caddy, J.; Wilanowski, T.; Ting, S.B.; Cunningham, J.M.; Jane, S.M. Spatial and temporal expression of the Grainyhead-like transcription factor family during murine development. Gene Exp. Patterns 2006, 6, 964–970. [Google Scholar] [CrossRef]
- He, J.; Feng, C.; Zhu, H.; Wu, S.; Jin, P.; Xu, T. Grainyhead-like 2 as a double-edged sword in development and cancer. Am. J. Transl. Res. 2020, 12, 310–331. [Google Scholar]
- Mlacki, M.; Kikulska, A.; Krzywinska, E.; Pawlak, M.; Wilanowski, T. Recent discoveries concerning the involvement of transcription factors from the Grainyhead-like family in cancer. Exp. Biol. Med. 2015, 1, 1–6. [Google Scholar] [CrossRef]
- Kotarba, G.; Taracha-Wisniewska, A.; Wilanowski, T. Grainyhead-like transcription factors in cancer–Focus on recent developments. Exp. Biol. Med. 2020, 245, 402–410. [Google Scholar] [CrossRef]
- Rifat, Y.; Parekh, V.; Wilanowski, T.; Hislop, N.R.; Auden, A.; Ting, S.B.; Cunningham, J.M.; Jane, S.M. Regional neural tube closure defined by the Grainy head-like transcription factors. Dev. Biol. 2010, 345, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Pawlak, M.; Kikulska, A.; Wrzesinski, T.; Rausch, T.; Kwias, Z.; Wilczynski, B.; Benes, V.; Wesoly, J.; Wilanowski, T. Potential protective role of Grainyhead-like genes in the development of clear cell renal cell carcinoma. Mol. Carcinog. 2017, 56, 2414–2423. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wu, H.; Daxinger, L.; Danen, E.H.J. Genome-wide identification of binding sites of GRHL2 in luminal-like and basal A subtypes of breast cancer. BioRxiv 2020. [Google Scholar]
- Chung, V.Y.; Tan, T.Z.; Tan, M.; Wong, M.K.; Kuay, K.T.; Yang, Z.; Ye, J.; Muller, J.; Koh, C.M.; Guccione, E.; et al. GRHL2-miR-200-ZEB1 maintains the epithelial status of ovarian cancer through transcriptional regulation and histone modification. Sci. Rep. 2016, 18, 19943. [Google Scholar] [CrossRef]
- Werth, M.; Walentin, K.; Aue, A.; Schönheit, J.; Wuebken, A.; Pode-Shakked, N.; Vilianovitch, L.; Erdmann, B.; Dekel, B.; Bader, M.; et al. The transcription factor grainyhead-like 2 regulates the molecular composition of the epithelial apical junctional complex. Development 2010, 137, 3835–3845. [Google Scholar] [CrossRef]
- Rose, A.B. Introns as Gene Regulators: A Brick on the Accelerator. Front. Genet. 2019, 9, 672. [Google Scholar] [CrossRef] [PubMed]
- Korfanty, J.; Stokowy, T.; Widlak, P.; Gogler-Piglowska, A.; Handschuh, L.; Podkowiński, J.; Vydra, N.; Naumowicz, A.; Toma-Jonik, A.; Widlak, W. Crosstalk between HSF1 and HSF2 during the heat shock response in mouse testes. Int. J. Biochem. Cell Biol. 2014, 57, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Kądziołka, B.; Leśniak, W.; Filipek, A. Regulation of CacyBP/SIP expression by NFAT1 transcription factor. Immunobiology 2017, 222, 872–877. [Google Scholar] [CrossRef] [PubMed]

| Stimulus | Transcription Factor | Reference |
|---|---|---|
| Transforming growth factor β (TGF β) | ND | [13] |
| Epidermal growth factor (EGF) | ND | [13] |
| Interleukin 1 β (IL-1 β) | ND | [13] |
| Brain-derived neurotrophic factor (BDNF); basic fibroblast growth factor 2 (FGF2) | c-Jun/c-Fos (AP-1) | [16] |
| Nitric oxide (NO) | Sp1, Sp3 | [17,18] |
| Gonadotrophin (GD) | ND | [13,22] |
| Hypoxia, anti-cancer drugs | HIF1 | [13,20,21] |
| Interferon γ | STAT1 | [19,20] |
| ND | NFκB | [13] |
| ND | CTF-NF1 | [13] |
| ND | GRHL2 | Present work |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Głowacka, A.; Bieganowski, P.; Jurewicz, E.; Leśniak, W.; Wilanowski, T.; Filipek, A. Regulation of S100A10 Gene Expression. Biomolecules 2021, 11, 974. https://doi.org/10.3390/biom11070974
Głowacka A, Bieganowski P, Jurewicz E, Leśniak W, Wilanowski T, Filipek A. Regulation of S100A10 Gene Expression. Biomolecules. 2021; 11(7):974. https://doi.org/10.3390/biom11070974
Chicago/Turabian StyleGłowacka, Aleksandra, Paweł Bieganowski, Ewelina Jurewicz, Wiesława Leśniak, Tomasz Wilanowski, and Anna Filipek. 2021. "Regulation of S100A10 Gene Expression" Biomolecules 11, no. 7: 974. https://doi.org/10.3390/biom11070974
APA StyleGłowacka, A., Bieganowski, P., Jurewicz, E., Leśniak, W., Wilanowski, T., & Filipek, A. (2021). Regulation of S100A10 Gene Expression. Biomolecules, 11(7), 974. https://doi.org/10.3390/biom11070974






